Abstract
The formation of appressoria, specialized plant penetration structures of Magnaporthe oryzae, is regulated by the MST11-MST7-PMK1 MAP kinase cascade. One of its downstream transcription factor, MST12, is important for penetration and invasive growth but dispensable for appressorium formation. To identify additional downstream targets that are regulated by Pmk1, in this study we performed phosphorylation assays with a protein microarray composed of 573 M. oryzae transcription factor (TF) genes. Three of the TF genes phosphorylated by Pmk1 in vitro were further analyzed by coimmunoprecipitation assays. One of them, MoSFL1, was found to interact with Pmk1 in vivo. Like other Sfl1 orthologs, the MoSfl1 protein has the HSF-like domain. When expressed in yeast, MoSFL1 functionally complemented the flocculation defects of the sfl1 mutant. In M. oryzae, deletion of MoSFl1 resulted in a significant reduction in virulence on rice and barley seedlings. Consistent with this observation, the Mosfl1 mutant was defective in invasive growth in penetration assays with rice leaf sheaths. In comparison with that of vegetative hyphae, the expression level of MoSFL1 was increased in appressoria and infected rice leaves. The Mosfl1 mutant also had increased sensitivity to elevated temperatures. In CM cultures of the Mosfl1 and pmk1 mutants grown at 30°C, the production of aerial hyphae and melanization were reduced but their growth rate was not altered. When assayed by qRT-PCR, the transcription levels of the MoHSP30 and MoHSP98 genes were reduced 10- and 3-fold, respectively, in the Mosfl1 mutant. SFL1 orthologs are conserved in filamentous ascomycetes but none of them have been functionally characterized in non-Saccharomycetales fungi. MoSfl1 has one putative MAPK docking site and three putative MAPK phosphorylation sites. Therefore, it may be functionally related to Pmk1 in the regulation of invasive growth and stress responses in M. oryzae.
Introduction
Rice blast caused by Magnaporthe oryzae is one of the most destructive diseases that affects rice production worldwide. In the last two decades, the M. oryzae-rice pathosystem has been developed as a model system for studying fungal-plant interactions [1], [2], [3]. The rice blast fungus can attack different tissues of rice plants although leaf blast and panicle blast cause the greatest yield losses. Plant infection is initiated when an asexual spore lands on the rice leaf surface and germinates. A specialized infection structure called an appressorium is formed at the tip of the germ tube. The fungus then uses enormous turgor pressure generated in the appressorium to penetrate the plant surface and underlying cells [4], [5]. Once inside plant cells, invasive hyphae grow biotrophically and spread to nearby cells, possibly via plasmodesmata [6], [7], before switching to necrotrophic growth later in the infection process.
Several signal transduction pathways have been implicated in the regulation of appressorium formation, penetration, and infectious growth in M. oryzae [3], [8]. The PMK1 mitogen-activated protein (MAP) kinase, an ortholog of Saccharomyces cerevisiae Fus3/Kss1, is essential for appressorium formation and infectious growth in rice tissues [9]. The pmk1 mutant fails to form appressoria and is nonpathogenic on healthy or wounded rice seedlings. However, germ tubes of the pmk1 mutant have no defect in surface recognition and produce subapical swollen bodies on hydrophobic surfaces. The expression of PMK1 is increased during appressorium formation and conidium development [10]. Pmk1 localizes to the nucleus in appressoria. In addition to the PMK1 pathway, cyclic AMP signaling is known to be involved in the surface recognition and generation of appressorium turgor in M. oryzae. Exogenous cAMP stimulates appressorium formation on hydrophilic surfaces and deletion of the MAC1 adenylate cyclase gene results in defects in appressorium formation [11], [12]. The M. oryzae genome has two genes encoding catalytic subunits of protein kinase A (PKA). The cpkA but not cpk2 mutant is delayed in appressorium formation and defective in appressorium penetration [8], [13], [14]. Studies in several other phytopathogenic fungi also have shown that both the cAMP signaling and MAP kinase pathways are involved in the regulation of various plant infection and differentiation processes [15], [16]. However, molecular mechanisms regulating the interaction or cross-talking between these two pathways are not clear. In the corn smut fungus Ustilago maydis, the Prf1 transcription factor is activated by both the cAMP-PKA and MAP kinase pathways [17], [18].
In M. oryzae, several components of the PMK1 pathway have been characterized, including MST11, MST50, and MST12 [19], [20], [21]. MST12, an ortholog of yeast STE12, encodes a transcription factor that interacts with Pmk1 in yeast two-hybrid assays. Although it is defective in appressorial penetration and invasive growth, the mst12 mutant still forms appressoria [22], [23]. To identify additional downstream targets that are regulated by Pmk1, here we performed in vitro phosphorylation assays with a protein microarray composed of recombinant proteins of 573 M. oryzae transcription factor (TF) genes that were individually expressed in the budding yeast [24]. Protein microarray has been shown as a powerful proteomics tool for profiling protein-protein, protein-DNA, protein-RNA, and protein-glycan interactions and for identification of substrates of various enzymes, such as protein kinases, acetyltransferases, ubiquitin E3 ligases [25], [26], [27]. Three of the putative Pmk1 targets identified in phosphorylation assays the M. oryzae TF microarray were further analyzed by coimmunoprecipitation assays. One of them, an ortholog of yeast SFL1, was found to interact with Pmk1 in vivo. In yeast, Sfl1 serves as a transcriptional repressor for the flocculation-related genes and a transcriptional activator for the stress-responsive genes [28], [29]. The Mosfl1 deletion mutant was reduced in virulence and invasive growth but still formed appressoria. Similar to the pmk1 mutant, it had increased sensitivity to elevated temperatures, suggesting that Sfl1 and Pmk1 may be functionally related in the regulation of invasive growth and stress responses in M. oryzae.
Results
MoSfl1 interacts with Pmk1
To identify transcription factors as the downstream effectors of Pmk1, a protein microarray composed of GST-fusion proteins of all predicted M. oryzae TF genes [24] was used for in vitro phosphorylation assays with purified recombinant proteins of Pmk1 [9], [30]. GST-Pmk1 fusion proteins are known to have protein kinase activities in vitro [9]. After incubating the protein microarray with GST-Pmk1 in the presence of r-33P-ATP for 30 min, the phosphorylation reaction was stopped and phosphorylation signals were analyzed with the GenePix Pro software [31], [32]. An autophosphorylation reaction without addition of any kinase was set up as a negative control. A total of 85 putative transcription factors were found to be phosphorylated by Pmk1.
Three of them, MGG_06971, MGG_09869, and MGG_04933, were selected for verification for their interactions with Pmk1 in vivo. MGG_06971 was named MoSFL1 because it is orthologous to yeast SFL1 [28]. MGG_09869 is homologous to SWI6 in yeast [33] and MGG_04933 is a putative transcription factor conserved in filamentous fungi. FLAG-tagged constructs were generated for these three genes by the yeast GAP repair approach [34] and transformed into the wild-type strain 70-15. Transformants expressing the MGG_06971-, MGG_09869-, or MGG_04933-3×FLAG fusion constructs (Table 1) were identified by PCR and confirmed by western blot analysis (Fig. S1).
