Abstract
The pigment cells of vertebrates serve a variety of functions and generate a stunning variety of patterns. These cells are also implicated in human pathologies including melanoma. Whereas the events of pigment cell development have been studied extensively in the embryo, much less is known about morphogenesis and differentiation of these cells during post-embryonic stages. Previous studies of zebrafish revealed genetically distinct populations of embryonic and adult melanophores, the ectotherm homologue of amniote melanocytes. Here, we use molecular markers, vital labeling, time-lapse imaging, mutational analyses, and transgenesis to identify peripheral nerves as a niche for precursors to adult melanophores that subsequently migrate to the skin to form the adult pigment pattern. We further identify genetic requirements for establishing, maintaining, and recruiting precursors to the adult melanophore lineage and demonstrate novel compensatory behaviors during pattern regulation in mutant backgrounds. Finally, we show that distinct populations of latent precursors having differential regenerative capabilities persist into the adult. These findings provide a foundation for future studies of post-embryonic pigment cell precursors in development, evolution, and neoplasia.
Author Summary
Understanding the biology of post-embryonic stem and progenitor cells is of both basic and translational importance. To identify mechanisms by which stem and progenitor cells are established, maintained, and recruited to particular fates, we are using the zebrafish adult pigment pattern. Previous work showed that embryonic and adult pigment cells have different genetic requirements, but little is known about the molecular or proliferative phenotypes of precursors to adult pigment cells or where these precursors reside during post-embryonic development. We show here that post-embryonic pigment cell precursors are associated with peripheral nerves and that these cells migrate to the skin during the larval-to-adult transformation when the adult pigment pattern forms. We also define morphogenetic and differentiative roles for several genes in promoting these events. Finally, we demonstrate that latent precursor pools persist into the adult and that different pools have different capacities for supplying new pigment cells in the context of pattern regeneration. Our study sets the stage for future analyses to identify additional common and essential features of pigment stem cell biology.
Introduction
A fundamental challenge for modern developmental biology is to determine how populations of stem and progenitor cells are established, maintained, and recruited to differentiate at particular times and places during post-embryonic development and in the adult organism. The significance of the problem cannot be overstated. Not only are these cells essential for normal development and homeostasis, but understanding their biology has profound translational importance. If we seek to evoke regenerative responses in a clinical content, then post-embryonic stem and progenitor populations may well supply the cells for doing so [1]–[3]. If we hope to delay natural tissue senescence, it is the life cycle of these cells that may need to be manipulated [4]–[7]. And if we aim to control malignancy, these cells or their transformed progeny will often be our targets of choice [8]–[10].
Pigment cells are of great utility for understanding the biology of post-embryonic stem and progenitor cells. Pigment cells are a classic and enduring system for studying morphogenesis and differentiation, and a century of effort has provided a firm understanding of many aspects of pigment cell development in the embryo [11]–[14]. These cells arise from neural crest cells, which migrate from the dorsal neural tube and contribute not only to pigment cells, but also glia and neurons of the peripheral nervous system, bone and cartilage of the craniofacial skeleton, and more. Despite the long-standing interest in these embryonic events, it is now clear that pigment cells of adults derive in large part from post-embryonic stem cells that are themselves of neural crest origin [15]–[18]. We know some of the mechanisms that underlie post-embryonic precursor development yet many outstanding questions remain. Foremost among these concern the genes and cellular behaviors by which pigment stem or progenitor cells are established during early development and subsequently maintained, whether there exist distinct subpopulations of such cells with different genetic requirements and potentials, and how these cells are recruited during normal development and homeostasis.
Answers to these questions will provide insights into the basic biology of the adult pigment cell lineage, and can inform our understanding of post-embryonic neural crest derivatives as well as stem and progenitor cells more generally. These answers are also of enormous biomedical significance, as the skin pigment cell of mammals, the melanocyte, is associated with a variety of human pathologies [19] and transformed cells of this lineage give rise to melanoma, one of the most common cancers [20], [21] and also one of the most deadly [21]–[25]. Poor outcomes reflect the inefficacy of non-surgical treatments and the highly invasive character of melanoma cells [26]–[29]. This invasiveness results in part from neural crest and melanocyte-specific factors that are already expressed by untransformed precursors, as well as lineage-specific factors that are re-expressed upon transformation [30]. Better understanding the genetic requirements and dynamics of melanocyte development and homeostasis can thus provide insights into the behaviors of transformed cells, and may suggest novel strategies for clinical intervention.
In recent years, the zebrafish has proven to be a tractable system for elucidating features of pigment cell development in the embryo and during the larval-to-adult transformation, a period of post-embryonic development analogous to later organogenesis, fetal and neonatal development of mammals [31]–[33]. In the embryo, neural crest cells differentiate into embryonic/early larval melanophores, the zebrafish homologue of mammalian melanocytes. Melanophores and melanocytes depend on many of the same genes and pathways [12], [14], [34], and melanomas with characteristics equivalent to those of human melanomas can be induced in zebrafish [18], [34], [35].
The development of zebrafish adult pigmentation involves a “pigment pattern metamorphosis” in which an embryonic/early larval pigment pattern is transformed into that of the adult [12], [33], [36]–[38]. Whereas the embryonic/early larval pigment cells and pattern develop by 3.6 SSL (standardized standard length [33]; about 4 days post-fertilization), new “metamorphic” melanophores begin to differentiate scattered over the flank by 5.9 SSL and ultimately migrate to form the adult stripes. Simultaneously, additional metamorphic melanophores appear already at sites of stripe formation, and many embryonic/early larval melanophores are lost. These events culminate in a juvenile pigment pattern by 11.0 SSL (4–5 weeks post-fertilization), consisting of two melanophore stripes bounding a lighter “interstripe”. Melanophores comprising these stripes reside in the “hypodermis” between the epidermis and the myotomes [39], [40]. Other adult melanophores are found in the epidermis, the dorsal scales, and the fins. Two additional classes of pigment cells also develop: yellow–orange xanthophores, which populate the interstripe and are required for organizing melanophores into stripes [38], [41], [42]; and iridescent iridophores, which are initially limited to the interstripe but later occupy melanophore stripes as well [33]. During later adult development, additional stripes and interstripes are added as the fish grows.
Mutants with pigment pattern defects limited to post-embryonic stages have suggested a model in which embryonic/early larval melanophores develop directly from the neural crest, whereas metamorphic melanophores develop from latent stem cells of presumptive neural crest origin. For example, picasso and puma mutants have normal embryonic/early larval melanophores, but profound deficiencies in their complements of metamorphic melanophores. picasso encodes the neuregulin receptor erbb3b, which acts both autonomously and non-autonomously to the metamorphic melanophore lineage. Pharmacological inhibition of ErbB signaling further revealed that erbb3b activity is required during neural crest migration for the later development of metamorphic melanophores, suggesting this locus is essential for establishing a pool of precursors that will differentiate only later during the larval-to-adult transformation [43]. By contrast, puma encodes tubulin alpha 8-like 3a (tuba8l3a) and acts autonomously to the metamorphic melanophore lineage. The temperature sensitivity of this allele allowed the identification of a critical period during pigment pattern metamorphosis, suggesting a role in maintaining or expanding a population of latent precursors, or recruiting these cells as melanophores [36], [44], [45].
To date it has not been known where latent precursors to metamorphic melanophores reside, how erbb3b, tuba8l3a or other loci promote the normal morphogenesis and differentiation of these cells and their progeny, or whether pigment cell precursors have indefinite or more limited re-population potential. Here we investigate these issues using molecular marker analyses, transgenesis, vital labeling, and time-lapse imaging in wild-type and mutant backgrounds. We show that during post-embryonic development, proliferative pigment cell precursors are associated with peripheral nerves and ganglia, and migrate to the hypodermis during pigment pattern metamorphosis where they differentiate as melanophores and iridophores. Nerve-associated pigment cell precursors are missing or reduced in ErbB-deficient and tuba8l3a mutant backgrounds. By contrast, these precursors persist in other mutants having less severe metamorphic melanophore deficiencies, though their subsequent development is marked both by defects, and partial regulation, of morphogenesis and differentiation. Finally, we show that latent precursors persist into the adult but that different precursor pools have different regenerative potentials. These findings provide a critical context for understanding the cellular bases of adult melanophore development, the mechanistic underpinnings of mutant phenotypes, and the roles for latent precursors in adult homeostasis, regeneration, and neoplasia.
Results
A melanogenic, erbb3b-dependent population of extra-hypodermal progenitor cells
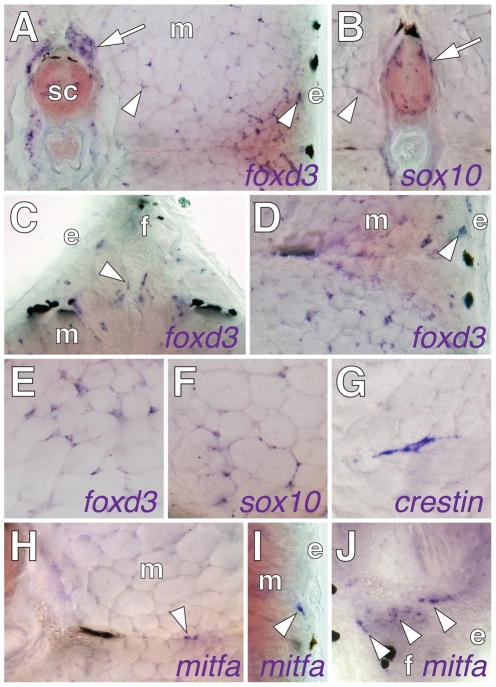
To identify tissues that might harbor latent precursors to adult melanophores, we examined post-embryonic zebrafish for transcripts expressed by embryonic neural crest cells, reasoning that some of the cells expressing such markers might comprise a population of undifferentiated melanophore precursors. We examined foxd3 and sox10, which are expressed by early neural crest cells, subpopulations of neural crest-derived glia, and some other cell types [46]–[49], as well as crestin, which is known only to be expressed by neural crest cells and their derivatives [50]. Cells expressing these loci were present in the hypodermis where the adult pigment pattern forms, but also in the myotomes, adjacent to the spinal cord, and at the bases of the fins (Figure 1A–1G), raising the possibility of both hypodermal and “extra-hypodermal” precursors for metamorphic melanophores.
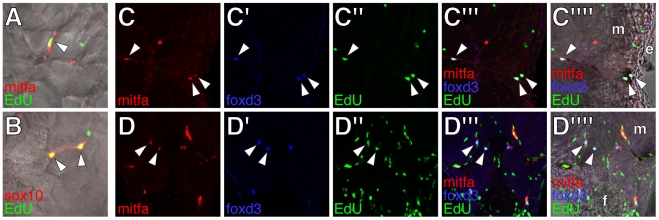
Figure 1. Post-embryonic expression of embryonic neural crest and glial markers.
Shown are in situ hybridizations performed on transverse sections of 7–9 SSL larvae. (A) foxd3 transcript was detected in dorsal root ganglia (arrow) and scattered cells (e.g., arrowheads) within the myotome (m) and near the epidermis (e). (B) sox10 expression by cells adjacent to the neural tube (arrow) and within the myotome (arrowhead). (C) foxd3+ cells (e.g., arrowheads) at the base of the dorsal fin (f). (D) foxd3+ cells within the myotomes and near the epidermis. (E–G) foxd3+, sox10+, and crestin+ cells within the myotomes. (H–J) Cells expressed mitfa (arrowheads), within the horizontal myoseptum (H), at the surface of the myotome (I), and at the base of anal fin (J) (see text for details).
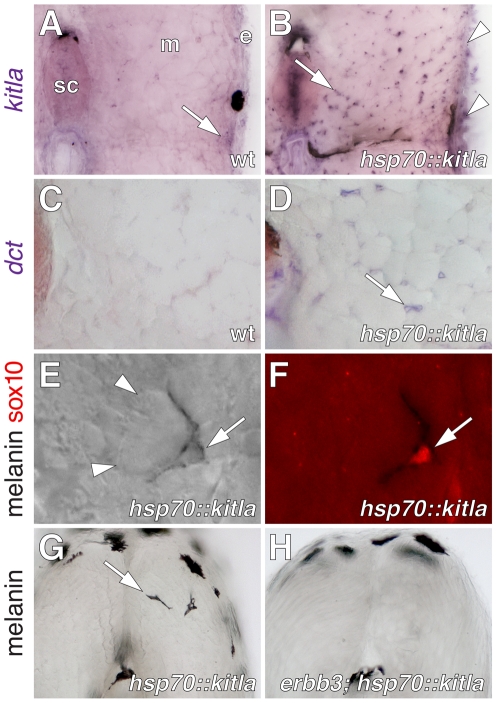
If extra-hypodermal, post-embryonic cells expressing genes typical of early neural crest cells contribute to metamorphic melanophores, we hypothesized that some of these cells should differentiate if supplied with appropriate trophic support. To test this idea, we used a heat-shock inducible transgenic line, Tg(hsp70::kitla)wp.r.t2, to misexpress the melanogenic factor kit ligand-a (kitla) [51] throughout the larva (Figure 2A, 2B). These larvae developed ectopic melanophores deep within the myotomes, which were never found in identically treated siblings lacking the transgene (Figure 2C–2G).
