Abstract
Intestinal microbiota plays an important role in human health, and its composition is determined by several factors, such as diet and host genotype. However, thus far it has remained unknown which host genes are determinants for the microbiota composition. We studied the diversity and abundance of dominant bacteria and bifidobacteria from the faecal samples of 71 healthy individuals. In this cohort, 14 were non-secretor individuals and the remainders were secretors. The secretor status is defined by the expression of the ABH and Lewis histo-blood group antigens in the intestinal mucus and other secretions. It is determined by fucosyltransferase 2 enzyme, encoded by the FUT2 gene. Non-functional enzyme resulting from a nonsense mutation in the FUT2 gene leads to the non-secretor phenotype. PCR-DGGE and qPCR methods were applied for the intestinal microbiota analysis. Principal component analysis of bifidobacterial DGGE profiles showed that the samples of non-secretor individuals formed a separate cluster within the secretor samples. Moreover, bifidobacterial diversity (p<0.0001), richness (p<0.0003), and abundance (p<0.05) were significantly reduced in the samples from the non-secretor individuals as compared with those from the secretor individuals. The non-secretor individuals lacked, or were rarely colonized by, several genotypes related to B. bifidum, B. adolescentis and B. catenulatum/pseudocatenulatum. In contrast to bifidobacteria, several bacterial genotypes were more common and the richness (p<0.04) of dominant bacteria as detected by PCR-DGGE was higher in the non-secretor individuals than in the secretor individuals. We showed that the diversity and composition of the human bifidobacterial population is strongly associated with the histo-blood group ABH secretor/non-secretor status, which consequently appears to be one of the host genetic determinants for the composition of the intestinal microbiota. This association can be explained by the difference between the secretor and non-secretor individuals in their expression of ABH and Lewis glycan epitopes in the mucosa.
Introduction
Growing evidence shows that the composition and diversity of the microbiota in the human intestine can have a surprisingly strong impact on the well-being and health of the host. For example, inflammatory bowel disease (IBD) has been associated with the disturbance of the intestinal microbiota, resulting in the modulation and dysregulation of the inflammatory responses in the intestine [1]. Microbiota composition has been shown to have an effect on the energy harvest and storage of the host [2] and thus, microbiota alterations associated with obesity may have a role in weight-associated health problems.
The microbiota composition in the human intestinal tract is determined by several factors, such as host genotype, health status, age, microbial interactions, and diet [3]. Based on numerous intervention studies, there is convincing evidence for the influence of the diet on the intestinal microbiota (e.g. [4], [5]). In contrast, although growing evidence indicates that host genetic background has a significant impact on the microbiota composition in the intestine, no specific genetic factors determining the intestinal microbiota composition have been established to date. Twin studies applying plate counts, PCR-DGGE fingerprinting or DNA microarrays have shown a higher similarity in the microbiota composition between monozygotic twins than between dizygotic twins, unrelated persons, marital couples and family members [6]–[7], thus clearly pointing to a strong effect of host genetics. In the study by Turnbaugh et al. [8], the pyrosequencing analysis also showed a higher level of similarity in the microbiota composition in twin pairs than between twins and their mothers or unrelated persons, although in their study the similarity of the microbiota between monozygotic twins did not differ from that of dizygotic twins.
The human intestinal tract is colonised with highly diverse and numerous microbiota which has an established role in maintaining the intestinal homeostasis. A particularly interesting group is bifidobacteria, which comprise the predominant intestinal microbiota in infants and are abundant also in the adult population comprising up to 6% of the normal intestinal microbiota [9]. An adult intestine is typically colonised with one to four bifidobacterial species [10], B. longum, B. adolescentis, B. bifidum and B. catenulatum being the most prevalent [11], [12]. Bifidobacteria have beneficial properties, such as immunomodulatory and pathogen inhibition effects (reviewed by [13]). They also are commonly incorporated in probiotic products.