Table 1. Wild-type and mutant strains of Magnaporthe oryzae used in this study.
| Strain | Genotype description | Reference |
| 70-15 | Wild type (MAT1-1, AVR-Pita) | [71] |
| Guy11 | Wild type (MAT1-2, avr-Pita) | [71] |
| Ku80 | Mgku80 deletion mutant of Guy11 | [37] |
| nn78 | pmk1 deletion mutant of Guy11 | [9] |
| I-27 | cpkA deletion mutant of 70-15 | [14] |
| GSF7 | Transformant of 70-15 expressing MoSFL1-3×FLAG fusion | This study |
| GSF9 | Transformant of 70-15 expressing MoSFL1-3×FLAG fusion | This study |
| GWF7 | Transformant of 70-15 expressing MGG_09869-3×FLAG fusion | This study |
| GWF9 | Transformant of 70-15 expressing MGG_09869-3×FLAG fusion | This study |
| WDF1 | Transformant of 70-15 expressing MGG_04933-3×FLAG fusion | This study |
| GK102 | Mosfl1deletion mutant of Ku80 | This study |
| GK116 | Mosfl1deletion mutant of Ku80 | This study |
| GK135 | Mosfl1deletion mutant of Ku80 | This study |
| C49 | Mosfl1/MoSFL1-GFP complemented transformant | This study |
| C51 | Mosfl1/MoSFL1-GFP complemented transformant | This study |
| P1 | MoSFL1 ΔT231A in GK102 | This study |
| P2 | MoSFL1 ΔS474A in GK102 | This study |
| P3 | MoSFL1 ΔT508A in GK102 | This study |
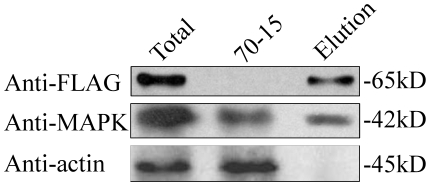
We then isolated total proteins from transformant GSF7 expressing the MoSFL1-3×FLAG construct and used the anti-FLAG antibody to pull down proteins interacting with MoSfl1 in M. oryzae. In both total proteins and proteins eluted from anti-FLAG M2 beads of transformant GSF7, a 64-kD band of the expected size of MoSfl1-3×FLAG fusion was detected with the anti-FLAG antibody (Fig. 1). When detected with an anti-MAPK antibody [10], the 42-kD Pmk1 band also was observed (Fig. 1). In the control experiment, the anti-actin antibody detected a 45-kD actin band only in total proteins isolated from 70-15 and transformant GSF7 but not in proteins eluted from anti-FLAG M2 beads. These data indicate that Pmk1 was co-immunoprecipitated with MoSfl1-3×FLAG.
Figure 1. MoSfl1 co-immunoprecipitates with Pmk1.
Western blots of total proteins (Total) and proteins eluted from the anti-FLAG M2 beads (Elution) of transformant GSF7 that expressed the MoSFL1-3×FLAG construct were detected with the anti-FLAG and anti-MAPK antibodies. Total proteins isolated from the wild-type strain (70-15) and detection with the anti-actin antibody was included as the control.
Similar co-immunoprecipitation (co-IP) assays were conducted with proteins isolated from transformants GWF9 and WDF1 that contained the MGG_09869- and MGG_04933-3×FLAG fusion constructs, respectively. However, co-immunoprecipitation of Pmk1 with FLAG-tagged MGG_09869 and MGG_04933 proteins was not observed (data not shown), indicating that MoSfl1 but not MGG_09869 or MGG_04933 physically interacts with Pmk1 in M. oryzae.
MoSFL1 functionally complements the yeast sfl1 mutant
MoSFL1 shared 29% and 32% similarity with S. cerevisiae SFL1 and CaSFL1 of Candida albicans, respectively. It is highly similar to its putative orthologs in other filamentous ascomycetes, including Fusarium graminearum (60% identity) and Neurospora crassa (53% identity). Like Sfl1 and CaSfl1, MoSfl1 contains a heat shock factor [HSF] domain that binds to the inverted repeats of AGAA-n-TTCT known as heat shock element [35].
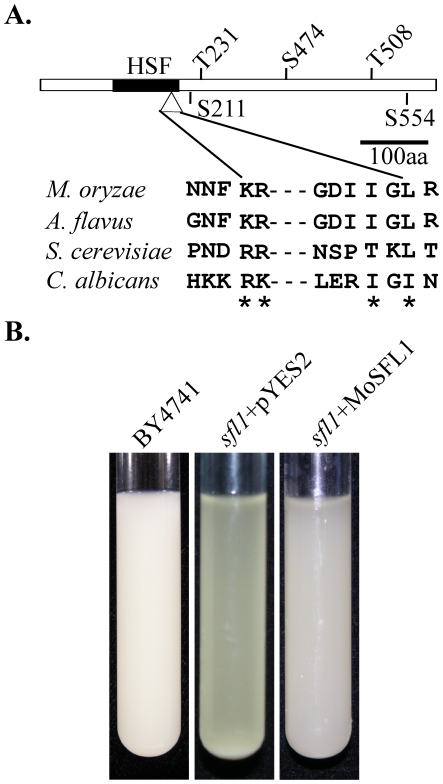
In comparison with CaSfl1, MoSfl1 does not possess glutamine-rich regions that are likely involved in protein-protein interactions. However, MoSfl1 has one MAP kinase docking site (amino acid residues KRGDIIGL) and three putative MAP kinase phosphorylation sites (Fig. 2A) that are conserved in the MoSfl1 orthologs from other fungi. To determine its function in S. cerevisiae, the MoSFL1 ORF was cloned into pYES2 as pYES2-MoSFL1 and transformed into the sfl1 mutant in the BY4741 yeast strain background [36]. Five Ura+ transformants were confirmed by PCR to contain the pYES2-MoSFL1 construct. Although only data with one of these transformants are provided, all of them were complemented in the flocculation defects. After overnight incubation in the galactose-containing medium, the sfl1 mutant cultures were clear due to the sediment of yeast cells. Like the wild type, cultures of the sfl1/MoSFL1 transformants were turbid (Fig. 2B). These results indicate that MoSFL1 can functionally complement the sfl1 mutant when expressed in S. cerevisiae.
Figure 2. Structural elements of MoSfl1 and complementation assays with the yeast sfl1 mutant.
A. The MoSfl1 protein contains a heat shock factor (HSF) domain (black box), a MAPK docking site (triangle), three putative MAP kinase phosphorylation sites (T231, S474, and T508), and two putative PKA phosphorylation sites (S211 and S554). The amino acid sequence of the MAPK docking site is conserved among Sfl1 and its orthologs from Magnaporthe oryzae, Aspergillus flavus, and Candida albicans. B. Expression of MoSFL1 suppressed the flocculation defect of the yeast sfl1 mutant. Cultures of Saccharomyces cerevisiae wild type strain BY4741 (SFL1) and transformants of Δsfl1 carrying pYES2 or pYES2-MoSFL1 were shaken at 30°C for 16 h and kept still for 15 min before being photographed.