Figure 2. kitla misexpression induced ectopic melanophores within the myotomes.
(A) kitla was normally expressed within the epidermis and hypodermis (arrow) during post-embryonic development of wild-type fish. sc, spinal cord. m, myotome, e, epidermis. (B) In sibling Tg(hsp70::kitla) larvae, heat shock resulted in increased kitla transgene expression within the epidermis (arrowheads) as well as ectopic expression within the myotomes (arrow), spinal cord, and elsewhere. (C) The late melanophore lineage marker dopachrome tautomerase (dct), encoding an enzyme required for melanin synthesis [96], [97], was not expressed within the myotomes of wild-type fish. (D) However, dct was expressed by scattered cells within the myotomes (arrow) in larvae misexpressing kitla. (E,F) Newly differentiated ectopic melanophores (arrow) were found between myotubes (arrowheads; E) and these cells continued to express sox10 protein (F). (G) Vibratome section revealing ectopic melanophores (arrow) within the myotome of a Tg(hsp70::kitla) larva 48 h after the initiation of kitla misexpression. Melanophores deep within the myotome were found only in Tg(hsp70::kitla), though melanophores were occasionally found within the horizontal myoseptum of both transgenic and wild-type larvae [ectopic melanophores per larva, Tg(hsp70::kitla): mean±SE = 1.3±0.15, range = 0–7 cells, n = 80 larvae; non-transgenic siblings: mean±SE = 0±0, range = 0, n = 69]. Longer durations of kitla misexpression resulted in more ectopic melanophores per larva. Suggesting that ectopic melanophores differentiated in situ rather than migrated into the myotomes from the hypodermis, labeling of hypodermal cells by photoconversion of mitfa::Eos+ [55] failed to reveal movement of cells away from enhanced kitla expression in the epidermis into the myotome (n = 10 larvae, 3–5 cells per individual). (H) In contrast to the wild-type, ectopic melanophores were significantly fewer in erbb3b; Tg(hsp70::kitla) mutants [Wilcoxon test, Z = 7.1, P<0.0001; ectopic melanophores per larva, erbb3b; Tg(hsp70::kitla): mean±SE = 0.04±0.03, range = 0–1, n = 50 larvae; non-transgenic siblings: mean±SE = 0±0, range = 0, n = 70] and in Tg(hsp70::kitla) larvae treated with AG1478 during the ErbB embryonic critical period [Wilcoxon test, Z = 2.9, P<0.005; ectopic melanophores per larva, AG1478-treated Tg(hsp70::kitla): mean±SE = 0.7±0.2, range = 0–4, n = 45 larvae; untreated Tg(hsp70::kitla) siblings: mean±SE = 1.6±0.2, range = 0–5, n = 35].
Previously we showed that erbb3b is essential for establishing precursors to metamorphic melanophores [43]. Accordingly, if extra-hypodermal kitla-responsive melanogenic cells and metamorphic melanophores arise from a common precursor pool, then ectopic melanophores should fail to develop in erbb3b mutants misexpressing kitla. Indeed, erbb3b; Tg(hsp70:kitla) larvae exhibited ∼30-fold fewer ectopic melanophores than wild-type Tg(hsp70::kitla) larvae (Figure 2H). A similar outcome was observed when wild-type larvae were treated with the ErbB inhibitor, AG1478, during the embryonic critical period for erbb3b activity in establishing metamorphic melanophore precursors [43]. Together, these results indicated that some extra-hypodermal cells are competent to differentiate as melanophores in the wild-type, but that most of these cells are missing when erbb3b activity is lost.
Extra-hypodermal melanophore precursors are nerve-associated, proliferative, and specified for the melanophore lineage during the larval-to-adult transformation
Our identification of extra-hypodermal cells expressing genes typical of early neural crest and glial cells, as well as kitla-responsive melanogenic cells within the myotomes, led us to ask whether any of these cells embark upon a melanophore differentiation program during normal post-embryonic development. Since precursors to metamorphic melanophores require erbb3b, we further predicted that any candidate extra-hypodermal precursors of these cells should be missing in larvae deficient for erbb3b activity.
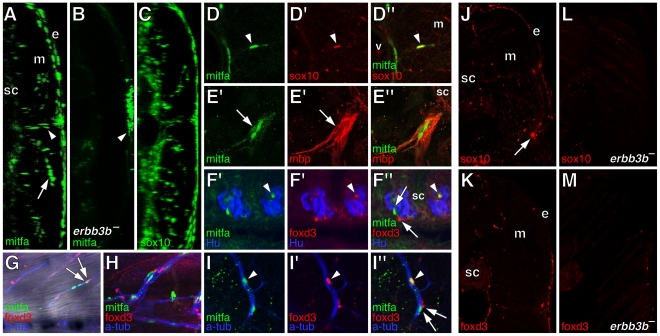
In the wild-type, we found extra-hypodermal cells expressing mitfa, encoding a master regulator of melanophore fate specification (Figure 1H–1J) [52], [53]. Cells expressing mitfa in extra-hypodermal locations typically did so at lower levels than cells within the hypodermis and were at the limit of detection given the sensitivity of in situ hybridization at post-embryonic stages. However, we also identified extra-hypodermal cells expressing GFP driven by the proximal mitfa promoter in the transgenic line, Tg(mitfa::GFP)w47, which faithfully recapitulates mitfa expression in the melanophore lineage and in bipotent precursors to melanophores and iridophores in the embryo [54], [55] (Figure 3A; Videos S1, S2). In contrast to the wild-type, mitfa::GFP+ cells were largely absent from both erbb3b mutants and wild-type larvae treated with AG1478 during the embryonic erbb3b critical period (Figure 3B and see below; Video S3). In neither genetic background could we detect extra-hypodermal cells expressing transcript for dct, encoding a melanin synthesis enzyme.
Figure 3. Extra-hypodermal cells expressing mitfa::GFP, foxd3, and sox10 in wild-type larvae and their deficiency in erbb3b mutants.
All images are from early metamorphic (6.2–8.0 SSL) wild-type larvae except for B, L, and M, from erbb3b mutant larvae. (A–C) Transverse confocal projections (collapsing ∼1.5 mm of trunk along the anterior-posterior axis) showing GFP+ cells in living larvae (left side of each larva is shown). Images correspond to Videos S1, S2, S3. (A) mitfa::GFP+ cells in a wild-type fish occur in the hypodermis, between the epidermis (e) and the myotome (m), within the the myotome itself (arrow), and above the spinal cord (sc). Arrowhead, location of the horizontal myoseptum. (B) In erbb3b mutants, most mitfa::GFP+ cells were missing. This image is intentionally overexposed compared to A, revealing faint reflected fluorescence from iridescent iridophores in the hypodermis (arrowhead), which are present in wild-type larvae as well. (C) sox10::GFP+ cells were found in extra-hypodermal locations of wild-type Tg(−4.9sox10:egfp)ba2 larvae [98]. (D–M) Immunohistochemical analyses of fixed specimens. (D) Co-expression of mitfa::GFP (green) and sox10 (red). v, vertebral column. (E) mitfa::GFP+ cells aligned with mbp+ glia (red) along ventral root motor fibers. Arrow, mitfa::GFP+ cells did not co-express mbp. (F) Lateral view showing mitfa::GFP+ cells and foxd3+ cells (red) between Hu+ neurons (blue) of dorsal root ganglia. mitfa::GFP+ and foxd3+ cells were often found close to one another (e.g., arrows) whereas other cells co-expressed mitfa::GFP and foxd3 (arrowhead). (G) Lateral view with superimposed brightfield and fluorescence images showing mitfa::GFP+ and foxd3+ cells along a peripheral nerve fiber stained for acetylated alpha tubulin (blue) within the myotome. Arrows, adjacent mitfa::GFP+ and foxd3+ cells. (H). A nerve plexus near the base of the caudal fin harbored numerous mitfa::GFP+ and foxd3+ cells. (I) Along a peripheral nerve within the myotome some cells co-expressed mitfa::GFP and foxd3 (arrowhead), whereas cells expressing either mitfa::GFP+ or foxd3+ were often juxtaposed (arrows). (J,K) Transverse sections through the dorsal trunk showing sox10+ cells (J) and foxd3+ cells (K) n the hypodermis, within the myotomes, and near the spinal cord. Arrow, lateral line nerve. (L,M) In erbb3b mutant larvae, very few sox10+ (L) or foxd3+ (M) cells were found.
The many extra-hypodermal mitfa::GFP+ cells in larvae contrasts with embryogenesis [54] and suggests that extra-hypodermal cells may be specified for the melanophore lineage during the larval-to-adult transformation. If so, we predicted that cells expressing markers typical of early neural crest cells (or glia) should occassionally be found to express mitfa::GFP as well. We therefore examined larvae for simultaneous expression of mitfa::GFP, sox10, and foxd3, and, to learn where these cells reside, we examined larvae with cell-type specific markers for surrounding tissues. These analyses revealed numerous mitfa::GFP+ cells associated with peripheral nerves and ganglia, including the doral root ganglia, ventral motor root fibers, lateral line nerve, and nerve fibers coursing through the myotomes (Figure 3E–3H; Figure S1). Ectopic melanophores within the myotomes of wild-type Tg(hsp70:kitla) larvae were likewise nerve-associated (Figure S2).
Double label analyses with markers of neural crest, glial, and melanophore lineages revealed that, in wild-type larvae, mitfa::GFP+ cells were often in close proximity to cells expressing sox10 and foxd3, and also co-expressed sox10, as expected given the direct regulation of mitfa by sox10 (Figure 3D, 3G, 3H) [56], [57]. We also found that 4–15% of mitfa::GFP+ cells co-expressed foxd3, with doubly labeled cells occurring most frequently during early metamorphosis, when the rate of melanophore population increase is maximal [36] (Figure 3F, 3I; Figure 4A, 4B). This frequency of double labeling is reminiscent of 15–18 h embryos, in which 9–12% of mitfa::GFP+ cells are foxd3+ [54]. In contrast to the co-labeling of mitfa::GFP and foxd3, we did not find mitfa::GFP expression by myelinating glia expressing myelin basic protein (mbp) (Figure 3E; Figure S1). As anticipated, however, some cells expressing foxd3 or sox10 co-expressed mbp. All of these cell types were deficient in erbb3b mutants and wild-type larvae treated with AG1478 during the embryonic erbb3b critical period (Figure 3J–3M; Figure 4A, 4B; Figure S4), though a few residual mitfa::GFP+ and foxd3+ cells occurred in anterior and posterior regions, corresponding to axial levels where a few residual melanophores develop during metamorphosis [43] (data not shown).
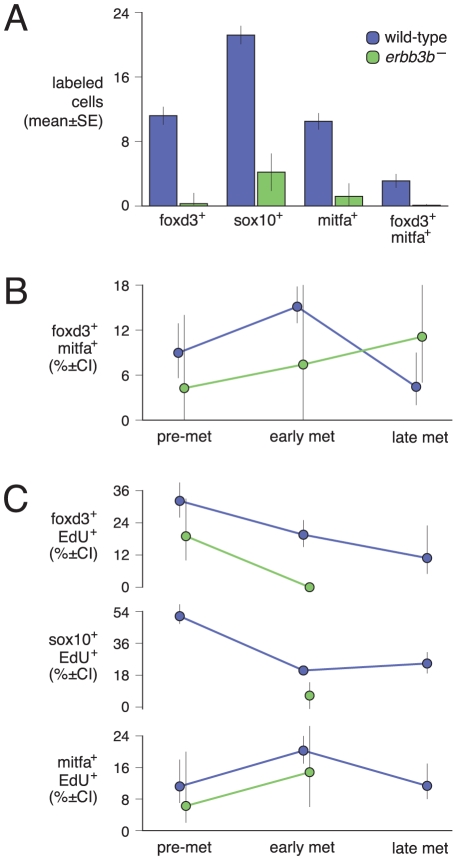
Figure 4. Missing extra-hypodermal precursor cells in erbb3b mutants, co-expression of molecular markers, and temporally regulated proliferation.
