Abstract
High affinity capture agents against protein targets are essential components for immunoassays, regardless of specific analysis format. Here we describe the use of DNA-encoded antibodies for rapidly screening the kinetic and equilibrium binding properties of twelve commercial antibodies in a parallel analysis format using a multiplexed array of microring optical resonators. We show that DNA-encoding offers advantages in terms of antigen binding capacity, compared to covalently tethered antibodies; we also demonstrate that this linkage modality facilitates the rapid self-assembly of multiplexed arrays on account of complementarily between the DNA sequences on the antibodies and sensor array, respectively. Furthermore, DNA-encoded antibodies also allow for sensor array regeneration and reprogramming, as chaotropic agents can be used to disrupt the DNA-DNA duplexes that link the capture agents to the sensor without harming the underlying DNA on the surface, which can subsequently be reloaded with antibodies either targeting the same or different antigens.
Introduction
A major challenge in developing sensitive and robust protein immunoassays is identifying appropriate antibody capture agents for the intended target antigen. Although assay performance is profoundly affected by the ultimate sensitivity of the analytical methods, an oft-encountered limitation is imposed by poor antibody performance. Furthermore, many ultra sensitive detection techniques acquire their sensitivity from the use of extremely high affinity capture agents rather than fundamentally more sensitive measurement technologies—a complication when performing head-to-head evaluation of different methodologies in the absence of more general comparables. Nonetheless, high affinity protein capture agents are absolutely essential for robust immunoassays, and many hurdles are often encountered in their pursuit. For example, among a selection of commercially available antibodies against a certain target, the equilibrium and kinetic binding constants can vary significantly from vendor to vendor, clone to clone, and even lot toot. Furthermore, these metrics are rarely available from vendors, making the direct evaluation of the performance of antibodies an important component of biosensor development.
Label-free, refractive index-sensitive sensor platforms,1–8 have been widely used for evaluating protein-protein binding kinetics. Typically, these methods utilize microspotting or microfluidic technologies to directly create arrays of protein capture agents on the sensor surface in a process that is completely separate from the subsequent interaction screening. Although these screening formats work well for many applications, in this paper we demonstrate an expansion upon these capabilities by utilizing DNA-encoded antibodies for the screening of antibody kinetics using arrays of microring optical resonators. Microring resonators are refractive index-responsive optical devices that our group has recently demo nstrated as a versatile tool for the sensitive detection of biomolecules.9–11 Beyond these detection applications, the modular multiplexing capability of the semiconductor-based platform make it an attractive technology for multiplexed and label-free interaction monitoring.8
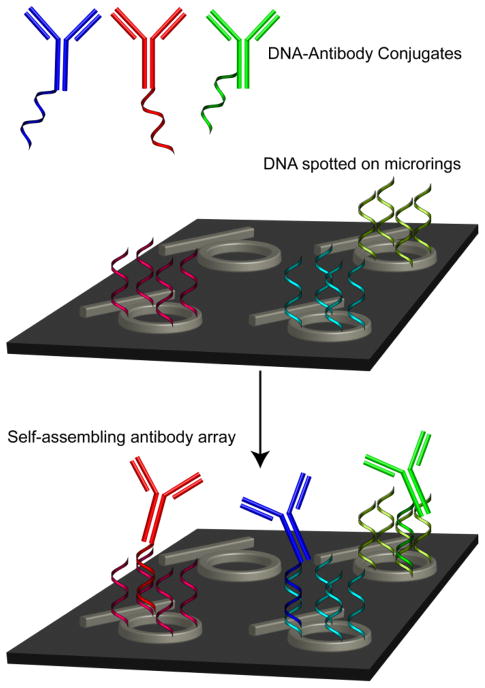
As described previously,12–18 DNA micro arrays can be converted into antibody arrays via a self-assembly process that involves conjugating antibodies with DNA strands which are complementary to DNA strands immobilized on the surface. Figure 1 shows an illustration of this concept whereby ssDNA-tagged antibodies are directed to specific cDNA-modified microrings via the Watson -Crick base pairing of the respective DNA sequences. Used for antigen detection, this sensor function alization strategy has been utilized in both fluorescent microarray 17–21 and label-free surface plasmon resonance analysis platforms.13, 22, 23
Figure 1.
Covalent DNA-antibody conjugates (blue, red, and green) are created in parallel with a microring resonator chip (not to scale) that has been functionalized with unique complementary DNA strands via microspotting. After flowing the conjugates over the surface, the conjugates self-assemble onto the chip surface as dictated by the complementary DNA -DNA base pairing interactions. Non-functionalized rings serve as controls since they do not have any DNA -antibody conjugates directed towards them.
Advantages of this approach—both for biomolecule detection as well as capture agent screening—come from several factors. First, DNA microarrays, are generally more robust than protein microarrays on account of the high sensitivity of proteins to denaturation on hydrophobic surfaces,24, 25 at air/water interface s,26 and under dehydrated storage conditions.27 To avoid these deleterious effects on protein microarrays, microfluidic deposition techniques can be used to create patterned arrays of antibodies in situ, immediately before analysis. For example, Nahsol et al. used the Prote On XPR36 with a 6 × 6 array of fluidics for screening applications.4 However, microspotting has an advantage over microfluidic approaches in that it can allow larger and higher density arrays to be created. A second advantage of the DNA-encoding strategy comes from the nature of the surface immobilization interaction. Although the DNA-DNA duplex is sufficient to provide a robust linkage under assay conditions, the base-pairing interaction can be disrupted using chaotropic or alkaline agents, as has been demonstrated previously.23 This allows for there generat ion of sensor surfaces even when the desired antibody -antigen interactions are irreversible under standard antibody regeneration conditions (e.g. low pH, high pH, chaotropic, highly chelating, and high ionic strength conditions).28 This capability can also be advantageously used to dynamically reprogram the specificity of sensor arrays by replacing one set of antibodies with others that have a different set of specificities but are encoded with the same DNA sequences.