The Mosfl1 mutant is reduced in conidiation but has a normal growth rate
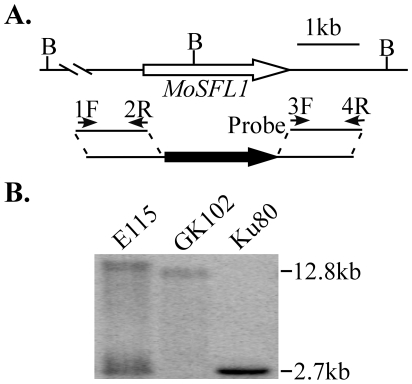
To knock out MoSFL1, its upstream and downstream flanking sequences were amplified with primer pairs 1F/2R and 3/4R (Fig. 3A). The MoSFL1 gene replacement construct (Fig. 3A) was then generated with the double-joint PCR method and transformed into M. oryzae strain Ku80 [37]. Around 140 hygromycin-resistant transformants were isolated and screened by PCR with primers ScF and ScR. Three putative Mosfl1 gene replacement mutants (Table 1) were identified and confirmed by Southern blot analysis (Fig. 3), indicating that the gene replacement frequency was approximately 2.1% for MoSFL1 even in the Ku80 [37] strain background. When genomic DNAs were digested with BamHI and hybridized with the downstream flanking sequence of the MoSFL1 gene as the probe, the Mosfl1 knockout mutant had a 12.8-kb band instead of the 2.7-kb wild-type band (Fig. 3B). The ectopic transformant E115 had the 2.7-kb wild-type band and an additional band (>12.8 kb) resulting from ectopic integration of the gene replacement cassette in the genome.
Figure 3. The MoSFL1 gene replacement vector and mutants.
A. The MoSFL1 genomic region and gene replacement construct. The upstream and downstream flanking sequences were amplified with primers 1F/2R and 3F/4R, respectively, and connected with the hph cassette by double joint PCR. B, BamHI. B. Southern blot of BamHI-digested DNA of Ku80, Mosfl1mutant GK102, and ectopic transformant E115 was hybridized with the downstream sequence of MoSFL1 amplified with primers 3F and 4R as the probe. Because of the BamHI site located in the middle of the MoSFL1 gene, the Mosfl1 deletion mutant had a 12.8-kb band instead of the wild-type 2.7-kb band.
In comparison with Ku80, the Mosfl1 mutant was reduced about 70% in conidiation (Table 2). In contrast, the growth rate of the Mosfl1 mutant was similar to that of Ku80 or ectopic transformant E115 (Table 2). To determine whether deletion of MoSFL1 had any effect on responses to different physiological stresses, we assayed vegetative growth of the mutant on various media. The Mosfl1 mutant had a growth rate similar to that of Ku80 under nitrogen- or carbon-starvation conditions, on V8 juice agar and minimal medium (MM), and on complete medium (CM) with 1 M sorbitol, 0.7 M NaCl, or different concentrations of H2O2 (Table S1). These results indicate that MoSFL1 is dispensable for responses to nutritional, hyperosmotic, and oxidative stresses.
Table 2. Phenotypes of the Mosfl1 mutant in growth, development, and differentiation.
| Strain | Conidiation(106 spores/plate) | Growth rate(mm/day)a | Conidial germination (%)b | Appressoriumformation (%) | Cytorrhysisc |
| Ku80 | 101.7±16.4 | 3.3±0.1 | 100±0.0 | 95.2±2.4 | 63±3.6 |
| E115 | 100.0±16.9 | 3.4±0.1 | N/Ad | N/A | N/A |
| GK102 | 28.7±8.9 | 3.4±0.1 | 100±0.0 | 96.9±1.2 | 73.8±0.5 |
| C49 | 105.3±1.2 | 3.5±0.1 | N/A | N/A | N/A |
The growth rate was measured with 14-day-old CM cultures grown in race tubes. Mean and standard deviation were calculated with results from three replicates.
Percentage of conidia germination and appressorium formation by 24 h. Means and standard errors were calculated from three independent repeats (at least 100 conidia were counted in each repeat).
Percentage of appressoria that underwent cytorrhysis in 35% PEG. Mean and standard deviations were calculated from four replicates. At least 100 appressoria were examined in each repetition.
Not assayed.
MoSFL1 is dispensable for conidium germination, appressorium formation, and turgor generation
To determine the role MoSFL1 in infection-related morphogenesis, we assayed appressorium formation with the Mosfl1 mutant. On hydrophobic surfaces, the Mosfl1 mutant had no defect in surface attachment or conidium germination. After 2 h of incubation at room temperature, over 95% of mutant conidia germinated and produced normal germ tubes (data not shown). A vast majority of the Mosfl1 germ tubes (>95%) formed appressoria within 24 h. We also examined appressorium formation at 8 h. The wild-type and mutant strains had no obvious difference in the percentage of germ tubes forming appressoria. These data indicate that MoSFL1 is dispensable for conidium germination and appressorium formation.
In M. oryzae, appressorium turgor plays a critical role in plant penetration. To determine whether the Mosfl1 mutant had defects in turgor generation, we conducted the cytorrhysis assay [4]. In the presence of 35% PEG, Ku80 and the Mosfl1 mutant strains had similar amounts of appressoria that underwent cytorrhysis (Table 2, P>0.1), suggesting that appressoria of the wild-type and mutant strains had similar turgor pressures.
The Mosfl1 mutant is reduced in virulence
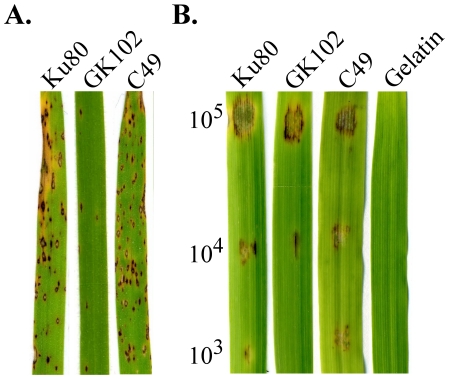
To determine the virulence of the Mosfl1 mutant, two-week-old seedlings of rice cultivar CO-39 were used for infection assays. On rice leaves sprayed with conidium suspensions of 5×104 spores/ml, strain Ku80 formed typical blast lesions (Fig. 4A). The Mosfl1 deletion mutant caused fewer and often smaller lesions on rice leaves. On average, 3.3±0.6 lesions/5-cm leaf tip were observed on leaves sprayed with the Mosfl1 mutant. Under the same conditions, the wild type caused 21.3±3.5 lesions/5-cm leaf tip. Even after prolonged incubation such as 10 dpi, lesions caused by the mutant were still restricted and normally lacked the necrosis zone on the edge (Fig. 4A). To confirm that the observed phenotypes are directly related to MoSFL1 deletion, we generated the MoSFL1-GFP fusion construct and transformed it into the Mosfl1 mutant GK102. The resulting transformant C49 (Table 1) was normal in conidiation (Table 2) and plant infection (Fig. 4).