(A) Occurrence of cells in sections from the mid-trunk of wild-type and erbb3b mutant larvae. Each class of cells was reduced in erbb3b mutants (all P<0.0001). (B,C) Cell frequencies in wild-type and erbb3b mutants across stages. pre-met, pre-metamorphosis (4.9–5.3 SSL); early met, early metamorphosis (6.2–8.0 SSL); late met, late metamorphosis (9.0–13.0 SSL). (B) The frequency of foxd3+; mitfa::GFP+ cells was greatest in wild-type larvae during early pigment pattern metamorphosis (doubly vs. singly labeled cells, difference among stages: χ 2 = 15.7, d.f. = 2, P<0.0005; N = 1217 total cells examined). Doubly labeled cells tended to be rarer and delayed in erbb3b mutants (N = 83 total cells examined). (C) The frequencies of EdU+ cells differed significantly among stages, with more foxd3+ and sox10+ cells labeled with EdU during the pre-metamorphic period, and more mitfa::GFP+ cells labeled with EdU during early metamorphosis (EdU labeling frequency variation among stages, foxd3+: χ 2 = 11.3, d.f. = 2, P<0.005, N = 450 cells; sox10+: χ 2 = 140.7, d.f. = 2, P<0.0001; N = 1679 cells; mitfa::GFP+: χ 2 = 14.4, d.f. = 2, P<0.001, N = 927 cells). In erbb3b mutants, EdU labeling frequencies were reduced in comparison to wild-type for foxd3+ cells (χ 2 = 3.4, d.f. = 1, P = 0.06; N = 44 cells) and sox10+ cells (χ 2 = 11.4, d.f. = 1, P<0.001; N = 77 cells), though not significantly so for mitfa::GFP+ cells (P>0.1; N = 59 cells). Asymmetric confidence intervals in B and C, Bayesian 95% upper and lower bounds.
If some foxd3+ and sox10+ cells are progenitors to post-embryonic melanoblasts, we predicted that a period of population expansion could precede the appearance of mitfa::GFP+ cells, which might themselves be proliferative. Consistent with this idea, we found EdU incorporation by all three cell types but EdU-labeling of post-embryonic foxd3+ and sox10+ cells was most frequent prior to pigment pattern metamorphosis, whereas EdU-labeling of mitfa::GFP+ cells was most frequent during the peak of pigment pattern metamorphosis (Figure 4C, Figure 5). Given these findings, we further asked if doubly labeled foxd3+; mitfa::GFP+ cells constitute an especially proliferative population. These analyses revealed EdU-incorporation in 55% of foxd3+; mitfa::GFP+ cells, a significantly higher frequency than for cells expressing only foxd3 (17% EdU+) or only mitfa::GFP (24% EdU+; χ 2 = 131.7, d.f. = 2, P<0.0001). The frequency of EdU labeling amongst foxd3+; mitfa::GFP+ cells did not vary significantly across stages (χ 2 = 2.3, d.f. = 2, P = 0.3). Finally, in erbb3b mutants sampled at selected stages, we found lower levels of EdU incorporation than in wild-type, though small numbers of cells overall resulted in correspondingly low statistical power (Figure 4C; Figure S4).
Figure 5. Proliferative extra-hypodermal cells revealed by post-embryonic EdU incorporation.
All images from transverse sections of wild-type larvae staged as in Figure 3 (see Figure S4 for comparisons with erbb3b mutant larvae). (A,B) Merged images showing cells (arrowheads) within the myotomes labeled for either mitfa::GFP (red in A) or sox10::GFP (red in B) as well as EdU (green). (C,D) Cells within the lateral myotomes (m) and near the hypodermis (C) or at the base of the anal fin (f in D) labeled for mitfa::GFP (red), foxd3 (blue), and EdU (green). Merged views show fluorescence images or fluorescence images with brightfield overlays. Arrowheads, triple-labeled cells. m, myotome. e, epidermis.
The foregoing analyses indicated that, during post-embryonic development, a proliferative population of extra-hypodermal, erbb3b-dependent foxd3+ and sox10+ cells associated with peripheral nerves and ganglia becomes specified as precursors to melanophores (or as bipotential precursors to melanophores and iridophores; see reference [55]).
Extra-hypodermal cells contribute to the hypodermal populations of metamorphic melanophores and iridophores
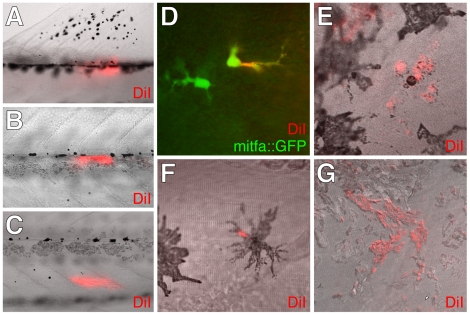
The development of post-embryonic, extra-hypodermal mitfa::GFP+ cells suggested the possibility that such cells migrate to the hypodermis during metamorphosis. To test this idea, we injected DiI into the myotomes, the horizontal myoseptum, or the base of the dorsal fin of wild-type or Tg(mitfa::GFP) larvae and we assessed after ≥4 d whether DiI-labeled cells were present within the hypodermis distant from the injection sites. In 10–30% of injected larvae we found hypodermal DiI-labeled melanized cells, mitfa::GFP+ cells, or iridophores (Figure 6), indicating that some extra-hypodermal cells migrated to the hypodermis during metamorphosis.
Figure 6. DiI-labeling showed extra-hypodermal contributions to metamorphic melanophores and iridophores.
(A–C) DiI labeled tissues imaged immediately after injection into the base of the dorsal fin (A), the vicinity of the horizontal myoseptum and lateral line nerve (B), and the inner myotome (C). Each site yielded hypodermal DiI+; mitfa::GFP+ cells or DiI+ melanophores (12 of 30 larvae, 3 of 30 larvae, 15 of 87 larvae, respectively). (D–F) DiI+ cells that expressed either mitfa::GFP (D) or contained melanin (E,F) found within the lateral hypodermis 4 d following injection into the base of the dorsal fin (D, F) or the inner myotome (E). (G) DiI-labeling was observed for additional cells including iridophores. Although the frequencies with which DiI labeled pigment cells were found differed between injection sites, each site gave rise to DiI+ iridophores at a frequency indistinguishable from that of DiI+ mitfa::GFP+ cells (χ 2 = 0.6, d.f. = 1, P = 0.4). We did not observe DiI-labeled xanthophores.
To further test the hypothesis that extra-hypodermal cells contribute to the hypodermal melanophore population, we examined cell behaviors by time-lapse imaging of trunks derived from Tg(mitfa::GFP) larvae. Movies revealed the differentiation of mitfa::GFP+ cells into melanophores as well as their migration (Figure 7A; Videos S4, S5, S6). We therefore assessed the migratory pathways by which mitfa::GFP+ cells had reached the hypodermis. Approximately half of all mitfa::EGFP+ cells arrived within the hypodermis during imaging. Some entered the hypodermis after migrating over the dorsal or ventral margins of the myotomes, whereas others originated from within the myotomes, emerging either from the vicinity of the horizontal myoseptum or along vertical myosepta (Figure 7B–7D, Figure 8; Videos S7, S8, S9, S10). Movies also revealed the movement of mitfa::GFP+ cells along nerves and their detachment from nerves to migrate more broadly through the fish (Videos S11, S12). Together then, DiI labeling and time-lapse imaging indicated that extra-hypodermal cells contribute to hypodermal mitfa::GFP+ cells, melanophores, and iridophores during the larval-to-adult transformation.
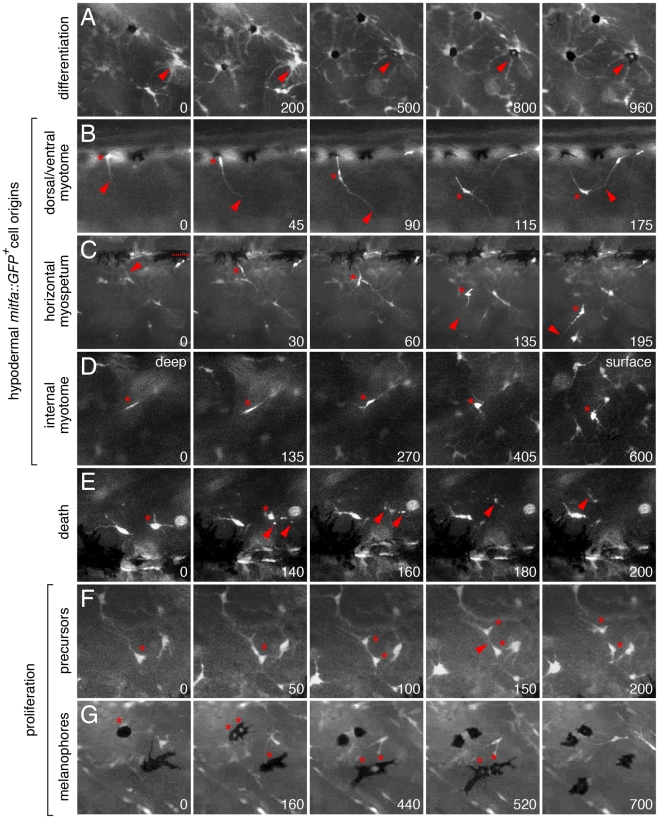
Figure 7. Ex vivo time-lapse imaging revealed extra-hypodermal origins and morphogenetic behaviors of hypodermal mitfa::GFP+ cells and melanophores.
All panels show lateral views of larval trunks and are derived from time-lapse movies of Tg(mitfa::GFP) larvae. Elapsed time (min) at lower right of each panel. (A) mitfa::GFP+ cells differentiated into melanophores (e.g., arrowhead). (B–D) mitfa::GFP+ cells entered the hypodermis during the larval-to-adult transformation. (B) A cell at the dorsal margin of the myotomes extended a long process (arrowhead) into the hypodermis and interacted with processes of a second cell. *, cell body. (C) A long process (arrowhead) preceded emergence of the cell body (*) from the level of the horizontal myoseptum (dotted line). This cell subsequently interacted with a neighboring cell, extended a processes ventrally, and moved in that direction. (D) A cell initially deep within the myotome (*) emerged into the hypodermis. The focal plane changes across panels, from deep within the myotome to the surface of the myotome and hypodermis, where other cells are found already. (E) Death of mitfa::GFP+ cell (*) revealed by fragmentation and cellular debris (arrowheads). (F,G) mitfa::GFP+ cells (F) and melanophores that retain some residual GFP expression (G) proliferating within the hypodermis. Melanophores in G are imaged in a kita mutant (see text for details). See Videos S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16.
Figure 8. Sources of mitfa::GFP+ cells that entered the hypodermis.
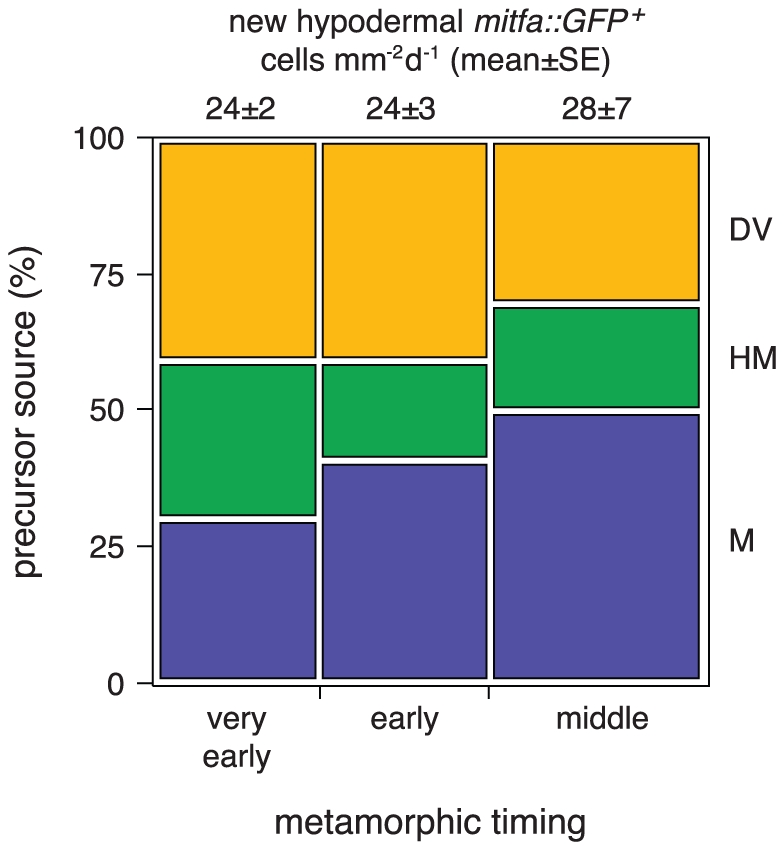
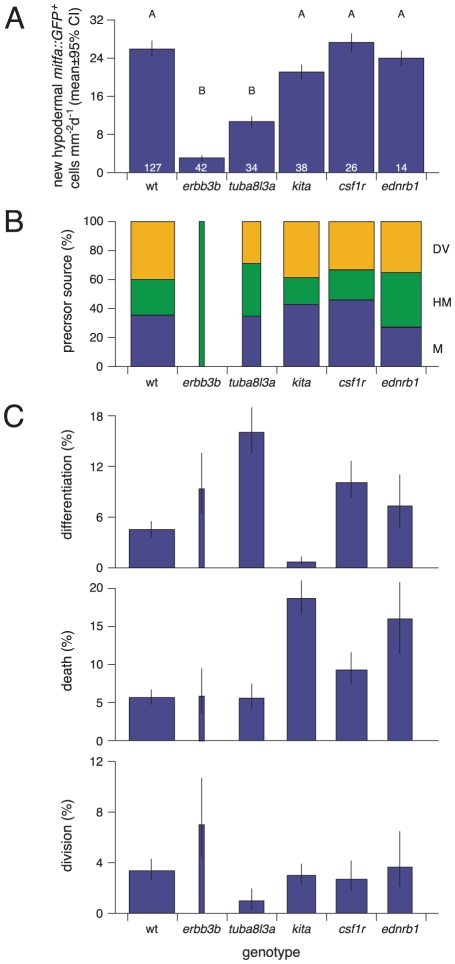
Shown are the relative frequencies of cells entering the hypodermis in time-lapse movies of trunks derived from wild-type larvae (N = 127 larvae, 1220 mitfa::GFP+ total cells examined) during different periods of pigment pattern metamorphosis (very early, 6.4–6.6 SSL; early, 6.6–6.8 SSL; middle, 6.8–7.6 SSL). Cells newly arrived in the hypodermis (n = 644) were classified as migrating over the dorsal or ventral margins of the myotomes (DV), emerging from the vicinity of the horizontal myoseptum (HM) and lateral line, or emerging from within the myotomes (M), tyically along vertical myosepta. Individuals were binned by stage and bar widths are proportional to the average numbers of newly appearing hypodermal mitfa::GFP+ cells per larva, normalized to cells mm−2 day−1 (mean±SE above bars). The relative frequencies of cells arising from different sources differed between stages (χ 2 = 20.0, d.f. = 4, P<0.0001), with cells increasingly likely to emerge from within the myotomes as compared to migrating over the dorsal or ventral margins of the myotomes.