In this paper we explored the combined utility of DNA-encoded antibodies and arrays of silicon photonic microring resonators as a versatile, multiplexed bioanalysis platform. We first confirmed the ability of the DNA-encoding strategy to direct antibodies to the appropriately cDNA-modified microring resonators and then validated the antigen-recognition capability of the immobilized capture agents. We next utilized DNA-encoded antibodies to evaluate the kinetics and secondary antibody recognition properties of 12 different commercial capture agents: six that recognized prostate specific antigen (PSA) and six specific for α-fetoprotein (AFP). For kinetic measurements we used the kinetic titration method described by Karlsson et al.,29 which streamlines the evaluation process by avoiding the need to regenerate the surface between each addition of antigen. We first evaluated the six PSA antibodies in parallel; then after only a 15 -minute rinse with 8 Murea, we reprogrammed the same microring resonator array substrate with the six AFP antibodies, which were subsequently interrogated in parallel. In addition to screening binding kinetics, we also evaluated antibody pairs for secondary antibody recognition to surface-bound antigen/antibody complexes. In this manner we were able to quickly determine which combinations of antibodies could function as a set for sandwich immunoassays. This also provided information about common binding epitopes among antibodies, since it is assumed that antibodies that form sandwich pairs are binding separate epitopes, whereas antibody pairs that bind in a mutually exclusive fashion are assumed to bind to proximal epitopes.
Although previous work has demonstrated the use of DNA-conjugated proteins for kinetic measurement,22 this paper is the first to demonstrate multiplex kinetic analysis using DNA-encoded antibodies. Overall, the combination of DNA-encoded antibodies and microring resonator arrays is shown to be a promising combination for not only the evaluation of capture agent binding kinetics, but also other key characteristics that are important in the development of robust protein immunoassays.
Experimental
Reagents and Materials
Succinimidyl 4-formylbenzoate (S-4FB), succinimidyl 6-hydrazinonicotinamide acetone hydrazone (S-HyNic), 3-N-((6-(N′-Isopropylidene-hydrazino))nicotinamide)propyltriethyoxy-silane (HyNic Silane), and antibody-oligonucleotide conjugation kits were purchased from SoluLink (San Diego, CA). Custom DNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). Monoclonal mouse anti-human PSA antibodies clones B732M, B731M, 5A6, 5G6, 8A6, monoclonal mouse anti-human AFP antibodies clones B491M, 131–12210, 057–11301, 1301, 1305, and purified human AFP and PSA were purchased from Meridian Life Science, Inc. (Saco, ME). Monoclonal mouse anti-human PSA antibody clone 6915780 and monoclonal mouse anti-human AFP antibody clone 2127435 were obtained from Fitzgerald Industries International (Concord, MA). For convenience, all of the antibodies will be referred to by their specificity and clone number for future reference (e.g. anti-AFP-B491M). However, modifications to this convention include: anti-AFP-210 for clone 131–12210, anti-AFP-301 for clone 057–11301, anti-PSA-780 for clone 6915780, and anti-AFP-435 for clone 2127435.
Zeba spin filter columns and Starting Block were purchased from Pierce (Rockford, IL). Vivaspin molecular weight cutoff filters (50,000 and 5,000 Da MWCO), were obtained from GE Healthcare (Waukesha, WI). Phosphate buffered saline (PBS), with a standard 10 mM phosphate ion concentration, was reconstituted from Dulbecco’s phosphate buffered saline packets purchased from Sigma-Aldrich (St. Louis, MO). Aniline was obtained from Acros Organics (Geel, Belgium). All other chemicals were obtained from Sigma-Aldrich and used as received.
All buffers were made with purified water (ELGA PURELAB filtration system; Lane End, UK), and the pH was adjusted as necessary with 1 M HCl or 1 M NaOH. A different PBS buffer with 100 mM phosphate (100 mM PBS) was made with 150 mM NaCl, 22.5 mM monobasic sodium phosphate, and 77.7 mM dibasic sodium phosphate and then pH-adjusted to either pH 7.4 or pH 6.0. PBS with 0.05% Tween-20 (PBST) was made by adding Tween-20 to standard PBS buffer (Dulbecco’s formulation), pH 7.4. All solutions were degassed under vacuum sonication before being flowed across the sensor surface.
Instrumental Setup and Chip Fabrication
The sensor chips and microring resonator interrogation instrumentation was acquired from Genalyte, Inc. (San Diego, CA), and have been previously described.30 Each 6 × 6 mm microchip contains thirty-two 30-μm diameter microrings that have adjacent linear waveguides with input and output diffractive grating couplers at each end to enable independent determination of the optical cavity spectrum of each microring using a tunable, external cavity diode laser (center frequency 1560 nm). Resonance wavelengths are measured as the wavelength at which the out-coupled light that propagates past the microring is negatively attenuated. A fluoropolymer cladding layer over the chip with etched annular openings enables exposure of active sensing rings while keeping thermal control rings buried under the cladding layer. In each experiment, twenty-four rings are responsive to surface chemistry and biological modification while eight are left under the cladding as thermal controls. All measurements for these experiments were made with the sensor chips loaded into a custom cell with microfluidic flow channels defined by a 0.007-inch thick Mylar gasket (both single and dual channel) with a previously described design.9 Solutions were flowed through the chips using an integrated autosampler that draws from Parafilm-coated (Pechiney Plastic Packaging Company; Chicago, IL)polystyrene 96 -well plates (Costar brand, Fisher Scientific).
Silane Functionalization
Microring array substrates were first cleaned with piranha solution31 (3:1 H2SO4:30% H2O2) for 30 seconds followed by rinsing with water and N2 drying. To introduce reactive functional groups, the substrates are immersed in a 1 mg/mL solution of HyNic Silane (20 mg/mL HyNic Silane in DMF stock solution diluted to 1 mg/mL with ethanol) for 30 minutes, followed by rinsing with ethanol and then water.
Oligonucleotide Functionalization
Oligonucleotide sequences, which were designed to minimize cross-reactivity between DNA probes, were previously reported for their utility in spatially localizing DNA-antibody conjugates.16 The exact sequences, named A, A′, B, B′, F, F′, K, K′, L, L′, and M, M′, (where a prime symbol indicates a complementary sequence), are listed in the Supporting Information in Table S1. All oligonucleotides were synthesized with a 5′ amino terminal group to facilitate attachment to either the substrate or antibody. Oligonucleotides were functionalized with S-4FB according to manufacturer (SoluLink) instructions. Briefly, oligonucleotides were buffer exchanged to 100 mM PBS pH 7.4 and then a 20-fold molar excess of S-4FB in DMF was added. Solutions were allowed to react overnight at room temperature and then were buffer exchanged into 100 mM PBS pH 6.0 using 5 kDa MWCO filters.