Figure 4. Infection assays with the Mosfl1 mutant.
A. Seedlings of rice cultivar CO-39 were sprayed with conidia of Ku80, Mosfl1 mutant GK102, and complemented transformant C49. Typical leaves were photographed 7 days post inoculation (dpi). B. Detached barley leaves were drop inoculated with conidia from the same set of strains. The concentrations of conidium suspensions (conidia/ml) were marked on the left. Inoculation with 0.25% gelatin was the negative control.
To further prove that the Mosfl1 mutant is reduced in virulence, detached barley leaves were drop inoculated with different concentrations of conidia. At 1×103 conidia/ml, only the sites inoculated with the wild type but not the Mosfl1 mutant had visible symptoms (Fig. 4B). At 1×104 conidia/ml, the Mosfl1 mutant caused limited necrosis at the inoculation sites. The wild type caused much more severe symptoms under the same conditions (Fig. 4B). Even at the sites inoculated with 1×105 conidia/ml, the wild type still appeared to be more virulent than the Mosfl1 mutant. Extensive necrosis surrounding the drop-inoculation sites was only observed in the wild type (Fig. 4B). These results indicate that the Mosfl1 mutant had a reduced virulence on rice and barley leaves and was defective in lesion expansion or development of the surrounding necrosis zone.
MoSFL1 plays a role in infectious growth after penetration
Because the Mosfl1 mutant formed smaller lesions than the wild type on rice and barley leaves, we examined the defects of the Mosfl1 mutant in infectious growth after penetration. In penetration assays with rice leaf sheath epidermal cells, invasive hyphae formed by Ku80 branched and spread to nearby rice leaf sheath epidermal cells by 48 h (Fig. 5). Under the same conditions, the mutant normally had only limited growth. At 48 h, invasive hyphae of the Mosfl1 mutant appeared to be confined to the penetrated plant cell. Extensive growth of invasive hyphae was only observed in plant tissues inoculated with Ku80 but not the Mosfl1 mutant.
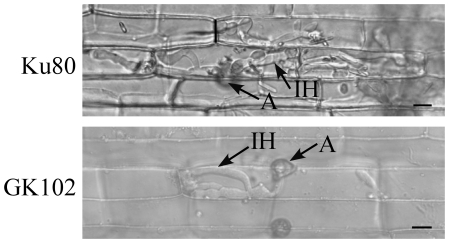
Figure 5. Penetration assays with the Mosfl1 mutant.
Rice leaf sheaths inoculated with conidia from Ku80 and Mosfl1 mutant GK102 were examined 48 hpi. The mutant was restricted in invasive growth compared with the wild type. A, appressorium; IH, invasive hyphae. Bar = 10 µm.
The Mosfl1 mutant is defective in heat tolerance
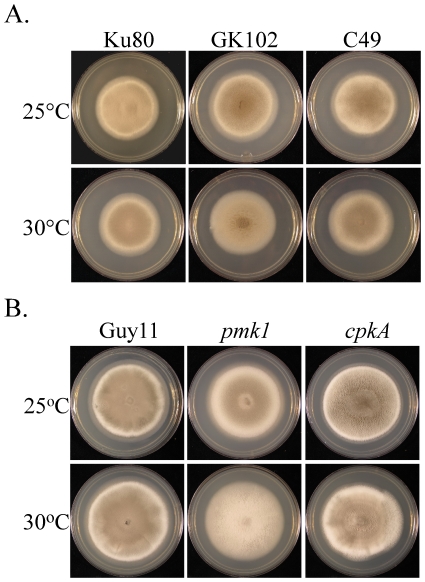
Similar to Sfl1 of S. cerevisiae, MoSf1 has a heat-shock factor domain and may be involved in regulating heat tolerance. When cultured at 25°C, the Mosfl1 mutant and the Ku80 strain had similar colony morphologies (Fig. 6A). However, colonies of the Mosfl1 mutant were significantly reduced in aerial hyphal growth compared with those of Ku80 when cultured at 30°C on CM (Fig. 6A) or oatmeal agar (Fig. S2) plates. Aerial hyphal growth of the complemented transformant C49 was similar to that of Ku80 at 30°C, suggesting that MoSfl1 plays a role in heat tolerance during the growth of aerial hyphae. We also assayed heat tolerance with the pmk1 and cpkA mutants. Similar to the Mosfl1 mutant, the pmk1 mutant was significantly reduced in the production of aerial hyphae at 30°C (Fig. 6B). The elevated temperature had no obvious effect on the cpkA mutant (Fig. 6B).
Figure 6. Increased heat sensitivity in the Mosfl1 and pmk1 mutants.
A. Five-day-old CM cultures of Ku80, Mosfl1 mutant GK102, and complemented transformant C49. B. Seven-day-old CM cultures of the wild type Guy11, pmk1 mutant nn78, and cpkA mutant I-27. All of the cultures were incubated at 25°C (top) or 30°C (bottom). While the growth rates of Ku80 and Guy11 were not affected, the production of aerial hyphae was reduced in the Mosfl1 and pmk1 mutants when cultured at 30°C.
To confirm that the observed phenotypes are directly related to MoSFL1 deletion, we generated the MoSFL1-GFP fusion construct and transformed it into mutant GK102. The resulting transformant C49 (Table 1) was normal in conidiation (Table 2), plant infection (Fig. 4), and growth at 30°C (Fig. 6A), indicating that expression of the wild-type MoSFL1 allele fully complemented the defects of the Mosfl1 mutant.
Downstream genes regulated by MoSFL1
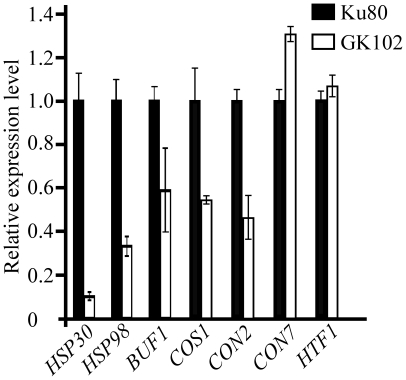
Because of the defect of the Mosfl1 mutant in heat tolerance and the regulation of heat shock-related genes by Sfl1 in yeast and C. albicans [29], we assayed the transcript abundance of M. oryzae genes homologous to yeast HSP30, HSP60, and HSP98 that encode heat shock proteins. RNA samples were isolated from cultures of Ku80 and mutant GK102. Compared to Ku80, the transcription levels of MoHSP30 and MoHSP98 were reduced 10- and 3-fold, respectively, in the Mosfl1 mutant (Fig. 7). The expression of MoHSP60 was not significantly changed in the mutant (data not shown).