Genetic requirements for extra-hypodermal precursor morphogenesis and differentiation
Our findings suggested that a normal complement of adult melanophores depends on contributions from a pool of extra-hypodermal precursors. If this is the case, we predicted that mutants with severe metamorphic melanophore deficiencies should have correspondingly severe deficiencies of extra-hypodermally derived mitfa:GFP+ cells. To test the contributions of extra-hypodermal cells to hypodermal mitfa::GFP+ cells and melanophores, we crossed the Tg(mitfa::GFP) transgene into erbb3b and tuba8l3a mutants, which exhibit severely reduced numbers of metamorphic melanophores [36], [43], [44].
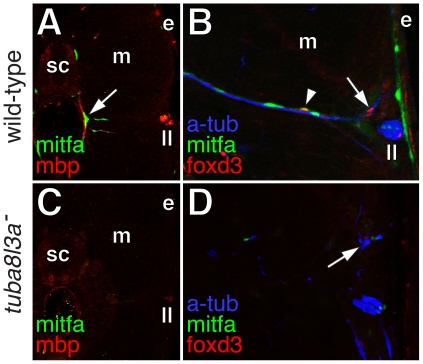
In comparison to the wild-type, and as predicted from the foregoing analyses, erbb3b mutants had dramatically fewer extra-hypodermally derived mitfa::GFP+ cells (Figure 9A; Video S13). erbb3b mutant mifa::GFP+ cells originated from the vicinity of the horizontal myoseptum (Figure 9B), and once in the hypodermis, these cells were more likely to differentiate and to divide (Figure 9C; in contrast to the somewhat reduced rates of EdU incorporation prior to reaching the hypodermis shown in Figure 4C). tuba8l3a mutants also had significantly fewer extra-hypodermally derived mitfa::GFP+ cells. These cells were more likely to differentiate, but divided at only one-third the frequency of the wild-type (Figure 9; Video S14). tuba8l3a mutants exhibit a post-embryonic demyelination of the peripheral nervous system [44], and we found that regions exhibiting mbp+ glial deficiencies and peripheral nerve defasciculation had fewer mitfa::GFP+ and foxd3+ cells (Figure 10). We did not observe cells doubly labeled for foxd3 and mitfa::GFP in tuba8l3a mutants.
Figure 9. Genetic controls over the origins, differentiation, and morphogenesis of hypodermal mitfa::GFP+ cells.
Shown are analyses of time-lapse movies for trunks derived from wild-type and mutant larvae (N = 281 larvae, 5241 total cells examined). (A) Total numbers of newly arising hypodermal mitfa::GFP+ cells differed among genotypes (square root transformed data, F 5,273 = 30.2, P<0.0001). Shown are least squares means (±95% confidence intervals) after controlling for significant differences among stages (F 2,273 = 3.9, P<0.0001) and normalized to cells mm−2 day−1. Letters above bars indicate means that differed significantly (P<0.05) by Tukey-Kramer post hoc comparisons. Numbers within bars indicate numbers of larval trunks examined. (B) The origins of new hypodermal mitfa::GFP+ cells differed among genotypes (χ 2 = 145.6, d.f. = 10, P<0.0001; n = 1582 total new cells). Bar widths are proportional to the total numbers of new hypodermal cells observed in each genotype (shown in A). DV, cells entering the hypodermis after migrating over the dorsal or ventral myotome margins; HM, cells entering from the vicinity of the horizontal myoseptum. M, cells entering from within the myotomes. The sources of mitfa::GFP+ cells did not differ significantly across stages overall (χ 2 = 0.003, d.f. = 4, P = 1), though different genotypes exhibited stage-dependent variation (stage x genotype interaction: χ 2 = 46.3, d.f. = 20, P<0.0001; not shown). (C) Frequencies of differentiation, death and proliferation differed among genotypes. Bar widths are proportional to the total numbers of hypodermal mitfa::GFP+ cells and melanophores observed per larva, after controlling for area and duration of imaging, and normalized to cells mm−2 day−1 (larva means±SE: wild-type, 116±9; erbb3b, 13±8; tuba8l3a, 62±9; kita, 72±10; csf1r, 78±14; ednrb1, 61±16). Differentiation, The likelihood of mitfa::GFP+ cells acquiring melanin during imaging differed among genotypes (χ 2 = 100.6, d.f. = 5, P<0.0001; n = 335 total differentiating cells): the relatively few erbb3b and tuba8l3a mutant cells were especially likely to differentiate whereas very few kita mutant cells differentiated. Additional effects were attributable to stage (χ 2 = 30.3, d.f. = 2, P<0.0001) and a stage x genotype interaction (χ 2 = 27.3, d.f. = 10, P<0.0001; not shown). Death, The incidence of mitfa::GFP+ cells dying during imaging differed among genotypes (χ 2 = 116.9, d.f. = 5, P<0.0001; n = 507 total dying cells) with particularly high rates of death in kita and ednrb1 mutants. Additional variation was attributable to differences among stages (χ 2 = 23.5, d.f. = 2, P<0.0001) and a stage x genotype interaction (χ 2 = 20.4, d.f. = 10, P<0.05) resulting from an increased likelihood of ednrb1 mutant cells dying at later stages (not shown) Division, The incidence of mitfa::GFP+ cells dividing differed significantly among genotypes (χ 2 = 23.6, d.f. = 5, P<0.0001; n = 142 total dividing cells). Asymmetric confidence intervals, Bayesian 95% upper and lower bounds.
Figure 10. Extra-hypodermal precursors were deficient in tuba8l3a mutant larvae.
(A) Wild-type larvae exhibited mitfa::GFP+ cells (green) associated with mbp+ glia (red) of peripheral nerves (arrow). sc, spinal cord; m, myotome; e, epidermis; ll, lateral line nerve. (B) mitfa::GFP+ cells (green), foxd3+ cells (red; arrow), and doubly labeled mitfa::GFP+; foxd3+ cells (arrowhead) were associated with nerve fibers stained for acetylated alpha tubulin (blue). (C) In tuba8l3a mutants, regions deficient for mbp+ glia were also deficient for mitfa::GFP+ cells. (D) Peripheral nerves were often defasciculated (arrow) and were deficient for foxd3+ and mitfa::GFP+ cells.
Amongst the erbb3b- and tuba8l3a-dependent metamorphic melanophore populations, are temporally and genetically distinct subpopulations, comprising early metamorphic melanophores that are initially dispersed but later migrate into stripes, and late metamorphic melanophores that develop already at sites of stripe formation [37], [43], [45] (Figure S5). Early metamorphic melanophores are ablated in kita mutants, but persist in colony stimulating factor-1 receptor (csf1r) and endothelin receptor b1 (ednrb1) mutants. By contrast, late metamorphic melanophores persist in kita mutants, but are ablated in csf1r and ednrb1 mutants [37], [38], [58], [59]. To test if these differences reflect differential persistence of distinct precursor pools, or differences in the subsequent morphogenesis and differentiation of cells arising from a common precursor pool, we crossed the Tg(mitfa::GFP) transgene into kitab5, csf1rj4e1 and ednrb1b140 mutants and examined the origins of hypodermal mitfa::GFP+ cells as well as their frequencies of differentiation, death and proliferation.
In contrast to erbb3b and tuba8l3a mutants, kita, csf1r, and ednrb1 mutants did not exhibit significantly fewer extra-hypodermally derived mitfa::GFP+ cells than the wild-type, though cells in kita mutants typically failed to differentiate and instead died at high frequency, whereas cells in csf1r and ednrb1 mutants were more likely both to differentiate and to die (Figure 9; Video S15). We did not observe gross defects in mitfa:GFP+ cell motility in any of the mutant backgrounds. Finally, in contrast to the proliferation of unmelanized mitf::GFP+ cells (Figure 9C), proliferation of differentiated melanophores was rare in the wild-type (0.1%; N = 3822 melanophores observed) and in most of the mutants (0.2–0.4%; N = 4358 melanophore observed). In kita mutants, however, the few melanophores that differentiated divided frequently (14%; N = 35 melanophores observed; variation among genotypes: χ 2 = 38.1, d.f. = 5, P<0.0001; Video S16). Together these data show that erbb3b and tuba8l3a each promote the development of extra-hypodermal mitfa::GFP+ cells, whereas all five loci promote the differentiation and morphogenesis of these cells once they reach the hypodermis.
Latent melanophore precursors in adult zebrafish
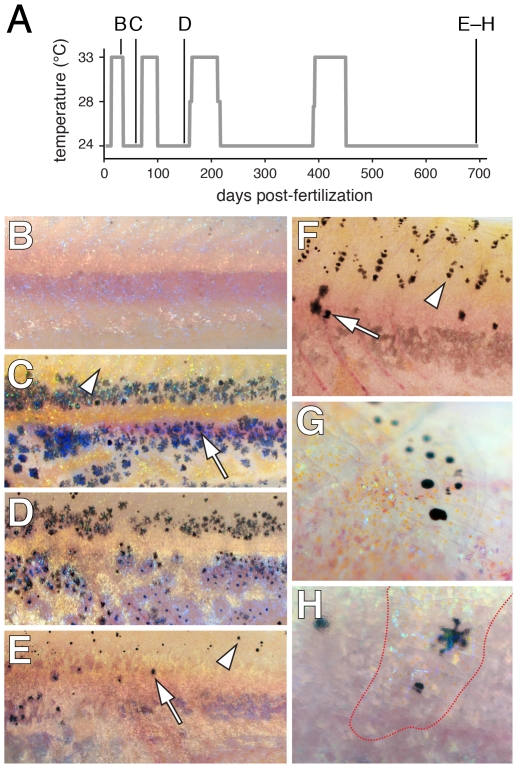
Our demonstration that extra-hypodermal precursors contribute to hypodermal melanophores led us to ask whether latent pigment cell precursors persist into the adult. Extra-hypodermal foxd3+ and mitfa::GFP+ cells were distributed in adult fish similarly to metamorphic stages and also were found associated with the scales (Figure S6). To test the capacity of latent precursors to supply new melanophores, we sought to ablate melanophores with the goal of provoking a regenerative response. Because fish doubly mutant for kitab5 and presumptive null alleles of csf1r lack body melanophores [38], we reasoned that fish doubly mutant for kitab5 and the temperature-sensitive allele csf1rut .r1e174 (csf1rTS) [60] should have fewer melanophores (equivalent to kitab5 single mutants) at permissive temperature, but should lack all melanophores at restrictive temperature. Repeated exposure to restrictive and permissive temperatures should thus allow for repeated cycles of ablation and regeneration of these kita-independent, csf1r-dependent melanophores. As predicted, kita; csf1rTS double mutants that were initially indistinguishable from kita single mutants lost body melanophores when shifted to restrictive temperature (Figure 11A, 11B). After returning to permissive temperature, fish initially recovered kita-independent hypodermal melanophores, though progressively fewer of these cells were regenerated in successive ablation–recovery cycles (Figure 11C, 11D). Surprisingly, ablations also resulted in the de novo development and regeneration of scale melanophores, which are normally absent from kita mutants (Figure 11F, 11G)[37]. The few later hypodermal melanophores that were recovered in kita; csf1rTS mutants were often located beneath scales populated with melanophores, iridophores and xanthophores (Figure 11H), raising the possibility that some of these regenerative hypodermal melanophores may have been scale-derived. Overall, these findings suggest that precursors to kita-independent, csf1r-dependent hypodermal melanophores persist in the adult yet have a finite regenerative potential, whereas an additional precursor pool associated with adult scales has a greater regenerative capability.