DNA Spotting
Multiplexed functionality was installed on the microring arrays by alternately spotting 4FB-functionalized DNA strands A, B, F, K, L, M onto a microchip that had been previously functionalized with HyNic Silane. For initial tests with ‘F -anti-PSA-5A6 and/or B′-anti-AFP-B491M, only strands B and F were added to the chips. In all cases, several rings on each chip were not functionalized with DNA and used as controls. Six-plex chip spotting was achieved using a Nanoe Nabler spotting system from BioForce Nanosciences (Ames, IA) using a concentration of at least 100 μM DNA in 100 mM PBS buffer pH 6.0 mixed in a 1:1 ratio with DMSO, to slow solvent evaporation. For chips with only one or two different DNA strands added, the DNA was spotted manually using a stereoscope to direct fluid placement. After spotting, the drops of solution were dried on a hot plate (~70 °C) and incubated in 80+% relative humidity (or higher) for 1–2 hours to allow rehydration of the DNA on the surface. Chips were then immersed into S-4FB-modified Starting Block. The Starting Block was modified following the same procedure as oligonucleotide functionalization but 100 μL of 5 mg/mL S-4FB was added to 1.5 mL Starting Block. The blocking solution was removed by rinsing with water, and then additional S-4FB modified blocking solution was added to the chip before incubating overnight in a humidity chamber at 4°C. Sensor chips were then rinsed with water, immersed in 8M urea for 20 minutes to remove excess blocking protein, and then finally rinsed with water and dried under nitrogen.
DNA-Antibody Conjugate Synthesis
To create DNA-antibody conjugates, antibodies were first functionalized with S-HyNic following the manufacturer’s guidelines.32 Briefly, S-HyNic in DMF was added in 5-fold molar excess to ~1 mg/mL antibody that had previously been buffer exchanged into 100 mM PBS pH 7.4 with a Zeba spin filter and reacted for at least two hours at room temperature. The antibody was then exchanged into 100 mM PBS pH 6.0 and concentrated using a 50 kDa MWCO filter, which also served to remove residual S-HyNic. The 4FB-modified DNA was then added in 20-fold molar excess to the HyNic-modified antibody solution and allowed to react overnight at 4 °C. DNA-antibody conjugates were then purified away from the excess DNA using a Superdex 200 10/300 GL column on an AKTA FPLC, both from GE Healthcare (Waukesha, WI). The separation was performed at 4 °C with a PBS isocratic elution. The collected fractions were concentrated with 50 kDa MWCO filters to yield purified solutions of DNA-antibody conjugates. The final conjugate concentration measured between 100–400 μg/mL, as determined by measuring the differential absorption at 260 versus 280 nm, corresponding to the DNA and IgG, respectively, using a NanoDrop UV-V is absorbance system (Thermo Fisher Scientific, Wilmington, DE). The ratios of DNA to antibody for each conjugate, as determined by UV-Vis absorbance, are given in Table S2. The conjugates had ratios from 0.4 to 1.5, but most fall around 0.5 moles of DNA to moles of antibody. As a result, most conjugates only have a single strand of DNA and are intermixed with non-functionalized antibodies. This ratio could be increased, of course, by increasing the ratio of S-HyNic to antibody; however, it is desirable to keep the number of DNA molecules in each conjugate at two or fewer in order to minimize potential interference with the binding region. Alternatively, an oligonucleotide-antibody conjugation kit from SoluLink was used to synthesize and purify the conjugates using a magnetic-bead-based separation, according to manufacturer’s instructions. Although both purification methods provided conjugates of identical analytical behavior, the conjugation kit is advantageous for the parallel preparation of multiple conjugates, as opposed to serial FPLC purification. The following conjugates were synthesized: A′-anti-PSA-8A6, B ′-anti-PSA-B732M, F′-anti-PSA-5A6, K′-anti-PSA-780, L′-anti-PSA-B731M, M′-anti-PSA-5G6, A′-anti-AFP-1301, B′-anti-AFP-B491M, F′-anti-AFP-435, K′-anti-AFP-1305, L′-anti-AFP-210, M′-anti-AFP-301.
Validation of DNA-Encoded Antibody Binding to cDNA-Modified Microring Resonators and Subsequent Antigen Recognition Capability
To validate the ability to localize DNA-antibody conjugates onto microrings presenting specific cDNAs (strands F and B), we tested F ′-anti-PSA-5A6 and B ′-anti-AFP-B491M. A solution of 20 μg/mL of the B ′-anti-AFP-B491M conjugate was first flowed across the surface followed by 5 μg/mL of the F′-anti-PSA-5A6 conjugate. Because the relative binding rates of the DNA-antibody conjugates varied, the concentrations were adjusted so that the so that binding would occur at a similar rate. Following addition of conjugates, 1 μg/mL of AFP and then 1 μg/mL of PSA were flowed across the chip surface.
To test the loading-response behavior of the anti-PSA-5A6 antibody, two methods were used. First, several chips functionalized with strand F had different surface densities of F′-anti-PSA-5A6 added to the surface by varying the concentration of the conjugate added to the chip as well as the incubation time of the conjugate. Following addition of the conjugate, 1 μg/mL of PSA was flowed across the surface, and the equilibrium, saturation response was measured after 5 minutes of exposure. Following this test, each chip was regenerated by flowing 8M urea over the chip surface for 15 minutes with water rinses before and after the urea. This enabled antibody conjugates loading to be repeated multiple times.
To compare the capture performance of DNA-encoded antibodies with those directly immobilized on the sensor surface via a covalent linkage with HyNic silane (as we have described previously33), we flowed 4FB-modified anti-PSA-5A6 (using the same procedure as the S-HyNic modification, but substituting S-4FB for S-HyNic) over HyNic-silane-functionalized sensor chips. We utilized a previously-described 4-channel flow cartridge8 to react rings with different concentrations of anti-PSA-5A6 from 0.7 μg/mL to 7 μg/mL. The variations in concentration enabled different amounts of capture antibody to be loaded on different rings on the same chip, as directly measured by monitoring the real-time shifts in microring resonance wavelength during antibody immobilization. Since these surfaces could not be regenerated to reload antibody, two separate chips (8 channels total) were used to collect the covalent immobilization data. After functionalizing the chip with anti-PSA-5A6, the chip was blocked overnight with 4FB-modified Starting Block and then loaded into a flow cartridge with a single channel to direct the antigen solution over all the sensor rings. Then a solution containing1 μg/mL of PSA was flowed over the chip and the saturation response was measured after 5 minutes of exposure.