Figure 7. qRT-PCR assay of MoHSP30, MoHSP98, BUF1, COS1, CON2,CON7, and HTF1 expression in the Ku80 and Mosfl1 mutant strains.
The relative expression levels of MoHSP30, MoHSP98, BUF1, COS1, CON2, CON7, and HTF1 were compared between the GK102 and Ku80 strains (arbitrarily set to 1) cultured at 30°C. Mean and standard error were calculated with data from three biological replicates.
On CM plates, colonies of the Mosfl1 mutant appeared to be whitish or less pigmented compared to the grayish colonies of the wild type, which may be related to a reduction in melanin synthesis. In M. oryzae, one of the major genes involved in the biosynthesis of melanin is BUF1 [38]. When assayed by qRT-PCR, the transcription level of BUF1 was reduced about 2-fold in the Mosfl1 mutant (Fig. 7). These results suggest that MoSfl1 is involved in the transcriptional regulation of melanin biosynthesis and some heat-shock related genes, such HSP30 and HSP98 homologs in M. oryzae.
Because the Mosfl1 mutant reduced in conidiation, we also assayed the expression levels of four genes that are known to be related to conidiation, COS1, CON2, CON7, and HTF1 [39], [40], [41]. While the expression of CON7 and HTF1 were not significantly altered in the Mosfl1 mutant, the transcription levels of COS1 and CON2 were reduced over 2-fold in the Mosfl1 mutant compared to Ku80 (Fig. 7). Because COS1 is essential for conidiophore development [39], a reduction in COS1 expression may be related to reduced conidiation in the mutant.
MoSFL1 expression is increased during plant infection compared with vegetative growth
Although expressing the MoSFL-GFP construct rescued the defects of the Mosfl1 mutant in plant infection, conidiation, and heat tolerance, we failed to detect GFP signals in the conidia, appressoria, vegetative hyphae, or invasive hyphae of transformant C49 (data not shown). MoSfl1 has a nuclear localization signal and is predicted to be a nuclear protein. In C. albicans, CaSfl1 was shown to be localized to the nucleus [42], [43]. However, we failed to detect GFP signals in the nucleus in transformant C49.
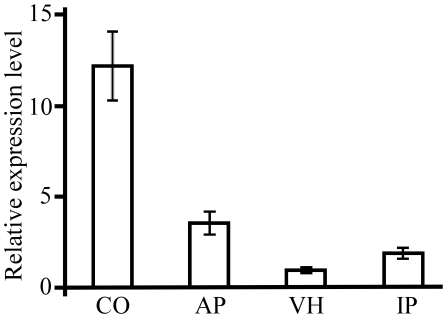
We then used the qRT-PCR assay to estimate the expression levels of MoSFL1 in conidia, mature appressoria, vegetative hyphae, and infected rice leaves (5 dpi). MoSFL1 was expressed in all four stages (Fig. 8). The expression of MoSFL1 was significantly increased in appressoria and infected rice leaves compared to vegetative hyphae. However, the highest transcription level of MoSFL1 was in conidia. Nevertheless, GFP signals were not observed in conidia or appressoria of the MoSFL1-GFP transformant, which suggests that the activation of MoSfl1 and its localization to the nucleus are too transient to be detected with the GFP marker at these developmental stages. However, it remains possible that the expression level of MoSFL1, even in conidia, is relatively low compared to other genes.
Figure 8. Expression of MoSFL1 in different developmental stages.
RNA samples used for qRT-PCR assays were isolated from conidia (CO), appressoria (AP), vegetative hyphae (VH), and infected plants (IP). The relative expression level of MoSFL1 was compared to that of the VH stage (arbitrarily set to 1). Mean and standard error were calculated with data from three biological replicates.
Discussion
The protein microarray of M. oryzae contains GST-fusion proteins of predicted TF genes in the genome [24]. In this study, we used the M. oryzae protein microarray for in vitro phosphorylation assays with GST-Pmk1 fusion proteins with kinase activities [9]. The PMK1 MAP kinase and its orthologs are important for appressorium formation and invasive growth in M. oryzae [9], [19], [20]. A total of 85 genes was found to be phosphorylated by Pmk1, including homologs of Pro1 [44], StuA [45], SFL1 [28], PPR1 [46], and SWI6 [47]. A number of fungal-specific Zn (2)–Cys (6) binuclear cluster and zinc finger proteins [48] also were found to be phosphorylated by Pmk1. However, the specificity of protein kinase-substrate interactions is likely compromised in in vitro phosphorylation assays. Many of these putative targets of Pmk1 identified in protein microarray experiments may be not functionally related with Pmk1 in vivo.
Three genes, MGG_04933, MGG_06971, and MGG_09869, were selected for co-IP assays. Only one of them, MoSFL1 (MGG_06971), was confirmed by co-IP assays to interact with Pmk1 in vivo. In S. cerevisiae, Sfl1 functions as a transcriptional repressor of flocculation-related genes [28], [49]. Although the overall sequence identity is only 29%, MoSFL1 could functionally complement the flocculation defects of the sfl1 mutant (Fig. 1). One of the yeast genes regulated by Sfl1 is FLO11, which encodes a cell surface glycoprotein required for flocculation and filamentation [49], [50]. Transcription factor Flo8 functions antagonistically with Sfl1 as a transcriptional activator of FLO11 [51], [52]. In C. albicans, CaFLO8 and CaSFL1 had opposite roles in hyphal development. CaSFL1 also appeared to function as a negative regulator of flocculation [42], [43]. However, like many other filamentous ascomycetes, the M. oryzae genome lacks distinct orthologs of FLO8 and FLO11. Therefore, the subset of genes regulated by MoSFL1 or its function in M. oryzae may differ significantly from that of Sfl1 in yeast.
Deletion of MoSFL1 had no obvious effect on responses to hyperosmotic, oxidative, and nutritional stresses. However, the Mosfl1 mutant had increased sensitivities to elevated temperatures for aerial hyphal growth. At 30°C, aerial hyphal growth was reduced in the mutant. Like other Sfl1 orthologs, MoSfl1 has a HSF domain that recognizes heat shock elements in the promoters of target genes [35]. In yeast, genes with heat shock elements, such as STA1 and SUC2, are repressed by Sfl1 [49], [53], [54]. However, Sfl1 also is involved in the activation of HSP30 transcription [29]. When assayed by qRT-PCR, the expression levels of MoHSP30 and MoHSP98 genes were significantly reduced in the Mosfl1 mutant. In C. albicans, CaSfl1 also is involved in the activation of heat-shock protein genes HSP30 and HSP90 under certain stress conditions [55]. SFL1 orthologs may be well conserved in the activation of heat shock related genes in ascomycetes.