Figure 11. Limited regeneration of adult hypodermal melanophores following genetic ablation.
(A) Time-course of temperature shifts, with letters corresponding to images in B–H. Final sample size at 698 days post-fertilization: n = 5. (B) A young adult kita; csf1rTS mutant at 33°C lacked melanophores as in kita; csf1rj4e1 mutants [38] (some melanized cellular debris resulting from melanophore death is evident dorsally). (C) Temperature downshift to 24°C allowed recovery of a hypodermal melanophore (e.g., arrow) complement initially indistinguishable from kita single mutants [59]. Arrowhead, the dorsal flank is initially devoid of scale-associated melanophores, as is typical of kita mutants. (D,E) Additional rounds of ablation and recovery yield progressively fewer hypodermal melanophores (arrow), though some melanophores develop on the dorsal scales (arrowhead). Hypodermal xanthophores and iridophores were depleted as well (data not shown). (F) Dorsal flank of another individual showing hypodermal melanophores (arrow) and scale melanophores (arrowhead). (G) Detail of scale-associated melanophores. Iridescent iridophores and yellow-orange xanthophores are apparent as well. (H) Detail of hypodermal melanophores viewed through an overlying scale containing a concentration of xanthophores and iridophores (outlined in red).
Discussion
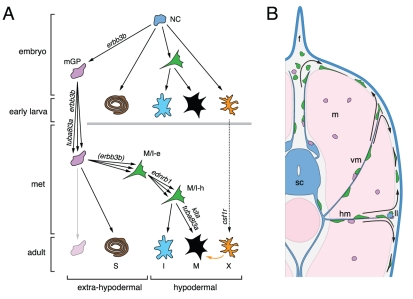
The results of this study and previous analyses [36], [43], [45] suggest a model for the development of adult melanophores in zebrafish (Figure 12). Pluripotent foxd3+ precursors to glia [46], [61], adult melanophores and iridophores are established in an erbb3b-dependent manner during embryogenesis, and thereafter are associated with post-embryonic peripheral nerves and ganglia. This precursor population is expanded and maintained during pre-metamorphic larval development in an erbb3b- and tuba8l3a-dependent manner, and cells within this pool become specified for pigment cell lineages beginning immediately before, and continuing through, pigment pattern metamorphosis. During the larval-to-adult transformation, these extra-hypodermal precursors migrate to the hypodermis, and there contribute to metamorphic melanophores and iridophores. Some enter the hypodermis after migrating over the dorsal or ventral margins of the myotomes, others emerge from vertical or horizontal myosepta; some may emigrate from the lateral line nerve. Once in the hypodermis, these cells require tuba8l3a for their proliferation, as well as kita, ednrb1, and, to a lesser extent, csf1r, for their survival and eventual differentiation. Later in adults, some latent precursors persist and can supply a limited number of new hypodermal melanophores, whereas other precursors associated with scales have a greater regenerative capacity. Below we discuss several aspects and implications of this model.
Figure 12. Model for establishment and maintenance of adult pigment cell precursors and their recruitment during development and regeneration.
(A) Hypothesized lineage relationships, showing neural crest (NC) cells in the early embryo that give rise to Schwann cells and pigment cells of the early larva as well as erbb3b-dependent progenitors to metamorphic glial and pigment cell lineages (mGP). mGP are maintained in association with peripheral nerves and ganglia, express foxd3, and their population expands (multiple arrowheads) in a tuba8l3a-dependent manner. During pigment pattern metamorphosis (met), some mGP differentiate as metamorphic Schwann cells (S) and the expansion of this lineage likely requires erbb3b (not shown). Other mGP become specified for metamorphic pigment cell lineages, as marked by mitfa::GFP expression. Some mitfa::GFP+ cells will give rise to melanophores or iridophores and are initially extra-hypodermally located in peripheral nerves and ganglia (M/I-e) but then migrate to the hypodermis (M/I-h). The expansion of this population requires ednrb1 [58]. Some M/I-h will differentiate as metamorphic iridophores (I), other M/I-h expand their population in a tuba8l3a- and kita-dependent manner and ultimately differentiate as metamorphic melanophores (M). Individual M/I-e or M/I-h may be bipotent for melanophore and iridophore fates, as in embryos [55], or their respective populations may harbor precursors already specified for either the melanophore or iridophore fate. csf1r-dependent metamorphic xanthophores (X) presumably arise from a different precursor population (dashed line) and promote the survival of metamorphic melanophores (orange arrow) [38], [42], [60]. Some mGP persist into the adult and have a limited re-population potential. (B) Schematic of metamorphic larva illustrating sources and migratory pathways of metamorphic melanophore and iridophore precursors. Shown are mGP and M/I-e (colors as in A) associated with nerves and beneath the dorsal fin (f). M/I-e enter the hypodermis (arrows) from the dorsal or ventral margins of the myotomes (m), or after migrating along nerves associated with the the vertical myosepta (vm) or the horizontal myoseptum (hm). Others may arise from the lateral line nerve (ll). Once in the hypodermis, these cells differentiate as melanophores (green cell with heavy black outline) or iridophores (not shown). sc, spinal cord. Additional populations of precursors that may give rise to LM melanophores and scale melanophores are not shown (see text).
Extra-hypodermal niches and the erbb3b- and tuba8l3a-dependence of latent precursors to adult melanophores
A major finding of our study is that post-embryonic mitfa::GFP+ cells are associated with peripheral nerves coursing through the myotomes as well as more medial nerves and ganglia. We further showed that nerve-associated cells could be induced to differentiate ectopically as melanophores, and that extra-hypodermal mitfa::GFP+ cells migrate to the hypodermis where some differentiate as melanophores during normal development. These observations suggest that peripheral nerves or ganglia serve as niches for post-embryonic precursors to adult melanophores and are broadly consistent with a recent study demonstrating a peripheral nerve origin for adult skin melanocytes of amniotes [62] as well as analyses revealing interconversion of glial and melanocyte fates in vitro [63]–[65]. Our study complements and extends recent lineage tracing studies of flounder larvae, in which adult pigment cell precursors were found to migrate to the hypodermis from dorsal and ventral regions during pigment pattern metamorphosis [66], [67].
Our analyses also provide insights into the molecular and proliferative phenotypes of metamorphic melanophore precursors. foxd3 often acts as a transcriptional repressor and is associated with the maintenance of pluripotency and pluripotent cells [68]–[71]. In the neural crest lineage, foxd3 is expressed by pluripotent cells and presumptive glia, and can inhibit mitfa transcription, favoring iridophore or glial over melanogenic fates [54], [55], [61], [72]. We found that some nerve-associated foxd3+ cells co-expressed mitfa::GFP just before and continuing through pigment pattern metamorphosis, and that such cells were especially likely to have incorporated EdU. These observations raise the possibility that nerve-associated foxd3+ cells are a pluripotent and proliferative population that can give rise to hypodermal melanophores and iridophores during pigment pattern metamorphosis. Our finding that DiI-labeled, extra-hypodermal tissues give rise to melanophores and iridophores with equal frequency is consistent with this idea. Because the mitfa::GFP transgene we employed is repressible by foxd3, we speculate that co-expression of mitfa::GFP reflects low levels of perduring foxd3 protein as precursors adopt a pigmentary fate, similar to observations of early neural crest morphogenesis [54], or that a balance between the anti-melanogenic and pro-melanogenic effects of foxd3 and mitfa, respectively, prevents specified cells from differentiating prematurely. Nevertheless, we note that our analyses revealed more extra-hypodermal mitfa::GFP+ cells than mitfa+ cells, which differs from the one-to-one corrrespondence of such cells in embryos [54]. Additional mitfa::GFP+ cells could reflect low levels of mitfa expression that fall below the threshold for detection by in situ hybridization at post-embryonic stages, yet are sufficient for accumulating detectable levels of relatively stable GFP. Or, mitfa::GFP expression in some cells could reflect a partial disregulation of the transgene, as might occur if regulatory elements for post-embryonic expression are missing. Distinguishing between these possibilities will require the production and analysis of additional transgenic reporter lines, but whichever the outcome, the mitfa::GFP reporter we have used will be a valuable tool for further dissecting the mechanisms of post-embryonic melanophore development.
Examination of erbb3b and tuba8l3a mutants provides additional support for the idea that extra-hypodermal, nerve-associated precursors are essential for metamorphic melanophore development. We found that presumptive precursors to glia and pigment cells were missing from erbb3b mutants and wild-type larvae in which ErbB activity had been inhibited during the embryonic critical period for adult melanophore development. An on-going requirement for erbb3b is suggested as well, by reduced complements of adult melanophores following acute inhibition of ErbB signaling in sensitized backgrounds during pigment pattern metamorphosis [43], and by reduced rates of EdU incorporation in erbb3b mutants during post-embryonic development (this study). Because ErbB signaling can repress melanocyte differentiation [62], [73], a further role in preventing the premature differentiation of nerve-associated precursors to hypodermal pigment cells seems likely. Notably, our finding that extra-hypodermal, kitla-responsive melanogenic cells were missing in ErbB-deficient backgrounds at post-embryonic stages contrasts with increased numbers of such cells at embryonic/early larval stages [74]. This difference likely reflects the pleiotropic nature of ErbB signals and a difference between the stages examined: blocking ErbB activity in the embryo presumably results in a de-repression of melanophore differentiation amongst transiently persisting precursor cells, whereas by post-embryonic stages, such precursors presumably have been lost.
In contrast to erbb3b mutants, tuba8l3a mutants exhibit a post-embryonic demyelination of peripheral nerves and a corresponding critical period for adult melanophore development [36], [44], [45]. In agreement with our model, tuba8l3a mutant larvae were deficient for mbp+ glia, nerve-associated foxd3+ cells and foxd3+; mitfa::GFP+ cells, and also had reduced rates of division amongst hypodermal mitfa::GFP+ cells. Because tuba8l3a acts autonomously to the metamorphic melanophore lineage [45], these findings suggest a defect in the maintenance or expansion of pluripotent precursors to mbp+ glia and mitfa:GFP+ pigment cell precursors. The post-embryonic onset of these phenotypes further suggests the existence of genetically distinct populations of embryonic and adult glia, analogous to embryonic and metamorphic melanophores.
Pattern development, regulation, and the roles of temporally and genetically distinct metamorphic melanophore populations
Time-lapse imaging comparisons of wild-type and mutant backgrounds further defined roles for genes previously known to function in pigment pattern development: kita, csf1r, and ednrb1 all promoted the survival of mitfa::GFP+ cells whereas kita also promoted the differentiation of these cells as melanophores. Yet, these analyses revealed compensatory responses of pigment cells and their precursors in mutant backgrounds as well. For example, residual mitfa::GFP+ cells in mutants with extra-hypodermal precursor deficiencies (erbb3b, tuba8l3a) or hypodermal defects in cell survival (csf1r, ednrb1) exhibited concomitantly greater rates of differentiation, and, in one instance (erbb3b), an increased rate of proliferation. Moreover, the reduced survival and differentiation of mitfa::GFP+ cells in kita mutants was coupled with a 70-fold increase in melanophore proliferation. These findings highlight the remarkably regulative nature of zebrafish pigment pattern development [36], [41], [42] and also the importance of direct imaging for understanding cellular behaviors that would not be predicted from terminal phenotypes alone.