DNA-antibody Conjugate Kinetic Screening Experiments
To load a DNA-antibody conjugate onto a chip, ~5 μg/mL (or higher concentration if DNA-DNA binding kinetics for a particular pair of complementary strands were slow) of conjugate was flowed over the chip surface and terminated with a buffer rinse of PBST after loading enough antibody to create a 30–50 pm shift. Saturation experiments with PSA were made by flowing 1 μg/mL PSA in PBST over the chip for 5 minutes. A chip surface could be regenerated by flowing 8M urea over the chip surface for 15 minutes with water rinses before and after the urea. Before performing the multiplexed kinetic evaluation experiments, the DNA-antibody conjugates were tested individually to ensure that no cross-reactivity occurred and to determine the relative binding rate of each conjugate. Thereafter, conjugates could be added simultaneously by adjusting the concentrations so that the rate of addition to the surface was similar for all of the DNA-antibody conjugates; thus, equivalent loadings were achieved for a single functionalization time period. Kinetic titration experiments were made by adding increasing concentrations of PSA or AFP for a 1.5-minuteantigen association period followed by a 4.5-minute rinse with PBST during which desorption of the antigen was observed.
Screening for secondary antibodies was accomplished by loading all six of the DNA-antibody conjugates for a particular antigen over the chip surface and then adding 1 μg/mL of the appropriate antigen (PSA or AFP) for 5 minutes. Following this step, each antibody (not conjugated to DNA) was flowed sequentially over the chip at a concentration of 1 μg/mL. Each chip was loaded into a flow cartridge with two separate microfluidic channels so that each channel could have a different sequence of antibodies flowed over the chip. Following sequential addition of all 6 antibodies for a particular antigen on both channels, the chip was then regenerated with 8M urea, and the experiment was repeated with different sequences of antibodies added to each channel.
Data Analysis
Raw microring resonance wavelength data, recorded as a function of time, was corrected for any thermal drift of bulk refractive index shifts using on-chip control rings (exposed to solution, but not functionalized with DNA). The signal from all of the control rings was averaged and then subtracted from each of the individual active sensor rings. Kinetic titration data was divided into association and dissociation traces and fit (Origin Pro 8.1; Origin Lab Corporation; Northampton, MA) using the integrated rate equation for one-to-one Langmuir binding, as described by Karlssonet al.29
Results and Discussion
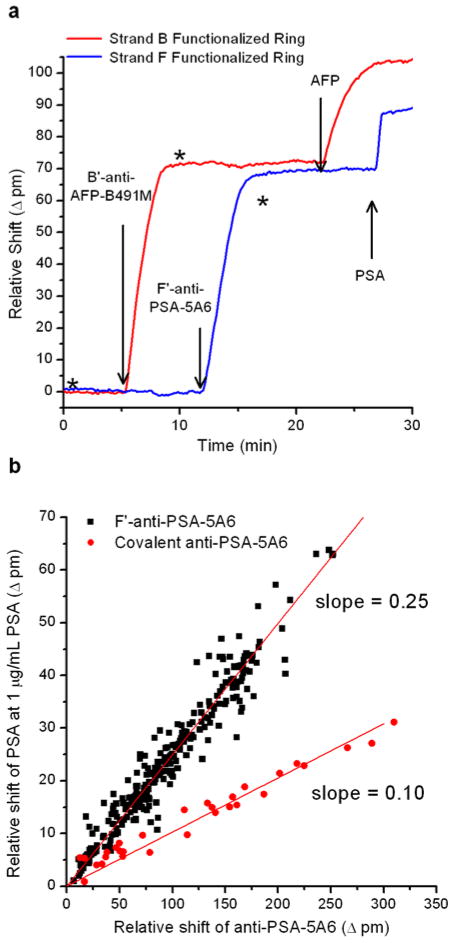
Before measuring the kinetics of binding to DNA-antibody conjugates, we first wanted to validate the functionality of the conjugates both in terms of loading onto ssDNA -presenting microrings as well as their subsequent ability to recognize the targeted antigen. Initially, both F ′ anti-PSA-5A6 and B′-anti-AFP-B491M were sequentially loaded onto a chip functionalized with DNA strands F and B. Next, a saturating concentration (1 μg/mL) of AFP and then PSA were flowed across the chip. As can be seen in Figure 2a, introduction of the antibody conjugates results in large, positive shifts in the resonance wavelength (Δpm), consistent with biomolecular deposition. The subsequent addition allows us to confirm that there is no non-complementary binding on rings to which a conjugate has not been specifically encoded in accordance with Watson-Crick base pairing. In addition, introduction of a high (1 μg/mL) concentration of AFP and then PSA to all the rings only results in a response on the rings functionalized with the specific DNA-antibody conjugates. From these results we can conclude that the DNA -antibody conjugates self-assemble to the appropriate sensor location and retain their specificity towards a target analyte. In addition, we observe no measurable non-specific binding of the AFP or PSA antigens to control rings that are blocked with Starting Block or on DNA -functionalized rings that have not had DNA-antibody conjugate added to them.
Figure 2.
(a) Initial validation of DNA -antibody conjugates showing orthogonal antibody loading and orthogonal antigen response. First B′-anti-AFP-B491M was added followed by F′-anti-PSA-5A6. Then 1 μg/mL of AFP was added followed by 1 μg/mL PSA. The red trace shows the response of a microring functionalized with DNA strand B and the blue trace shows the response of a microring functionalized with DNA strand F. For reference, an arrow is positioned at the time points of injection and asterisks (*) indicate the switch to running buffer. (b) Shift in resonance wavelength for F3′-anti-PSA-5A6-presenting microring resonators to 1 μg/mL PSA versus the amount(in units of Δpm)of F3 ′-anti-PSA-5A6 originally loaded onto the sensors before antigen interaction. The slope of the DNA -encoded antibody loading versus antigen response is ~2.5-foldgreater than that for covalently bound antibody, indicating an increased ability to bind to antigen.