To date, SFL1 orthologs have not been functionally characterized in plant pathogenic fungi. Deletion of MoSFL1 had no effect on hyphal growth in vitro, which is different from the Agsfl1 mutant in Ashbya gossypii [56]. Compared to the parental Ku80 strain, deletion of MoSFL1 had no obvious effect on appressorium formation or turgor pressure production. The Mosfl1 mutant was reduced in conidiation (Table 2) and virulence (Fig. 4A). In comparison with Ku80, the Mosfl1 mutant produced fewer lesions and the lesions caused by the mutant were usually smaller than those of the wild type. Reduced pathogenicity of the Mosfl1 mutant is likely related to its defects in invasive hyphal growth as shown in penetration assays with rice leaf sheaths (Fig. 5). In C. albicans, CaSfl1 represses the expression of several hypha-specific genes, including HWP1, ECE1, ALS1, ALS3, and FLO8 [43], [55]. Either overexpression or deletion of CaSFL1 attenuated the virulence of C. albicans in the mouse model. In M. oryzae, MoSFL1 may be involved in the repression of genes that are detrimental to in planta biotrophic growth, such as genes encoding cell wall degrading enzymes or enzymes responsible for the synthesis of phytotoxic compounds. It is also possible that MoSFL1 negatively regulates genes only required for vegetative growth. Deletion of MoSFL1 may result in improper regulation of subsets of genes and a reduction in conidiation and virulence.
Similar to MST12, MoSFL1 is involved in invasive hyphal growth. Although the Mosfl1 mutant was reduced in BUF1 expression in vegetative hyphae, it still formed melanized appressoria. Melanin biosynthesis is regulated differently in appressoria and in vegetative hyphae in M. oryzae and Colletotrichum lagenarium [57]. Our results indicated that melanization was normal in appressoria but reduced in vegetative hyphae in the Mosfl1 mutant. In M. oryzae, PTH12 and CON7 are two transcription factor genes that are reported to be essential for appressorium formation [58], [59], suggesting that they may be functionally related with the Pmk1 pathway. However, none of them were identified in the phosphorylation assays with the M. oryzae protein arrays with Pmk1. Also, we have generated the pth12 and con7 deletion mutants. Unlike the pmk1 mutant [41], hyphal tips of the pth12 and con7 mutants, had no defects in appressorium formation (Kong and Xu, unpublished).
MoSfl1 has one putative MAPK docking site, three putative MAPK phosphorylation sites, and two PKA phosphorylation sites that are conserved in Sfl1 orthologs from other filamentous ascomycetes. In S. cerevisiae, FLO11 was regulated by both MAP kinase and cAMP filamentation signaling pathways through transcription factors Ste12, Tec1, Flo8, and Sfl1 [49], [52]. There are no distinct orthologs of Flo8 and Tec1 in M. oryzae. Therefore, the MoSfl1-related regulatory network in M. oryzae must be different from that of S. cerevisiae. To test whether MoSfl1 functions downstream from Pmk1, we generated MoSFL1 mutant alleles in which three putative MAPK phosphorylation sites were deleted, and transformed these constructs into mutant GK102. The resulting transformants were normal in virulence (data not shown), suggesting that deletion of individual MAPK phosphorylation sites had no effect on its complementation of the Mosfl1 mutant. Therefore, these putative MAP phosphorylation sites are either not important or play redundant roles with other MAPK or PKA phosphorylation sites in the regulation of MoSfl1. In S. cerevisiae and C. albicans, Sfl1 proteins are phosphorylated by the Tpk2 catalytic subunit of PKA [51]. Although MoSfl1 was not phosphorylated by CpkA in vitro and the Mosfl1 and cpkA mutants had distinct phenotypes, it remains possible that the two putative PKA phosphorylation sites of MoSfl1 play a role in its activation. In C. albicans, the nuclear localization of CaSfl1 was not altered in the tpk2 null mutant or in response to exogenous cAMP, suggesting that other signaling pathways (possibly a MAP kinase pathway) also are involved in the activation of CaSfl1 [43]. In U. maydis, the Prf1 transcription factor is functionally related to the cAMP-PKA and Kpp2/Kpp6 MAP kinase pathways [17], [18]. It is possible that MoSfl1 is regulated by both the Pmk1 MAP kinase and cAMP signaling pathways in M. oryzae. Further characterization of the activation of MoSfl1 may lead to better understanding of the interaction between these two important signal transduction pathways.
Materials and Methods
Strains and growth conditions
The M. oryzae wild-type and mutant strains (Table 1) were routinely cultured on oatmeal agar (OTA) and complete medium (CM) as described previously [60]. For the heat tolerance assay, CM cultures were incubated at 30°C or 25°C for 7 days. Assays for growth rate and conidiation were conducted with OTA cultures [61]. Protoplast preparation and transformation were performed as described [62]. Transformants were selected on medium with 250 µg/ml of hygromycin B or 200 µg/ml zeocin (Invitrogen, CA).
Phosphorylation assays with the M. oryzae protein microarray
The protein microarray composed of all predicted M. oryzae transcription factors [24] were used for in vitro phosphorylation assays following the procedure described in phosphorylation assays with the S. cerevisiae protein microarray [31], [32]. GST-Pmk1 fusion proteins were expressed in the budding yeast. Purified GST-Pmk1 was tested for protein kinase activity in vitro using myelin basic protein (MBP) as the substrate [9], [31]. The M. oryzae protein microarray was incubated with GST-Pmk1 in the presence of r-33P-ATP at 30°C for 30 min [31]. The phosphorylation reaction was stopped with 0.5% SDS. After washing and drying, the microarray was exposed to an X-ray film. Phosphorylation signals were analyzed with the GenePix Pro software (www.moleculardevices.com). Positives were identified with a cutoff value of 3 [31], [32].
Complementation of the yeast sfl1 mutant
The MoSFL1 ORF was amplified with primers YCF/YCR (Table S2) from the first strand cDNA of Guy11 and cloned into plasmid pGEM-T Easy (Promega, WI). Among the resulting clones sequenced, clone pGT25 contains the entire ORF of MoSFL1, which was then sub-cloned into pYES2 as pMoSFL1. Competent cells of the sfl1 mutant of S. cerevisiae (Open Biosystems, AL) were prepared and transformed with pMoSFL1 using the alkali-cation yeast transformation kit (MP Biomedicals, OH). Ura3+ transformants were confirmed by PCR and assayed for flocculation in YPGal liquid medium as described previously [43], [63]. The yeast strain BY4741 (MATa his3 leu2 met15 ura3) used to generate the sfl1 mutant was used as the control.
Molecular manipulations
Putative MoSfl1-regulating genes were assayed by qRT-PCR. RNAs from conidia, appressoria, vegetative hyphae, and infected rice tissues were extracted with TRIzol (Invitrogen, CA) and reverse transcription was done with the AccuScript High Fidelity 1st Strand cDNA Synthesis Kit (Agilent Technologies, CA). RT-PCR was performed with the Stratagene Gene MX 3000 PM using the 2×Brilliant SYBR Green QPCR master mix (Agilent Technologies, CA). The relative quantification of each transcript was calculated by the 2-ΔΔCT method [64] using the tubulin gene (MGG_00604) as the internal control.