A particularly dramatic example of pattern regulation occurs during regeneration. Zebrafish larval melanophores regenerate following laser ablation or the administration of melanocytotoxic drugs [74]–[76], adult fin melanophores regenerate along with other tissues after fin amputation [77]–[79], and hypodermal body melanophores regenerate after localized laser ablations [80], [81]. To test the capacity of latent precursors to supply new hypodermal melanophores in the adult, we used fish doubly mutant for kita and csf1rTS in which loss of residual melanophores at restrictive temperature likely reflects the withdrawal of trophic support provided by csf1r-dependent xanthophores [42], [60]. Our finding that progressively fewer hypodermal melanophores were recovered after repeated ablations implies a limited re-population potential for latent precursors that give rise to these kita-independent, csf1r-dependent late metamorphic melanophores. Whether the same is true of kita-dependent early metamorphic melanophores remains to be determined. Nevertheless, our finding that scale melanophores regenerated repeatedly-in contrast to late metamorphic, hypodermal melanophores-suggests a more highly regulative precursor pool associated with the adult scales, and highlights the possibility of spatially and temporally distinct pools of precursors having different morphogenetic and differentiative potentials. That scale melanophores developed de novo in these fish, despite their absence from kita single mutants, likely reflects a priority effect; e.g., initially abundant xanthophores may repress melanophore development in kita mutants [42], [82], [83] but simultaneous development of both melanophores and xanthophores during regeneration allows for a stable pattern comprising both cell types. The similar re-population potential of scale and fin melanophores, and the previous observation that basonuclin-2 mutants, which are deficient for hypodermal melanophores, nevertheless retain both scale and fin melanophores [84], also suggest the possibility of more extensive similarities between scale- and fin-associated precursor pools.
Finally our study provides new insights into the temporally distinct populations of melanophores in zebrafish. We found severe deficiencies in the numbers of extra-hypodermally derived mitfa::GFP+ cells in erbb3b and tuba8l3a mutants, illustrating the critical role of such cells in supplying metamorphic melanophores overall. By contrast, we found no evidence for reduced numbers of extra-hypodermally derived mitfa::GFP+ cells in kita, csf1r, or ednrb1 mutants, indicating that different genetic requirements of early and late metamorphic melanophores do not reflect differences in the establishment or maintenance of these cells. These findings suggest either of two interpretations. Early and late metamorphic melanophores could arise from a common precursor pool with differences in residual melanophore complements among mutants reflecting specific requirements for kita, csf1r, and ednrb1 in downstream events of morphogenesis and differentiation. For example, our data indicate that mitfa::GFP+ cells require kita for their surival, and presumably, terminal differentiation: the development of late metamorphic melanophores in kita mutants could thus reflect the late appearance of factors able to substitute for kita activity in promoting melanophore differentiation. Conversely, we found that mitfa::GFP+ cells were only marginally dependent on csf1r: the failure of late metamorphic melanophores to develop in csf1r mutants could, in turn, reflect a late-arising, post-differentiation requirement for trophic support from csf1r-dependent xanthophores [38], [42], [60]. An alternative interpretation is that early and late metamorphic melanophores do arise from distinct, but still cryptic, precursor pools that simply were not revealed by our methods. In other systems, niches initially assumed to have just one type of stem or progenitor cell have sometimes been found to harbor distinct classes of cells with disparate proliferative or differentiative potentials [85]–[87]. Additional time-lapse imaging analyses at later stages and genetically based lineage analyses that are now being conducted should provide further insights into these possibilities.
Post-embryonic stem cells in development, evolution, and neoplasia
The identification of extra-hypodermal nerve-associated precursors to adult melanophores in zebrafish (this study) and amniotes [62] indicates that a fuller understanding of adult pigment cell development and pattern formation requires a focus on post-embryonic precursors, as distinct from embryonic neural crest cells. The existence of genetically separable populations of embryonic pigment cells, derived directly from neural crest cells, and adult pigment cells, derived from post-embryonic precursors, further suggests that species-differences in adult pigment patterns may be explicable by evolutionary changes in the establishment, maintenance or recruitment of post-embryonic latent precursors, with few if any consequences for earlier pigment patterns [88]. Lastly, a peripheral nerve origin for adult pigment cells also raises the possibility that the frequent and generally fatal metastases of melanoma cells to the central nervous system [26]–[29],[89],[90] may reflect the continued or reiterated expression of genes that favor proliferation and migration in a nerve microenvironment.
Materials and Methods
Fish rearing, staging, and genetic stocks
Fish were reared at 28–29°C, 14L:10D. Post-embryonic stages are reported as standardized standard length (SSL) measurements, which indicate developmental progress of free-feeding larvae more reliably than days post-fertilization [33]. Tg(mitfa::GFP)w47 and Tg(−4.9sox10:egfp)ba2 fish were generously provided by D. Raible and R. Kelsh, respectively. Tg(TrDct::mCherry)wp.r.t3 and Tg(hsp70::kitla)wp.r.t2 fish were produced using tol2kit Gateway vectors and Tol2-mediated transgenesis [91], [92] and heat shocks with the latter strain were administered at 37°C for 1 hr three times daily for two days. Experiments with erbb3b mutants used either of two presumptive null alleles, erbb3but.r2e1 or erbb3bwp.r2e2 [93]. Experiments with csf1r mutants used either the presumptive null allele csf1rj4blue or the temperature-sensitive allele csf1rut .r1e174 [60]. In temperature shift experiments, fish were shifted repeatedly between restrictive temperature (33°C) and permissive temperature (24°C). All experiments with the kita mutant used the presumptive null allele kitab5 [59]. tuba8l3aj115e1 encodes a missense substitution with temperature-sensitive effects. Experiments with this allele were performed at standard rearing temperature, intermediate between restrictive (33°C) and permissive (24°C) temperatures [36], [44], [45], to allow analyses of a fuller complement of mitfa::GFP+ cells. Quantitative analyses here are thus likely to underestimate effects of the tuba8l3a mutation. Animal use conformed to University of Washington IACUC guidelines.
Imaging and image analysis
Fish were viewed with Olympus SZX-12 or Zeiss Discovery epifluorescence stereomicroscopes or with a Zeiss Observer inverted compound epifluorescence microscope with Apotome. Images were collected in Axiovision software using Axiocam HR or MR3 cameras. For thick specimens, stacks of images were collected and processed using Zeiss Axiovision 6D Acquisition or Extended Focus modules and some fluorescence images were deconvolved using the Zeiss Axiovision Deconvolution module. Alternatively, specimens were viewed and images collected on Zeiss 510 META or Olympus FV1000 laser confocal microscopes.
Histology
For immunohistochemistry, larvae were fixed in 4% paraformaldehyde containing 1% DMSO in PBS, rinsed, embedded in agarose, then sectioned by vibratome at 150–200 µm. Sections were washed in PBS/1% DMSO/0.3% Triton-X pH 7.4 (PDTX), blocked in PDTX containing 10–20% heat inactivated goat serum then incubated overnight at 4°C with primary antibody. We used polyclonal antisera raised in rabbit against zebrafish sox10 (1:500; provided by B. Appel [94]), zebrafish foxd3 (1:400; D. Raible [47]), zebrafish mbp (1:50; W. Talbot [95]), and GFP (1:200; A11122, Invitrogen) as well as monoclonal antibodies against GFP (1:200; 3E6 A11120, Invitrogen) and acetylated α-tubulin (1:200; 6-11B-1, T6793 Sigma). After washing, sections were incubated with secondary antibodies (AlexaFluor 405, 488, 568, 647; Invitrogen), washed and imaged.
In situ hybridization of post-embryonic zebrafish followed [44], [84]. For some analyses larvae were sectioned at 100–300 µm by vibratome. Detailed methods for in situ hybridization are available online at http://protist.biology.washington.edu/dparichy/.
DiI injection
Cell Tracker CM-DiI (Invitrogen) was prepared as a stock solution in DMSO then diluted to 0.025–0.05% in 0.3 M sucrose just before use. Larvae were anesthetized briefly and injected with 1–2 nl of DiI using a borosilicate needle, imaged immediately to ascertain the specificity of staining in target tissues, then reared individually until analyzed.
Ex vivo time-lapse imaging
Larvae were rinsed with 10% Hanks medium, anesthetized and then sacrificed by decapitation using a razor blade. After removing the anterior portion of the trunk and discarding the tails, larval trunks were placed on 0.4 µm transwell membranes (Millipore) in glass-bottom dishes containing L-15 medium, 3% fetal bovine serum, and penicillin/streptomycin. Trunks were equilibrated at 28.5°C for 3 h then imaged for 18–24 h (20 or 30 minute intervals between images) on a Zeiss Observer inverted epifluorescence microscope. Comparisons between isolated trunks imaged continuously for 24 h and repeatedly anesthetized intact larvae did not reveal gross differences in the survival of mitfa::GFP+ cells, though average maximal estimated velocities of mitfa::GFP+ cells were reduced by ∼22% in cultured trunks as compared to intact larvae (P<0.05; N = 67 cells examined). Imaging over longer durations resulted in increased rates of cell death throughout the explant and thus were not used for analyses.
Pharmacological inhibition of ErbB activity
A stock solution of AG1478 [4-(3-chloroanilino)-6,7-dimethoxyquinazoline; Calbiochem] was diluted in DMSO. Embryos were treated with 3 µM AG1478 in 10% Hanks for through either 72 or 96 hours post-fertilization. To facilitate penetration, 0.5% DMSO was added to all media and embryos were dechorionated prior to treatment. Fish were reared in glass Petri dishes and solutions were changed daily.
EdU labeling
Larvae were incubated with 0.005% 5-ethynyl-2′-deoxyuridine (EdU; Intvitrogen) in 10% Hank's medium containing 1% DMSO for 36 h. Larvae were then sacrificed, fixed with 4% PFA/1% DMSO, and vibrotome sectioned (150–200 µm) for immunohistochemistry followed by histochemical detection of EdU according to manufacturer's recommendations.
Statistical analyses
Quantitative data were analyzed with JMP 8.0.2 (SAS Institute, Cary NC). Frequency data were examined using multiple logistic regression or contingency table analyses, and tested for effects of genotype, stage, and genotype x stage interactions. Significance of effects were assessed by likelihood ratio tests and non-significant factors were removed from the final models. Analyses of variance were used for continuous variables including counts. Residuals were examined for normality and homogeneity of variances, conditions that were achieved for some variables after transformation by square root or natural logarithm. Further details of statistical analyses are available upon request.
Supporting Information
mitfa::GFP+ cells amongst glia of the lateral line nerve. Sagittal view of a wild-type larva showing mitfa::GFP+ cells (mitfa, green; arrow) aligned with mbp+ glia (red) of the main trunk lateral line nerve, near the horizontal myoseptum.
(TIF)
Ectopic kitla-responsive melanogenic cells were nerve-associated. Shown is an ectopic sox10+ (red) melanophore (arrow) within the myotome adjacent to a nerve fiber stained for acetylated tubulin (green).
(TIF)
Defects in wild-type larvae treated with ErbB inhibitor AG1478 during the erbb3b embryonic critical period. (A,B) Post-embryonic mbp+ glia (red; arrowheads) were reduced though not eliminated in AG1478-treated larvae. Inset, mitfa::GFP+ cell (green) aligned on mbp+ glia of a peripheral nerve at the level of the horizontal myoseptum. sc, spinal cord; m, myotome; e, epidermis; ll, lateral line nerve. (C,D) foxd3+ cells (red; arrowheads) within the myotome were missing from AG1478-treated larvae.
(TIF)
Deficiencies in mitfa::GFP+ cells, foxd3+ cells, and EdU incorporation in erbb3b mutants. (A,B) Views and annotations correspond to those for wild-type larvae in main text Figure 5. Arrow, a rare EdU+; mitfa::GFP+ cell at the base of the ventral fin.
(TIF)
erbb3b-dependence of kita-independent hypodermal melanophores. (A) kitab5 presumptive null allele [59] with stripes of kita-independent hypodermal melanophores. (B) Fish doubly mutant for kitab5 and the presumptive null allele erbb3but.r2e1 showing loss of many kita-independent melanophores as well as gaps in the “interstripes” (e.g., arrowhead) reflecting an iridophore deficiency. Patches of residual melanophores may be of clonal origin.
(TIF)
mitfa::GFP+ cells in adult fish. Shown are cross-sections through ∼20 SSL (∼80 days post-fertilization) adult wild-type fish. (A) A persisting nerve-associated mitfa::GFP+ cell (green; arrow). sc, spinal cord; m, myotome. (B) mitfa::GFP+ cells (green; arrowhead) within the lateral line nerve. e, epidermis (C) mitfa::GFP+ cells (green; arrow) and foxd3+ cells (red; arrowhead) in the ventral myotomes and base of the anal fin (f). (D) mitfa::GFP+ cell (green; arrow) and doubly labeled mitfa::GFP+; foxd3+ cell (arrowhead) associated with an adult scale (s).
(TIF)
mitfa::GFP+ cells in a living wild-type larva. Images were collected in sagittal planes from the skin to the midline of the trunk. Image rotation reveals extra-hypodermal cells within the myotomes, that typically expressed mitfa::GFP at levels lower than in hypodermal cells.
(MOV)
mitfa::GFP+ and dct::mCherry+ melanophores in a living wild-type larva. In contrast to mitfa::GFP+ cells (green), cells expressing the later melanophore lineage marker dct were detected only in the hypodermis. Shown here are cells expressing mCherry driven by the Takifugu rubripes dct promoter.