We also sought to investigate batch-to-batch and chip-to-chip robustness of DNA-antibody conjugates, particularly in regards to the correlation between antigen binding response and the amount of antibody bound to the sensor. Using F′-anti-PSA-5A6, we performed multiple tests measuring the binding capacity of the antibody on the surface as a function of the amount of conjugate added to the DNA-functionalized surface. This was done by first adding F′-anti-PSA-5A6 to a chip functionalized with the complementary DNA strand F, a process that was measured in real-time to establish the relative amount of antibody bound to each microring. Following the addition of the conjugate, 1 μg/mL PSA was flowed over the surface and the saturating antigen response signal elicited was recorded. Using 8M urea to disrupt the DNA duplex formed between conjugate and ring surface, a single sensor chip could easily be reloaded multiple times with different amounts of conjugate via regeneration after measurement of antigen binding responses. In this way, it was very easy to rapidly accumulate a large amount of data establishing a correlation between antibody loading and antigen binding response.
Figure S1 in the Supporting Information demonstrates the efficacy of urea regeneration of a DNA surface loaded with DNA -antibody conjugates. In general, the ability to load DNA-antibody conjugate onto a chip was not significantly compromised by multiple regenerations. However, we have observed that the addition of multiple strands of DNA (more than two, depending on the strand) per antibody inhibits the full regeneration of surfaces due to multivalent binding effects. As a result we have kept the ratio of DNA to antibody in our synthesized conjugates at 1.5 or less.
We also measured the response from multiple sensor chips and several batches of F′-anti-PSA-5A6 conjugates, and the entirety of these data sets are compiled together as the black squares in Figure 2b. Impressively, the amount of antigen binding response is very well-correlated to antibody loading, a linear trend that holds across multiple sensor chips and independently synthesized batches of DNA-antibody conjugate.
For the sake of comparison, we performed a similar study using antibodies that were covalently tethered to the sensor surface, using a previously reported hydrazone linkage-based bioconjugate strategy.33 The same anti-PSA-5A6 antibody used to make the DNA-antibody conjugates was directly attached to the sensor chips while recording the accompanying shift in resonance wavelength. The sensor chip was then exposed to 1 μg/mL PSA and the response was measured. This process was repeated for a second sensor chip, and all of the results are plotted as the circular red data points in Figure 2b. A linear relationship between antibody loading and antigen binding response is again observed, but the slope is 2.5times greater in the case of the DNA-conjugate. Although the mechanism of this increase in antigen recognition capacity is still under investigation, we preliminarily attribute it to the fact that the underlying DNA functionalized microring surface is sufficiently hydrophilic to minimize the denaturation of attached antibodies through non-specific interactions. Furthermore, the flexibility afforded by the DNA linker might allow the antibody to sample more optimal orientations for antigen capture, as opposed to the shorter and more rigid covalent linkage.
Having validated the performance and established the utility of DNA-antibody conjugates for antigen binding, we then created multiplexed substrates that presented different DNA strands (named A, B, F, K, L, M) on each sensor chip. Following the determination that all six conjugates could be loaded without any non-specific antibody localization (Figure S2 in the Supporting Information), these 6-plex chips were then used to simultaneously evaluate the kinetic binding characteristics of multiple, commercially-available protein capture agents recognizing either PSA or AFP.
In order to expedite the data collection process for screening the binding characteristics of multiple antibodies, we followed the strategy of Karlsson and coworkers,29 who previously reported a kinetic titration method. This method enables subsequent additions of antigen solutions without having to fully regenerate in between.
Starting with a sensor chip spotted with 6 different ssDNAs at unique spatial locations, six different DNA-antibody conjugates were simultaneously loaded onto the sensor chip by flowing a mixture of all the conjugates simultaneously over the entire chip surface. In an effort to avoid the deleterious effects of steric crowding on kinetic measurements,34,35 the amount of each bound antibody was maintained in the range of 450–750 pg/mm2, (30–50 pm resonance wavelength shift, as establishedp reviously36). The six anti-PSA antibodies were evaluated initially before the surface was regenerated with 8M urea, and the six anti-AFP antibodies were added to the chip. Each titration consisted of 1.5-minute association phases of solutions containing 10, 20, 50, 100, 300, 1000, 5000 ng/mL concentrations of antigen (either PSA or AFP) with4.5 -minute dissociation rinses with buffer.
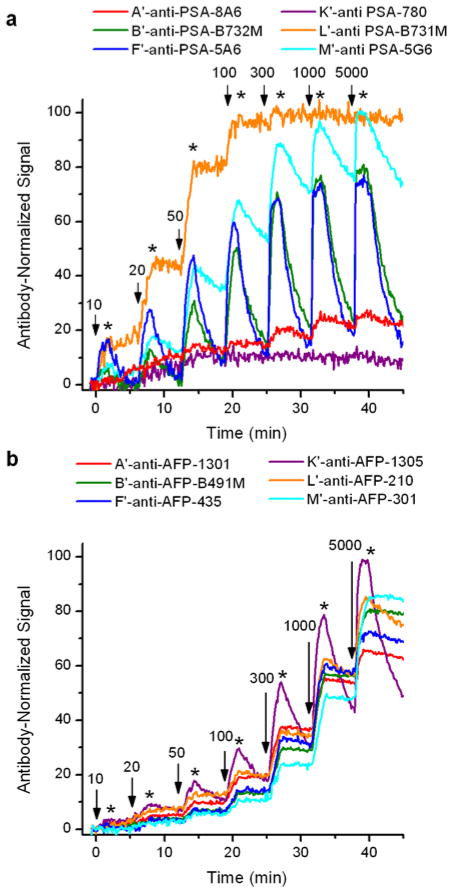
Figures 3a and 3b show the real-time shifts in resonance wavelength during the aforementioned kinetic titrations for both anti-PSA and anti-AFP antibodies, respectively. Each set of traces was normalized according to the amount of each antibody loaded onto the sensor array. Measurements were made in triplicate for each antibody, with six control rings comprising the rest of the 24-element sensor array. However, for clarity, data from only a single representative microring per antibody is shown.