Co-immunoprecipitation assays
The MoSFL-3×FLAG fusion was constructed by cloning the PCR products amplified with primers 6971flag/1F and 6971flag/3R from genomic DNA and co-transformed with XhoI-digested pHZ126 into XK1-25 [34] as pGL12. Plasmid pGL12 was transformed into the wild-type strain 70-15. Transformants expressing the MoSFL1-3×FLAG construct were identified by PCR screens and confirmed by western blot analysis using anti-FLAG antibodies. Total proteins were extracted from these confirmed transformants and incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich, MO) at 4°C overnight. Proteins bound to anti-FLAG beads were eluted and detected with anti-FLAG (Sigma-Aldrich, MO), anti-actin (Sigma-Aldrich, MO), or anti-MAPK (Cell Signaling Technology, MA) as described previously.
Generation of the MoSFL1 gene replacement construct and mutants
The double-joint PCR method [65] was used to generate the MoSFL1 gene replacement vector. A 0.8-kb upstream and a 0.8-kb downstream flanking sequences of MoSFL1 were amplified with primer pairs 1F/2R and 3F/4R, respectively. The hph cassette was amplified with primers HYG/F and HYG/R from pCB1003. The products of double-joint PCR were transformed into protoplasts of Ku80 [37]. Putative Mosfl1 deletion mutants were screened by PCR and confirmed by Southern blot analysis. For complementation assays, the wild-type allele of MoSFL1 was amplified with primers SFG/1F and NG/R and cloned into pYF1 plasmid [66] with bleomycin resistance. The same yeast GAP repair approach [34] was used to generate the T231A, S474A, and T508A mutant alleles of MoSFL1 that carried substitution mutations in three putative MAPK phosphorylation sites. The complementation vector and mutant alleles of MoSFL1 were transformed into protoplasts of GK102.
Germination, appressorium formation, cytorrhysis and penetration assays
Conidia were collected from one-week-old oatmeal agar cultures and resuspended in sterile distilled water to the concentration of 5×104 conidia/ml. Conidial germination and appressorium formation were assayed with plastic cover slips (Fisher Scientific Co.) as described [67]. Appressorium turgor pressure was estimated by the incipient cytorrhysis method with solutions of PEG-8000 [4]. For penetration assays with rice leaf sheaths, conidia were suspended to 105/ml and used to inoculate leaf sheaths of 3-week-old plants [6], [68]. Penetration and invasive hyphae were examined 48 h post inoculation by DIC microscopy.
Plant infection assays
Two-week-old seedlings of the rice cultivar CO-39 and 10-day-old seedlings of the barley cultivar Golden Promise were used for infection assays. Conidia were collected from 10-day-old oatmeal agar cultures and resuspended to 5×104/ml in 0.25% gelatin. Plant incubation, inoculation, and lesion examination were performed as described [13], [69]. The average number of lesions formed on the 5-cm tip regions of the second leaves of rice seedlings were counted as described [21], [70]. For drop-inoculation assays with detached barley leaves, conidia were diluted to 105, 104, and 103/ml. A 15-µl drop of each dilution was pipetted onto barley leaves placed over 2% (w/v) water agar and kept in a sealed moisture chamber. Symptom development was examined after incubating at 25°C (12 h light/12 h dark) for 5 days.
Supporting Information
Western blot analysis with transformants GWF9 and WDF1. Total proteins were isolated from a wild-type strain (70-15), transformant GWF9 expressing MGG_09869-3×FLAG, and transformant WDF1 expressing MGG_04933-3×FLAG fusion.
(TIF)
Increased heat sensitivity of the Mosfl1 mutant. Five-day-old oatmeal agar cultures of Ku80, Mosfl1 mutant GK102, and complemented transformant C49 grown at 30°C. The production of aerial hyphae was reduced in the Mosfl1 mutant.
(TIF)
Stress responses in the Mosfl1 mutants
(DOC)
PCR primers used in this study
(DOC)
Acknowledgments
We thank Drs. Steve Goodwin and Morris Levy at Purdue University for critical reading of this manuscript. We also thank Dr. Wende Liu for insightful discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the USDA Integrated Research Program (#2007-35319-102681). Guotian Li was partially supported by a China Scholarship Council fellowship, #2008630021, via Northwest A & F University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 2.Valent B. Rice blast as a model system for plant pathology. Phytopathology. 1990;80:33–36. [Google Scholar]
- 3.Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 4.Howard RJ, Ferrari MA, Roach DH, Money NP. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA. 1991;88:11281–11284. doi: 10.1073/pnas.88.24.11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thines E, Weber RW, Talbot NJ. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell. 2000;12:1703–1718. doi: 10.1105/tpc.12.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21:1273–1290. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Mehrabi R, Xu J-R. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot Cell. 2007;6:1701–1714. doi: 10.1128/EC.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 10.Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell. 2004;3:1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi WB, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi K, Hamer JE. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell. 1998;10:1361–1373. doi: 10.1105/tpc.10.8.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu JR, Urban M, Sweigard JA, Hamer JE. The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol Plant-Microbe Interact. 1997;10:187–194. [Google Scholar]
- 14.Mitchell TK, Dean RA. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell. 1995;7:1869–1878. doi: 10.1105/tpc.7.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Souza CA, Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev. 2001;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 16.Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, et al. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol. 2009;46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann HA, Kruger J, Lottspeich F, Kahmann R. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator prf1. Plant Cell. 1999;11:1293–1305. doi: 10.1105/tpc.11.7.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaffarnik F, Muller P, Leibundgut M, Kahmann R, Feldbrugge M. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 2003;22:5817–5826. doi: 10.1093/emboj/cdg554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park G, Xue C, Zhao X, Kim Y, Orbach M, et al. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell. 2006;18:2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Kim Y, Park G, Xu JR. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 2005;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue CY, Park G, Choi WB, Zheng L, Dean RA, et al. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14:2107–2119. doi: 10.1105/tpc.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu JR. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol. 2004;53:1695–1707. doi: 10.1111/j.1365-2958.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 23.Park G, Xue GY, Zheng L, Lam S, Xu JR. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol Plant-Microbe Interact. 2002;15:183–192. doi: 10.1094/MPMI.2002.15.3.183. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell TK, Dean RA, Xu JR, Zhu H, Oh YY, et al. Protein chips and chromatin immunoprecipitation - emerging technologies to study macromolecule interactions in Magnaporthe grisea. In: Wang GL, Valent B, editors. Advances in genetics, genomics and control of rice blast disease. Berlin: Springer-Verlag; 2009. pp. 73–82. [Google Scholar]