(MOV)
mitfa::GFP+ cells are missing from a live erbb3b mutant larva. Larva imaged as in Video S1 but over-exposed to provide spatial context. Fluorescent cells forming a stripe in the hypodermis are iridophores, which reflect under epifluorescence in multiple channels (these cells are present in the larva shown in Video S1, but their reflectance falls beneath the threshold for detection at the exposure shown).
(MOV)
mitfa::GFP+ cells differentiate as melanophores in the hypodermis. mitfa::GFP+ cells acquiring melanin (arrows) over ∼18 h.
(MOV)
:GFP+ cells. Low magnification view showing highly migratory mitfa::GFP+ cells over ∼18 h. This movie loops three times. mitfa::GFP+ cells are highly migratory and extend long probing processes. These cells can be seen migrating widely over the flank and also migrating from the body into the anal fin at the lower edge of the frame. In contrast to mitfa::GFP+ cells, xanthophores are larger, less motile and autofluoresce weakly in the GFP channel, particularly from accumulations of pteridine-containing organelles around the nucleus.
(MOV)
Migration of mitfa::GFP+ cells between body and fin. Detail at the margin between body and anal fin, showing movements of mitfa::GFP+ cells between these regions.
(MOV)
mitfa::GFP+ cells emerging from within the myotome. Several mitfa::GFP+ cells (arrows) enter the hypodermis from within the myotome; also see Video S8.
(MOV)
mitfa::GFP+ cells emerging from within the myotome. High magnification view showing a single mitfa::GFP+ cell (arrow) emerging from within the myotome. In this movie, only selected planes of focus from a complete z-series are shown, illustrating the progressive movement of this cell from the interior myotome to the surface. As the cell approaches the more superficial planes, additional mitfa::GFP+ cells already in the hypodermis come into focus.
(MOV)
mitfa::GFP+ cell emerging from the vicinity of the horizontal myoseptum. The movie loops three times.
(MOV)
mitfa::GFP+ cells entering the hypodermis after migrating over the dorsal myotome. One cell already at the dorsal myotome and another extending a process over the flank at the beginning of the movie (red arrows). A third cell extends a long filopodium (yellow arrow) over the flank before entering into the hypodermis.
(MOV)
mitfa::GFP+ cells migrating within the caudal nerve plexus. Shown are mitfa::GFP+ cells along a nerve plexus near the base of the caudal fin in the medial region of a larva. This particular individual is a kita mutant. Similar behaviors were observed in wild-type larvae.
(MOV)
mitfa::GFP+ cells migrating along and dispersing from peripheral nerves. Shown is an enhanced detail of Video S11. Arrowhead indicates a cell traversing along a nerve fiber then migrating away.
(MOV)
Morphogenesis of residual mitfa::GFP+ cells in erbb3b mutants. Residual mitfa::GFP+ cells were often patchily distributed and typically expressed GFP at lower levels than in wild-type. Red arrowhead, mitfa::GFP+ cell dividing. Arrow, mitfa::GFP+ cell emerging from the vicinity of the horizontal myoseptum. Yellow arrowhead, death of mitfa::GFP+ cell.
(MOV)
Proliferation of mitfa::GFP+ cells. A mitfa::GFP+ cell (arrow) divides within the hypodermis over ∼14 h in a wild-type larva.
(MOV)
Death of mitfa::GFP+ cells. Fragmentation and loss of three mitfa::GFP+ cells (arrows) in a kita mutant larva over ∼16 h (see text for details).
(MOV)
Proliferation of differentiated melanophores. Shown is the posterior tail of a kita mutant larva, illustrating successive melanophore divisions. mitfa::GFP+ cells also can be observed migrating on the body and between the body and caudal fin.
(MOV)
Acknowledgments
Thanks to Dave Raible, Kevin Curran, and other members of the Raible lab for providing reagents and for helpful discussions; members of the Parichy lab for assistance with fish rearing; and David Lyons, Will Talbot, Bruce Appel, and Robert Kelsh, who generously provided reagents.
Footnotes
The authors have declared that no competing interests exist.
This research supported by National Institute of General Medical Sciences (http://www.nigms.nih.gov/), National Institutes of Health grant R01 GM062182 to DMP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bedelbaeva K, Snyder A, Gourevitch D, Clark L, Zhang XM, et al. Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc Natl Acad Sci U S A. 2010;107:5845–5850. doi: 10.1073/pnas.1000830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salewski RP, Eftekharpour E, Fehlings MG. Are induced pluripotent stem cells the future of cell-based regenerative therapies for spinal cord injury? J Cell Physiol. 2010;222:515–521. doi: 10.1002/jcp.21995. [DOI] [PubMed] [Google Scholar]
- 3.Guo JK, Cantley LG. Cellular maintenance and repair of the kidney. Annu Rev Physiol. 2010;72:357–376. doi: 10.1146/annurev.physiol.010908.163245. [DOI] [PubMed] [Google Scholar]
- 4.Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6:103–115. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 6.Mayack SR, Shadrach JL, Kim FS, Wagers AJ. Systemic signals regulate ageing and rejuvenation of blood stem cell niches. Nature. 2010;463:495–500. doi: 10.1038/nature08749. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307:720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- 8.Alison MR, Islam S, Wright NA. Stem cells in cancer: instigators and propagators? J Cell Sci. 2010;123:2357–2368. doi: 10.1242/jcs.054296. [DOI] [PubMed] [Google Scholar]
- 9.Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12:299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 10.Le LQ, Shipman T, Burns DK, Parada LF. Cell of origin and microenvironment contribution for NF1-associated dermal neurofibromas. Cell Stem Cell. 2009;4:453–463. doi: 10.1016/j.stem.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Douarin NM. Cambridge: Cambride University Press; 1999. The Neural Crest. [Google Scholar]
- 12.Kelsh RN, Harris ML, Colanesi S, Erickson CA. Stripes and belly-spots-A review of pigment cell morphogenesis in vertebrates. Semin Cell Dev Biol. 2009;20:90–104. doi: 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas AJ, Erickson CA. The making of a melanocyte: the specification of melanoblasts from the neural crest. Pigment Cell Melanoma Res. 2008;21:598–610. doi: 10.1111/j.1755-148X.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- 14.Cooper CD, Raible DW. Semin Cell Dev Biol; 2008. Mechanisms for reaching the differentiated state: Insights from neural crest-derived melanocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 16.Wong CE, Paratore C, Dours-Zimmermann MT, Rochat A, Pietri T, et al. Neural crest-derived cells with stem cell features can be traced back to multiple lineages in the adult skin. J Cell Biol. 2006;175:1005–1015. doi: 10.1083/jcb.200606062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson KC, Fisher DE. Specification and loss of melanocyte stem cells. Semin Cell Dev Biol. 2009;20:111–116. doi: 10.1016/j.semcdb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 18.White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3:242–252. doi: 10.1016/j.stem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Nordlund JJ, Boissy RE, Hearing VJ, King RA, Oetting WS, et al., editors. 2nd Edition ed:Blackwell Publishing, Inc; 2006. The Pigmentary System: Physiology and Pathophysiology. [Google Scholar]
- 20.Rigel DS. Malignant melanoma: incidence issues and their effect on diagnosis and treatment in the 1990s. Mayo Clin Proc. 1997;72:367–371. doi: 10.4065/72.4.367. [DOI] [PubMed] [Google Scholar]
- 21.Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 22.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Ding S, et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J Clin Oncol. 2010;28:2452–2459. doi: 10.1200/JCO.2009.27.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 25.Uong A, Zon LI. Melanocytes in development and cancer. J Cell Physiol. 2010;222:38–41. doi: 10.1002/jcp.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouawad R, Sebert M, Michels J, Bloch J, Spano JP, et al. Treatment for metastatic malignant melanoma: old drugs and new strategies. Crit Rev Oncol Hematol. 2010;74:27–39. doi: 10.1016/j.critrevonc.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia S, Tykodi SS, Thompson JA. Treatment of metastatic melanoma: an overview. Oncology (Williston Park) 2009;23:488–496. [PMC free article] [PubMed] [Google Scholar]
- 28.Jilaveanu LB, Aziz SA, Kluger HM. Chemotherapy and biologic therapies for melanoma: do they work? Clin Dermatol. 2009;27:614–625. doi: 10.1016/j.clindermatol.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Barth A, Wanek LA, Morton DL. Prognostic factors in 1,521 melanoma patients with distant metastases. J Am Coll Surg. 1995;181:193–201. [PubMed] [Google Scholar]
- 30.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devoto SH, Stoiber W, Hammond CL, Steinbacher P, Haslett JR, et al. Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol Dev. 2006;8:101–110. doi: 10.1111/j.1525-142X.2006.05079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkes JW. The structure of fish skin. I. General organization. Cell Tissue Res. 1974;149:147–158. doi: 10.1007/BF00222270. [DOI] [PubMed] [Google Scholar]
- 33.Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Developmental Dynamics. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceol CJ, Houvras Y, White RM, Zon LI. Melanoma Biology and the Promise of Zebrafish. Zebrafish. 2008;5:247–255. doi: 10.1089/zeb.2008.0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 36.Parichy DM, Turner JM. Zebrafish puma mutant decouples pigment pattern and somatic metamorphosis. Developmental Biology. 2003;256:242–257. doi: 10.1016/s0012-1606(03)00015-0. [DOI] [PubMed] [Google Scholar]
- 37.Johnson SL, Africa D, Walker C, Weston JA. Genetic control of adult pigment stripe development in zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 38.Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 39.Hirata M, Nakamura K, Kanemaru T, Shibata Y, Kondo S. Pigment cell organization in the hypodermis of zebrafish. Dev Dyn. 2003;227:497–503. doi: 10.1002/dvdy.10334. [DOI] [PubMed] [Google Scholar]
- 40.Hawkes JW. The structure of fish skin. II. The chromatophore unit. Cell Tissue Res. 1974;149:159–172. doi: 10.1007/BF00222271. [DOI] [PubMed] [Google Scholar]
- 41.Parichy DM Turner JM. Cellular interactions during adult pigment stripe development in zebrafish; Academic Press Inc Elsevier Science. 2003. 486
- 42.Nakamasu A, Takahashi G, Kanbe A, Kondo S. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc Natl Acad Sci U S A. 2009;106:8429–8434. doi: 10.1073/pnas.0808622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budi EH, Patterson LB, Parichy DM. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development. 2008;135:2603–2614. doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson TA, Gordon TN, Lau HE, Parichy DM. Defective adult oligodendrocyte and Schwann cell development, pigment pattern, and craniofacial morphology in puma mutant zebrafish having an alpha tubulin mutation. Dev Biol. 2010;346:296–309. doi: 10.1016/j.ydbio.2010.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parichy DM, Turner JM, Parker NB. Essential role for puma in development of postembryonic neural crest-derived cell lineages in zebrafish. Dev Biol. 2003;256:221–241. doi: 10.1016/s0012-1606(03)00016-2. [DOI] [PubMed] [Google Scholar]
- 46.Kelsh RN, Dutton K, Medlin J, Eisen JS. Expression of zebrafish fkd6 in neural crest-derived glia. Mech Dev. 2000:161–164. doi: 10.1016/s0925-4773(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 47.Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, et al. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Developmental Biology. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 48.Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, et al. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Developmental Biology. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 49.Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 50.Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 51.Hultman KA, Bahary N, Zon LI, Johnson SL. Gene Duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3:e17. doi: 10.1371/journal.pgen.0030017. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 53.Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–1176. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- 54.Curran K, Raible DW, Lister JA. Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev Biol. 2009;332:408–417. doi: 10.1016/j.ydbio.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curran K, Lister JA, Kunkel GR, Prendergast A, Parichy DM, et al. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev Biol. 2010;344:107–118. doi: 10.1016/j.ydbio.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- 57.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parichy DM, Mellgren EM, Rawls JF, Lopes SS, Kelsh RN, et al. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev Biol. 2000;227:294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- 59.Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- 60.Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003;130:817–833. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- 61.Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–1858. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 63.Nichols DH, Weston JA. Melanogenesis in cultures of peripheral nervous tissue. I. The origin and prospective fate of cells giving rise to melanocytes. Dev Biol. 1977;60:217–225. doi: 10.1016/0012-1606(77)90120-8. [DOI] [PubMed] [Google Scholar]