Figure 3.
Real -time shifts in resonance wavelength from representative microring resonators functionalized with unique antibodies upon exposure to a) PSA, and b) AFP in a kinetic titration assay format. In both panels, solutions containing the targeted antigen at the listed concentrations (in units of zng/mL) are introduced for 1.5 minutes at time points indicated by black arrow. After this association phase, the solution was switched to buffer (as indicated with asterisks) and a 4.5 minute dissociation phase was recorded.
As shown in Figure 3a, A′-anti-PSA-8A6 and K′-anti-PSA-780 conjugates show very low binding activity. To verify that the poor antigen recognition was not due to the covalently-attached DNA, we tested all of the antibodies without pendant DNA by flowing them over a surface with immobilized rabbit-anti mouse IgG-Fc (RAM-Fc). Because the RAM-Fc binds mouse IgG in the Fc region, away from the antigen recognition sites, this method does not require antibody modification and has been used to orient antibodies on biosensor surfaces.37 Comparing the RAM-Fc immobilization approach to the DNA-assembled approach, the antigen binding behavior remained consistent between methods, with anti-PSA-8A6 and anti-PSA-780 displaying poor antigen binding performance in either case. Figure S3 in the Supporting Information shows data for the RAM-Fc experiments for all of the antibodies.
Figure 3b shows the real-time shifts in resonance wavelength for the six anti-AFP antibodies in a manner identical to the anti-PSA antibodies (Figure 3a). In this case, all the antibodies responded well and most of the antibodies had similar association and dissociation rates. Only K′-anti-AFP-1305 displayed markedly different behavior, in contrast to the collection of anti-PSA antibodies, which showed greater variation in binding kinetics.
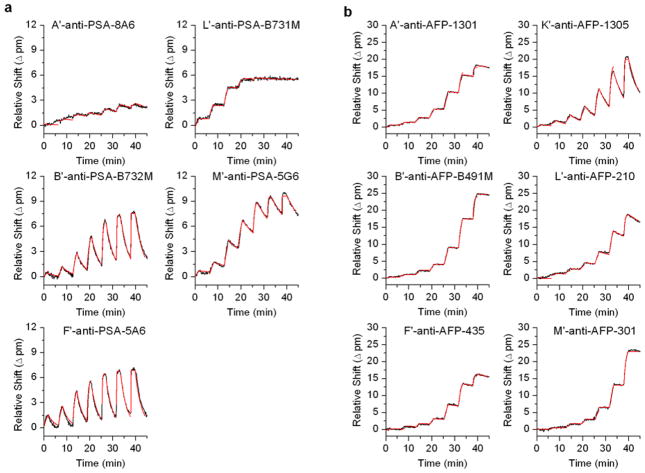
In order to quantitatively evaluate the binding kinetics of the arrayed antibodies, the real-time shifts in resonance wavelength from the kinetic titration were fit using the integrated rate equation for the one-to-one Langmuir binding model. Figure 4 shows the results of this fitting to a representative data set for each antibody. All fitting results are provided in the Supporting Information as Figures S4 and S5. As noted above, the A′-anti-PSA-8A6 and K′-anti-PSA -780 antibodies showed poor binding activity, which presented complications in fitting these data sets. For A′-anti-PSA-8A6, the poor binding translated to a poor fit, whereas we were unable to fit binding parameters for K′-anti-PSA −780, thus its omission from the figure.
Figure 4.
Kinetic fits (red) overlaid on the binding curve traces (black) for a representative kinetic titration of each of the DNA-antibody conjugates for both a) anti -PSA antibodies and b) anti-AFP antibodies. Due to the exceptionally poor performance of K′-anti-PSA-780, fitting could not be performed for this antibody, thus its omission from the figure.
Table 1 summarizes the average values for the kinetic association (ka) and dissociation = rates (kd) and equilibrium dissociation constant ( ) determined from the fitting the resonance wavelength shifts for the microring array during the kinetic titration assays. The determination of extremely slow rates of dissociation, which requires lengthy measurements of slowly diminishing response, can be complicated by non-ideal sensor performance (random noise, drift, etc.). For DNA-antibody conjugates, dissociation of DNA duplex is an additional concern as it could artificially hasten the observed off rate. To address this concern we determined that for the DNA sequences and the respective DNA-antibody conjugates, the rate of dissociation occurs at 0.05 Δpm/min or less. In addition, given our previously determined slope noise over a 5-minute time frame as 0.01–0.02 Δpm/min,9 we established a reasonable lower bound for measurable dissociation rates at ~0.05 pm/min. Consequently, this rate of dissociation for a 50 pm loading of antibodies is equivalent to a dissociation rate of 2 × 10−5 s −1. Thus, Table 1 lists any kd values that are too low to be measured with certainty as “≤ 2 × 10 −5 s −1,” and the value for KD is also given the appropriate upper bound. For our sets of antibodies, only B′-anti-AFP-B491Mand L′-anti-PSA-B731Mshow dissociation rates slower than 2 × 10−5 s −1. Although we are unable to benchmark these values against other, independent measurements, it is apparent that these antibodies have the highest affinities of the groups. As mentioned earlier the poor performance of the K′-anti-PSA -780 antibody prohibited accurate fitting to the model, and thus did not allow for determination of kinetic or equilibrium binding constants.
Table 1.