- 25.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 26.Lin YY, Lu JY, Zhang JM, Walter W, Dang WW, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu SH, Xie Z, Onishi A, Yu XP, Jiang LZ, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita A, Kikuchi Y, Kuhara S, Misumi Y, Matsumoto S, et al. Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins or yeast heat-shock transcription factor. Gene. 1989;85:321–328. doi: 10.1016/0378-1119(89)90424-1. [DOI] [PubMed] [Google Scholar]
- 29.Galeote VA, Alexandre H, Bach B, Delobel P, Dequin S, et al. Sfl1p acts as an activator of the HSP30 gene in Saccharomyces cerevisiae. Curr Genet. 2007;52:55–63. doi: 10.1007/s00294-007-0136-z. [DOI] [PubMed] [Google Scholar]
- 30.Xu JR, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci USA. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu XW, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 32.Mok J, Im H, Snyder M. Global identification of protein kinase substrates by protein microarray analysis. Nature Protocols. 2009;4:1820–1827. doi: 10.1038/nprot.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breeden L, Nasmyth K. Cell-cycle control of the yeast Ho gene: cis-and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 34.Bourett TM, Sweigard JA, Czymmek KJ, Carroll A, Howard RJ. Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet Biol. 2002;37:211–220. doi: 10.1016/s1087-1845(02)00524-8. [DOI] [PubMed] [Google Scholar]
- 35.Conlan RS, Tzamarias D. Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J Mol Biol. 2001;309:1007–1015. doi: 10.1006/jmbi.2001.4742. [DOI] [PubMed] [Google Scholar]
- 36.Giaever G, Chu AM, Ni L, Connelly C, Riles L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 37.Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, et al. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol. 2008;45:68–75. doi: 10.1016/j.fgb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Chumley FG, Valent B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol Plant-Microbe Interact. 1990;3:135–143. [Google Scholar]
- 39.Zhou Z, Li G, Lin C, He C. Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae. Mol Plant-Microbe Interact. 2009;22:402–410. doi: 10.1094/MPMI-22-4-0402. [DOI] [PubMed] [Google Scholar]
- 40.Shi ZX, Leung H. Genetic analysis of sporulation in Magnaporthe grisea by chemical and insertional mutagenesis. Mol Plant-Microbe Interact. 1995;8:949–959. [Google Scholar]
- 41.Liu WD, Xie SY, Zhao XH, Chen X, Zheng WH, et al. A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol Plant-Microbe Interact. 2010;23:366–375. doi: 10.1094/MPMI-23-4-0366. [DOI] [PubMed] [Google Scholar]
- 42.Bauer J, Wendland J. Candida albicans Sfl1 suppresses flocculation and filamentation. Eukaryot Cell. 2007;6:1736–1744. doi: 10.1128/EC.00236-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Su C, Mao X, Cao F, Chen J. Roles of Candida albicans Sfl1 in hyphal development. Eukaryot Cell. 2007;6:2112–2121. doi: 10.1128/EC.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masloff S, Poggeler S, Kuck U. The pro1(+) gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics. 1999;152:191–199. doi: 10.1093/genetics/152.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura M, Fukada J, Moriwaki A, Fujikawa T, Ohashi M, et al. Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci Biotechnol Biochem. 2009;73:1779–1786. doi: 10.1271/bbb.90146. [DOI] [PubMed] [Google Scholar]
- 46.Patzold AJ, Lehming N. Why Ppr1p is a weak activator of transcription. FEBS Lett. 2001;494:64–68. doi: 10.1016/s0014-5793(01)02312-2. [DOI] [PubMed] [Google Scholar]
- 47.Taba MRM, Muroff I, Lydall D, Tebb G, Nasmyth K. Changes in a Swi4,6-DNA-binding complex occur at the time of Ho gene activation in yeast. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- 48.Flor-Parra I, Vranes M, Kamper J, Perez-Martin J. Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell. 2006;18:2369–2387. doi: 10.1105/tpc.106.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci USA. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan X, Heitman J. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol Cell Biol. 2002;22:3981–3993. doi: 10.1128/MCB.22.12.3981-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim TS, Lee SB, Kang HS. Glucose repression of STA1 expression is mediated by the Nrg1 and Sfl1 repressors and the Srb8-11 complex. Mol Cell Biol. 2004;24:7695–7706. doi: 10.1128/MCB.24.17.7695-7706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song W, Carlson M. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 1998;17:5757–5765. doi: 10.1093/emboj/17.19.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang TT, Li D, Li WJ, Wang Y, Sang JL. CaSfl1 plays a dual role in transcriptional regulation in Candida albicans. Chin Sci Bull. 2008;53:2624–2631. [Google Scholar]
- 56.Grunler A, Walther A, Lammel J, Wendland J. Analysis of flocculins in Ashbya gossypii reveals FIG2 regulation by TEC1. Fungal Genet Biol. 2010;47:619–628. doi: 10.1016/j.fgb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Tsuji G, Kenmochi Y, Takano Y, Sweigard J, Farrall L, et al. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol Microbiol. 2000;38:940–954. doi: 10.1046/j.1365-2958.2000.02181.x. [DOI] [PubMed] [Google Scholar]
- 58.Kim S, Park SY, Kim KS, Rho HS, Chi MH, et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2009;5:e1000757. doi: 10.1371/journal.pgen.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Odenbach D, Breth B, Thines E, Weber RW, Anke H, et al. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol Microbiol. 2007;64:293–307. doi: 10.1111/j.1365-2958.2007.05643.x. [DOI] [PubMed] [Google Scholar]
- 60.Ding SL, Liu W, Iliuk A, Ribot C, Vallet J, et al. The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell. 2010;22:2495–2508. doi: 10.1105/tpc.110.074302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehrabi R, Ding S, Xu JR. MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot Cell. 2008;7:791–799. doi: 10.1128/EC.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, et al. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 66.Liu W, Zhou X, Li G, Li L, Kong L, et al. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 2011;7:e1001261. doi: 10.1371/journal.ppat.1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou X, Liu W, Wang C, Xu Q, Wang Y, et al. A MADS box transcription factor MoMcm1 is required for male fertility, microconidium production, and virulence in Magnaporthe oryzae. Mol Microbiol. 2011;80:33–53. doi: 10.1111/j.1365-2958.2011.07556.x. [DOI] [PubMed] [Google Scholar]
- 68.Koga H, Dohi K, Nakayachi O, Mori M. A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol Mol Plant Pathol. 2004;64:67–72. [Google Scholar]
- 69.Valent B, Farral L, Chumley FG. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 1991;127:87–101. doi: 10.1093/genetics/127.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talbot NJ, Kershaw MJ, Wakley GE, deVries OMH, Wessels JGH, et al. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell. 1996;8:985–999. doi: 10.1105/tpc.8.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chao CCT, Ellingboe AH. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can J Bot. 1991;69:2130–2134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blot analysis with transformants GWF9 and WDF1. Total proteins were isolated from a wild-type strain (70-15), transformant GWF9 expressing MGG_09869-3×FLAG, and transformant WDF1 expressing MGG_04933-3×FLAG fusion.
(TIF)
Increased heat sensitivity of the Mosfl1 mutant. Five-day-old oatmeal agar cultures of Ku80, Mosfl1 mutant GK102, and complemented transformant C49 grown at 30°C. The production of aerial hyphae was reduced in the Mosfl1 mutant.
(TIF)
Stress responses in the Mosfl1 mutants
(DOC)
PCR primers used in this study
(DOC)