- 64.Dupin E, Glavieux C, Vaigot P, Le Douarin NM. Endothelin 3 induces the reversion of melanocytes to glia through a neural crest-derived glial-melanocytic progenitor. Proc Natl Acad Sci U S A. 2000;97:7882–7887. doi: 10.1073/pnas.97.14.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dupin E, Real C, Glavieux-Pardanaud C, Vaigot P, Le Douarin NM. Reversal of developmental restrictions in neural crest lineages: transition from Schwann cells to glial-melanocytic precursors in vitro. Proc Natl Acad Sci U S A. 2003;100:5229–5233. doi: 10.1073/pnas.0831229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe K, Washio Y, Fujinami Y, Aritaki M, Uji S, et al. Adult-type pigment cells, which color the ocular sides of flounders at metamorphosis, localize as precursor cells at the proximal parts of the dorsal and anal fins in early larvae. Dev Growth Differ. 2008;50:731–741. doi: 10.1111/j.1440-169X.2008.01071.x. [DOI] [PubMed] [Google Scholar]
- 67.Yamada T, Okauchi M, Araki K. Origin of adult-type pigment cells forming the asymmetric pigment pattern, in Japanese flounder (Paralichthys olivaceus). Dev Dyn. 2010;239:3147–3162. doi: 10.1002/dvdy.22440. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 2002:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mundell NA, Labosky PA. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011 doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ignatius MS, Moose HE, El-Hodiri HM, Henion PD. colgate/hdac1 Repression of foxd3 expression is required to permit mitfa-dependent melanogenesis. Dev Biol. 2008;313:568–583. doi: 10.1016/j.ydbio.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buac K, Xu M, Cronin J, Weeraratna AT, Hewitt SM, et al. NRG1/ERBB3 signaling in melanocyte development and melanoma: inhibition of differentiation and promotion of proliferation. Pigment Cell Melanoma Res. 2009;22:773–784. doi: 10.1111/j.1755-148X.2009.00616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hultman KA, Budi EH, Teasley DC, Gottlieb AY, Parichy DM, et al. Defects in ErbB-dependent establishment of adult melanocyte stem cells reveal independent origins for embryonic and regeneration melanocytes. PLoS Genet. 2009;5:e1000544. doi: 10.1371/journal.pgen.1000544. doi: 10.1371/journal.pgen.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang CT, Johnson SL. Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development. 2006;133:3563–3573. doi: 10.1242/dev.02533. [DOI] [PubMed] [Google Scholar]
- 76.Yang CT, Sengelmann RD, Johnson SL. Larval melanocyte regeneration following laser ablation in zebrafish. J Invest Dermatol. 2004;123:924–929. doi: 10.1111/j.0022-202X.2004.23475.x. [DOI] [PubMed] [Google Scholar]
- 77.Rawls JF, Johnson SL. Temporal and molecular separation of the kit receptor tyrosine kinase's roles in zebrafish melanocyte migration and survival. Dev Biol. 2003;262:152–161. doi: 10.1016/s0012-1606(03)00386-5. [DOI] [PubMed] [Google Scholar]
- 78.Rawls JF, Johnson SL. Requirements for the kit receptor tyrosine kinase during regeneration of zebrafish fin melanocytes. Development. 2001;128:1943–1949. doi: 10.1242/dev.128.11.1943. [DOI] [PubMed] [Google Scholar]
- 79.Lee Y, Nachtrab G, Klinsawat PW, Hami D, Poss KD. Ras controls melanocyte expansion during zebrafish fin stripe regeneration. Dis Model Mech. 2010;3:496–503. doi: 10.1242/dmm.004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kondo S, Shirota H. Theoretical analysis of mechanisms that generate the pigmentation pattern of animals. Semin Cell Dev Biol. 2009;20:82–89. doi: 10.1016/j.semcdb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 81.Yamaguchi M, Yoshimoto E, Kondo S. Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proc Natl Acad Sci U S A. 2007;104:4790–4793. doi: 10.1073/pnas.0607790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodrich HB, Greene JM. An experimental analysis of the development of a color pattern in the fish Brachydanio albolineatus Blyth. J exp Zool. 1959;141:15–45. doi: 10.1002/jez.1401410103. [DOI] [PubMed] [Google Scholar]
- 83.Goodrich HB, Marzullo CM, Bronson WR. An analysis of the formation of color patterns in two fresh-water fish. J exp Zool. 1954;125:487–505. [Google Scholar]
- 84.Lang MR, Patterson LB, Gordon TN, Johnson SL, Parichy DM. Basonuclin-2 requirements for zebrafish adult pigment pattern development and female fertility. PLoS Genet. 2009;5:e1000744. doi: 10.1371/journal.pgen.1000744. doi: 10.1371/journal.pgen.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greco V, Guo S. Compartmentalized organization: a common and required feature of stem cell niches? Development. 2010;137:1586–1594. doi: 10.1242/dev.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zammit PS. All muscle satellite cells are equal, but are some more equal than others? J Cell Sci. 2008;121:2975–2982. doi: 10.1242/jcs.019661. [DOI] [PubMed] [Google Scholar]
- 88.Quigley IK, Turner JM, Nuckels RJ, Manuel JL, Budi EH, et al. Pigment pattern evolution by differential deployment of neural crest and post-embryonic melanophore lineages in Danio fishes. Development. 2004;131:6053–6069. doi: 10.1242/dev.01526. [DOI] [PubMed] [Google Scholar]
- 89.Bafaloukos D, Gogas H. The treatment of brain metastases in melanoma patients. Cancer Treat Rev. 2004;30:515–520. doi: 10.1016/j.ctrv.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Palmieri D, Chambers AF, Felding-Habermann B, Huang S, Steeg PS. The biology of metastasis to a sanctuary site. Clin Cancer Res. 2007;13:1656–1662. doi: 10.1158/1078-0432.CCR-06-2659. [DOI] [PubMed] [Google Scholar]
- 91.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, et al. The Tol2kit: A multisite gateway-based construction kit forTol2 transposon transgenesis constructs. Developmental Dynamics. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 93.Budi EH, Patterson LB, Parichy DM. Morphogenesis of post-embryonic neural crest-derived pigment cell precursors revealed by time lapse imaging during zebrafish adult pigment pattern formation. Dev Biol. 2010;344:448. [Google Scholar]
- 94.Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J Neurosci. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, et al. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 96.Kelsh RN, Schmid B, Eisen JS. Genetic analysis of melanophore development in zebrafish embryos. Dev Biol. 2000;225:277–293. doi: 10.1006/dbio.2000.9840. [DOI] [PubMed] [Google Scholar]
- 97.Guyonneau L, Murisier F, Rossier A, Moulin A, Beermann F. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol Cell Biol. 2004;24:3396–3403. doi: 10.1128/MCB.24.8.3396-3403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, et al. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mitfa::GFP+ cells amongst glia of the lateral line nerve. Sagittal view of a wild-type larva showing mitfa::GFP+ cells (mitfa, green; arrow) aligned with mbp+ glia (red) of the main trunk lateral line nerve, near the horizontal myoseptum.
(TIF)
Ectopic kitla-responsive melanogenic cells were nerve-associated. Shown is an ectopic sox10+ (red) melanophore (arrow) within the myotome adjacent to a nerve fiber stained for acetylated tubulin (green).
(TIF)
Defects in wild-type larvae treated with ErbB inhibitor AG1478 during the erbb3b embryonic critical period. (A,B) Post-embryonic mbp+ glia (red; arrowheads) were reduced though not eliminated in AG1478-treated larvae. Inset, mitfa::GFP+ cell (green) aligned on mbp+ glia of a peripheral nerve at the level of the horizontal myoseptum. sc, spinal cord; m, myotome; e, epidermis; ll, lateral line nerve. (C,D) foxd3+ cells (red; arrowheads) within the myotome were missing from AG1478-treated larvae.
(TIF)
Deficiencies in mitfa::GFP+ cells, foxd3+ cells, and EdU incorporation in erbb3b mutants. (A,B) Views and annotations correspond to those for wild-type larvae in main text Figure 5. Arrow, a rare EdU+; mitfa::GFP+ cell at the base of the ventral fin.
(TIF)
erbb3b-dependence of kita-independent hypodermal melanophores. (A) kitab5 presumptive null allele [59] with stripes of kita-independent hypodermal melanophores. (B) Fish doubly mutant for kitab5 and the presumptive null allele erbb3but.r2e1 showing loss of many kita-independent melanophores as well as gaps in the “interstripes” (e.g., arrowhead) reflecting an iridophore deficiency. Patches of residual melanophores may be of clonal origin.
(TIF)
mitfa::GFP+ cells in adult fish. Shown are cross-sections through ∼20 SSL (∼80 days post-fertilization) adult wild-type fish. (A) A persisting nerve-associated mitfa::GFP+ cell (green; arrow). sc, spinal cord; m, myotome. (B) mitfa::GFP+ cells (green; arrowhead) within the lateral line nerve. e, epidermis (C) mitfa::GFP+ cells (green; arrow) and foxd3+ cells (red; arrowhead) in the ventral myotomes and base of the anal fin (f). (D) mitfa::GFP+ cell (green; arrow) and doubly labeled mitfa::GFP+; foxd3+ cell (arrowhead) associated with an adult scale (s).
(TIF)
mitfa::GFP+ cells in a living wild-type larva. Images were collected in sagittal planes from the skin to the midline of the trunk. Image rotation reveals extra-hypodermal cells within the myotomes, that typically expressed mitfa::GFP at levels lower than in hypodermal cells.
(MOV)
mitfa::GFP+ and dct::mCherry+ melanophores in a living wild-type larva. In contrast to mitfa::GFP+ cells (green), cells expressing the later melanophore lineage marker dct were detected only in the hypodermis. Shown here are cells expressing mCherry driven by the Takifugu rubripes dct promoter.
(MOV)
mitfa::GFP+ cells are missing from a live erbb3b mutant larva. Larva imaged as in Video S1 but over-exposed to provide spatial context. Fluorescent cells forming a stripe in the hypodermis are iridophores, which reflect under epifluorescence in multiple channels (these cells are present in the larva shown in Video S1, but their reflectance falls beneath the threshold for detection at the exposure shown).
(MOV)
mitfa::GFP+ cells differentiate as melanophores in the hypodermis. mitfa::GFP+ cells acquiring melanin (arrows) over ∼18 h.
(MOV)
:GFP+ cells. Low magnification view showing highly migratory mitfa::GFP+ cells over ∼18 h. This movie loops three times. mitfa::GFP+ cells are highly migratory and extend long probing processes. These cells can be seen migrating widely over the flank and also migrating from the body into the anal fin at the lower edge of the frame. In contrast to mitfa::GFP+ cells, xanthophores are larger, less motile and autofluoresce weakly in the GFP channel, particularly from accumulations of pteridine-containing organelles around the nucleus.
(MOV)
Migration of mitfa::GFP+ cells between body and fin. Detail at the margin between body and anal fin, showing movements of mitfa::GFP+ cells between these regions.
(MOV)
mitfa::GFP+ cells emerging from within the myotome. Several mitfa::GFP+ cells (arrows) enter the hypodermis from within the myotome; also see Video S8.
(MOV)
mitfa::GFP+ cells emerging from within the myotome. High magnification view showing a single mitfa::GFP+ cell (arrow) emerging from within the myotome. In this movie, only selected planes of focus from a complete z-series are shown, illustrating the progressive movement of this cell from the interior myotome to the surface. As the cell approaches the more superficial planes, additional mitfa::GFP+ cells already in the hypodermis come into focus.
(MOV)
mitfa::GFP+ cell emerging from the vicinity of the horizontal myoseptum. The movie loops three times.
(MOV)
mitfa::GFP+ cells entering the hypodermis after migrating over the dorsal myotome. One cell already at the dorsal myotome and another extending a process over the flank at the beginning of the movie (red arrows). A third cell extends a long filopodium (yellow arrow) over the flank before entering into the hypodermis.
(MOV)
mitfa::GFP+ cells migrating within the caudal nerve plexus. Shown are mitfa::GFP+ cells along a nerve plexus near the base of the caudal fin in the medial region of a larva. This particular individual is a kita mutant. Similar behaviors were observed in wild-type larvae.
(MOV)
mitfa::GFP+ cells migrating along and dispersing from peripheral nerves. Shown is an enhanced detail of Video S11. Arrowhead indicates a cell traversing along a nerve fiber then migrating away.
(MOV)
Morphogenesis of residual mitfa::GFP+ cells in erbb3b mutants. Residual mitfa::GFP+ cells were often patchily distributed and typically expressed GFP at lower levels than in wild-type. Red arrowhead, mitfa::GFP+ cell dividing. Arrow, mitfa::GFP+ cell emerging from the vicinity of the horizontal myoseptum. Yellow arrowhead, death of mitfa::GFP+ cell.
(MOV)
Proliferation of mitfa::GFP+ cells. A mitfa::GFP+ cell (arrow) divides within the hypodermis over ∼14 h in a wild-type larva.
(MOV)
Death of mitfa::GFP+ cells. Fragmentation and loss of three mitfa::GFP+ cells (arrows) in a kita mutant larva over ∼16 h (see text for details).
(MOV)
Proliferation of differentiated melanophores. Shown is the posterior tail of a kita mutant larva, illustrating successive melanophore divisions. mitfa::GFP+ cells also can be observed migrating on the body and between the body and caudal fin.
(MOV)