List of kinetic and equilibrium binding parameters for DNA-antibody conjugates interacting with their target antigen
| ka (s−1 M−1) | kd (s−1) | KD (nM) | ||
|---|---|---|---|---|
| PSA antibody conjugates | A′-anti-PSA-8A6 | 8.7 × 105 | 9.2 × 10−4 | 1.1 |
| B′-anti-PSA-B732M | 4.1 × 106 | 3.8 × 10−3 | 0.91 | |
| F′-anti-PSA-5A6 | 9.4 × 106 | 6.3 × 10−3 | 0.67 | |
| L′-anti-PSA-B731M | 5.4 × 106 | ≤2 × 10−5 | ≤0.004 | |
| K′-anti-PSA-780 | - | - | - | |
| M′-anti-PSA-5G6 | 2.5 × 106 | 1.0 × 10−3 | 0.40 | |
| AFP antibody conjugates | A′-anti-AFP-1301 | 8.3 × 105 | 7.5 × 10−5 | 0.091 |
| B′-anti-AFP-B491M | 4.8 × 105 | ≤2 × 10−5 | ≤0.04 | |
| F′-anti-AFP-435 | 1.1 × 106 | 1.7 × 10−4 | 0.15 | |
| K′-anti-AFP-1305 | 1.8 × 106 | 2.6 × 10−3 | 1.4 | |
| L′-anti-AFP-210 | 6.7 × 105 | 3.9 × 10−4 | 0.58 | |
| M′-anti-AFP-301 | 3.4 × 105 | 4.3 × 10−5 | 0.12 |
Due to subtle variations in experimental conditions and measurement methodology, as well as uncertainties in fitting, experimentally derived kinetic and equilibrium binding values often have variances from group-to-group or technology-to-technology, making absolute, cross-platform comparisons of protein-protein interactions difficult. However, by comparing six antibodies side by side in identical assay conditions within the same microfluidic volume, we demonstrate a direct, head-to-head comparison of antibodies which can enable accurate selection of an antibody for a particular assay. For example, a fast association rate may desirable for certain applications where kinetics are of utmost importance. However, a slow dissociation rate may be preferred when the tightest possible binding (smallest KD) is needed, or if antigen needs to stay bound for additional rinse or recognition steps, such as with a sandwich-type assay. The parallel screening method presented herein provides a straightforward approach to rationally selecting antibodies with optimal characteristics for the desired assay.
In addition to characterizing the binding kinetics of each individual antibody, the easily programmable and readily regenerable DNA -encoded microring sensor array also providesa mechanism to rapidly screen for antibody sandwich pairs. The six DNA-antibody conjugates recognizing either PSA or AFP were loaded onto the chip surface, and then a high concentration of antigen was flowed over the sensor array, giving a near-saturating binding response. Each individual antibody (non-DNA conjugated) was sequentially flowed over the chip without regenerating the surface in between antibody additions (see Figures S6 and S7 in the Supporting Information for real time data). We determined that a “sandwich pair” was formed, when the response to the secondary antibody was greater than 10% of the antigen binding response after being exposed to the secondary antibody for 5 minutes or less. The results of these screening efforts are summarized in Table 2 (PSA) and Table 3 (AFP). Because the secondary antibodies were added without regeneration between additions, it was apparent that the addition of certain secondary antibodies interfered with subsequent addition of other secondary antibodies. Although this information is useful for determining overlapping binding epitopes for different antibodies, it also prevented determination of all possible sandwich pairs after a single run. To fully determine all combinations of antibody sandwich pairs, a urea regeneration step was used to strip the surface down to single-stranded DNA capture probes. The sensors were then reloaded with primary antibody conjugates, and the secondary screening process was repeat ed using a different sequence of secondary antibodies. This reordering ensured that if the binding of one sandwich pair interfered with the binding of a second antibody, there would be an opportunity for the second antibody to be tested without the first antibody having been flowed over the surface already. By using a fluidic system that featured two identically-encoded channels, all sandwich pairs were unambiguously assessed using only a single regeneration (4 tests overall; data presented in Figures S6 and S7).
Table 2.
Anti-PSA sandwich pair screening results showing sandwich-pair interactions (“X”) between surface-immobilized primary antibodies (row headings) and solution-phase secondary antibodies (column headings)
| anti-PSA-8A6 | anti-PSA-B732M | anti-PSA-5A6 | anti-PSA-780 | anti-PSA-B731M | anti-PSA-5G6 | |
|---|---|---|---|---|---|---|
| A′-anti-PSA-8A6 | - | - | X | - | X | X |
| B′-anti-PSA-B732M | - | - | - | X | - | - |
| F′-anti-PSA-5A6 | X | - | - | X | X | X |
| K′-anti-PSA-780 | - | X | X | - | X | X |
| L′-anti-PSA-B731M | X | - | X | X | - | - |
| M′-anti-PSA-5G6 | X | - | X | X | - | - |
Table 3.
Anti-AFP sandwich pair screening results showing sandwich-pair interactions (“X”) between surface-immobilized primary antibodies (row headings) and solution-phase secondary antibodies (column headings)
| anti-AFP-1301 | anti-AFP-B491M | anti-AFP-435 | anti-AFP-1305 | anti-AFP-210 | anti-AFP-301 | |
|---|---|---|---|---|---|---|
| A′-anti-AFP-1301 | - | X | - | X | - | X |
| B′-anti-AFP-B491M | X | - | X | X | X | X |
| F′-anti-AFP-435 | - | X | - | X | - | X |
| K′-anti-AFP-1305 | X | X | X | - | X | X |
| L′-anti-AFP-210 | - | X | - | X | - | X |
| M′-anti-AFP-301 | X | X | X | X | X | - |
As expected for monoclonal antibodies, capture agents did not show secondary recognition of antigens already bound to themselves, as the epitope was blocked during primary binding. Furthermore, in all cases successful sandwich pairs work in both configurations where either antibody is surface-immobilized and the other acting as the solution -phase secondary recognition agent. Interestingly, both the anti-PSA and a nti-AFP libraries show a large number of valid sandwich pairs. Anti-AFP antibodies exhibit a somewhat larger selection of sandwich pairs than the anti-PSA antibodies; this is not totally unexpected given that AFP is a larger protein than PSA, and would therefore possess more potential epitopes. Similarly, non-compatible sandwich pairs suggest that the antibodies bind to identical or proximal recognition domains and these types of analyses represent a form of epitope mapping. For example, anti-AFP-1301, anti-AFP-780, and anti-AFP-210 all seem to bind to a similar location on the antigen since their binding is mutually exclusive, as indicated in Tables 2 and 3 as well as from binding interference shown in Figures S6 and S7.
Overall, the results show how readily secondary binding agent compatibility can be determined using a multiplexed sensor array of DNA-encoded antibodies, and suggest a straightforward and efficient method for identifying pairs of capture agents that might be useful in sandwich-type immunoassays.
Conclusions
In this study we demonstrated the robust nature and utility of DNA-encoded antibodies for the multiplexed screening of capture agent binding properties, in terms of screening both their kinetic and equilibrium binding metrics, as well as their compatibility for forming sandwich pairs around a single antigen. Furthermore, this antibody attachment scheme appears to offer multiple advantages over covalent immobilization in terms of antibody binding capacity and array regeneration and reprogramming capabilities, thus reducing the need for complex microfluidic designs. Coupled with the inherent long term solution-phase storage stability of the DNA-antibody conjugates and ssDNA-functionalized sensor array, we envision that DNA-encoded antibodies will continue to be a valuable tool for both protein detection and the multiplexed evaluation of capture agent binding characteristics on a variety of transduction methodologies.
Supplementary Material
References
- 1.Krishnamoorthy G, Beusink JB, Schasfoort RBM. Anal Methods. 2010;2:1020–1025. doi: 10.1039/b9ay00176j. [DOI] [PubMed] [Google Scholar]
- 2.Krishnamoorthy G, Carlen ET, Bomer JG, Wijnperle D, deBoer HL, Berg Avd, Schasfoort RBM. Lab Chip. 2010;10:986–990. doi: 10.1039/c000705f. [DOI] [PubMed] [Google Scholar]
- 3.Bravman T, Bronner V, Lavie K, Notcovich A, Papalia GA, Myszka DG. Anal Biochem. 2006;358:281–288. doi: 10.1016/j.ab.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Nahshol O, Bronner V, Notcovich A, Rubrecht L, Laune D, Bravman T. Anal Biochem. 2008;383:52–60. doi: 10.1016/j.ab.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Hosse RJ, Tay L, Hattarki MK, Pontes-Braz L, Pearce LA, Nuttall SD, Dolezal O. Anal Biochem. 2009;385:346–357. doi: 10.1016/j.ab.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Özkumur E, Needham JW, Bergstein DA, Gonzalez R, Cabodi M, Gershoni JM, Goldberg BB, Ünlü MS. Proc Natl Acad Sci U S A. 2008;105:7988–7992. doi: 10.1073/pnas.0711421105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olkhov RV, Shaw AM. Anal Biochem. 2010;396:30–35. doi: 10.1016/j.ab.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Byeon J-Y, Bailey RC. Analyst. 2011 doi: 10.1039/c1030an00853b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washburn AL, Gunn LC, Bailey RC. Anal Chem. 2009;81:9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal Chem. 2010;82:69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luchansky MS, Bailey RC. Anal Chem. 2010;82:1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boozer C, Ladd J, Chen S, Yu Q, Homola J, Jiang S. Anal Chem. 2004;76:6967–6972. doi: 10.1021/ac048908l. [DOI] [PubMed] [Google Scholar]
- 13.Boozer C, Ladd J, Chen SF, Jiang ST. Anal Chem. 2006;78:1515–1519. doi: 10.1021/ac051923l. [DOI] [PubMed] [Google Scholar]
- 14.Ladd J, Boozer C, Yu Q, Chen S, Homola J, Jiang S. Langmuir. 2004;20:8090–8095. doi: 10.1021/la049867r. [DOI] [PubMed] [Google Scholar]
- 15.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J Am Chem Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder H, Adler M, Gerigk K, Müller-Chorus B, Götz F, Niemeyer CM. Anal Chem. 2009;81:1275–1279. doi: 10.1021/ac802228k. [DOI] [PubMed] [Google Scholar]
- 18.Wacker R, Schröder H, Niemeyer CM. Anal Biochem. 2004;330:281–287. doi: 10.1016/j.ab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Niemeyer CM, Sano T, Smith CL, Cantor CR. Nucleic Acids Res. 1994;22:5530–5539. doi: 10.1093/nar/22.25.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wacker R, Niemeyer CM. Chem BioChem. 2004;5:453–459. doi: 10.1002/cbic.200300788. [DOI] [PubMed] [Google Scholar]
- 21.Bano F, Fruk L, Sanavio B, Glettenberg M, Casalis L, Niemeyer CM, Scoles G. Nano Lett. 2009;9:2614–2618. doi: 10.1021/nl9008869. [DOI] [PubMed] [Google Scholar]
- 22.Fruk L, Kuhlmann J, Niemeyer CM. Chem Commun. 2009;230–232 doi: 10.1039/b817206d. [DOI] [PubMed] [Google Scholar]
- 23.Niemeyer CM, Boldt L, Ceyhan B, Blohm D. Anal Biochem. 1999;268:54–63. doi: 10.1006/abio.1998.3017. [DOI] [PubMed] [Google Scholar]
- 24.Maruyama T, Katoh S, Nakajima M, Nabetani H, Abbott TP, Shono A, Satoh K. J Membr Sci. 2001;192:201–207. [Google Scholar]
- 25.Vermeer AWP, Giacomelli CE, Norde W. Biochim Biophys Acta, Gen Subj. 2001;1526:61–69. doi: 10.1016/s0304-4165(01)00101-5. [DOI] [PubMed] [Google Scholar]
- 26.Sen P, Yamaguchi S, Tahara T. J Phys Chem B. 2008;112:13473–13475. doi: 10.1021/jp8061288. [DOI] [PubMed] [Google Scholar]
- 27.MacBeath G, Schreiber SL. Science (Wash) 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 28.Andersson K, Hamalainen M, Malmqvist M. Anal Chem. 1999;71:2475–2481. doi: 10.1021/ac981271j. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson R, Katsamba PS, Nordin H, Pol E, Myszka DG. Anal Biochem. 2006;349:136–147. doi: 10.1016/j.ab.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE J Sel Top Quantum Electron. 2010;16:654–661. [Google Scholar]
- 31.Caution!Piranha solutions are extraordinarily dangerous, reacting explosiv ely with trace quantities of organics.
- 32.Solulink; Vol. 2010.
- 33.Byeon JY, Limpoco FT, Bailey RC. Langmuir. 2010;26:15430–15435. doi: 10.1021/la1021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myszka DG. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson R, Michaelsson A, Mattsson L. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 36.Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Biosens Bioelectron. 2010;26:1283–1291. doi: 10.1016/j.bios.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nice E, Layton J, Fabri L, Hellman U, Engstrom A, Persson B, Burgess AW. J Chromatogr. 1993;646:159–168. doi: 10.1016/s0021-9673(99)87017-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.