The A, B and H blood group antigens are α1,2-linked fucose containing glycans present on glycoproteins and glycolipids of erythrocytes (red blood cells) in individuals representing A, B and H blood groups, respectively. The enzyme fucosyltransferase 1 encoded by the FUT1 gene is responsible for the synthesis of ABH antigens on erythrocytes. The ABH antigens are also expressed in mucus and other secretions, where their expression is generated by another enzyme, fucosyltransferase 2 (secretor type α1,2-fucosyltransferase) encoded by the FUT2 gene. In ABH secretor individuals (80% of Caucasians) fucosyltransferase 2 converts type 1 N-acetyllactosamine glycan chains to H antigen, which functions as a precursor for the A, B and Lewis b antigens. Non-secretor individuals do not express active fucosyltransferase 2 enzyme due to a non-sense mutation in the FUT2 gene and therefore they are not able to express the ABH antigens in their mucus and other secretions. The FUT2 gene together with the FUT3 gene encoding fucosyltransferase 3 (Lewis type α1,3/4-fucosyltransferase), is also required for the synthesis of Lewis b histo-blood group (Le a−b+) antigens in secretions. In non-secretor individuals, the FUT3 gives rise to the Lewis a histo-blood group (Le a+b−) antigen due to non-functional fucosyltransferase 2. Lewis negative (Le a−b−) individuals have a mutation in FUT3 gene leading to Lewis null phenotype, irrespective of the secretor status or FUT2 gene. These mucosal ABH and Lewis histo-blood group antigens are known to serve as an energy source [14] and adhesion receptors for many microbes [15], and thus could play a role in shaping the microbiota composition of the host.
In the present study, we report that the genetic variation in the human fucosyltransferase 2 (FUT2) gene determining the presence of mucosal α1,2-fucosylated glycan structures, such as ABH and Lewis b histo-blood group antigens, is strongly associated with the microbiota composition, in particular that of bifidobacteria, in the human intestinal tract. We studied the association of the secretor status (determined by the FUT2 gene) with the intestinal microbiota composition by comparing the dominant bacterial and bifidobacterial populations in faecal samples of non-secretor and secretor individuals. The denaturing gradient gel electrophoresis (PCR-DGGE) and qPCR analysis showed that the composition of the intestinal microbiota and particularly bifidobacteria was strongly associated with the host's secretor status. Secretor status determining the expression of the ABH and Lewis b glycan epitopes in the human intestine seems to be one of the host features significantly shaping the composition of bifidobacteria in the intestine.
Results
Blood group analysis
Fourteen of the study individuals were non-secretors and 57 secretors. Twelve of the individuals had Lewis a blood group and 48 had Lewis b blood group (Table 1). In addition, 11 individuals represented Lewis negative blood group, expressing neither Lewis a nor Lewis b antigens. For the Lewis negative samples, secretor status could not be determined by the hemagglutination assay. The secretor status determination of the Lewis negative individuals was based on the sequencing of the coding exon of the FUT2 gene.
Table 1. Distribution of ABH and Lewis Blood groups in the studied individuals.
| Blood group | Non-secretor (14) | Secretor(57) | All (71) |
| A (%) | 8 (57%) | 20 (34%) | 28 (39%) |
| AB (%) | 2 (14%) | 9 (16%) | 11 (15%) |
| B (%) | 1 (7%) | 9 (16%) | 10 (14%) |
| O (%) | 3 (21%) | 19 (33%) | 22 (31%) |
| Lewis a (Le a+b−) | 12 | 0 | 12 |
| Lewis b (Le a−b+) | 0 | 48 | 48 |
| Lewis negativeA (Le a−b−) | 2 | 9 | 11 |
Secretor status of the Lewis negative individuals was determined by sequencing the coding exon of the FUT2 gene.
The sequencing of the FUT2 exon showed that 9 of the Lewis negative individuals with unknown secretor phenotype turned out to be secretors and two were non-secretors, that is, they were homozygous for the 428G>A mutation leading to a FUT2 null-allele (428G>A, se428, W143X, rs601338) (Table 2). All individuals with the non-secretor phenotype were homozygous for the same FUT2 null-allele caused by the 428G>A mutation. Individuals with a secretor phenotype were either homozygous (GG) or heterozygous (GA) at the position 428 (Table 2) generating a functional FUT2 gene. There were no discrepancies between the serological and gene level determinations of the secretor status (Table 2).
Table 2. Comparison of FUT2 genotypedeterminations with secretor phenotype determinations.
| FUT2 genotype based on 428G>A SNP | Phenotype | Total | ||
| Non-secretor | Secretor | UnknownA | ||
| Non-secretor (AA) | 12 | 0 | 2 | 14 |
| Secretor (GA) | 0 | 27 | 6 | 33 |
| Secretor (GG) | 0 | 21 | 3 | 24 |
Secretor phenotype could not be determined for the Lewis negative individuals by hemagglutination assay (phenotypic assay) applied here.
PCR-DGGE gel-to-gel variation
In order to estimate the gel-to-gel variation in the PCR-DGGE analysis, we repeated 18 samples two or three times in the bifidobacterial–DGGE. The correlation of the replicate samples was 0.99 when PCR-DGGE band intensity values were used and 0.91 when the presence/absence of the bands was used, indicating that profiles of the replicate samples were highly similar and the gel-to-gel variation was low.
PCR-DGGE of dominant intestinal bacteria
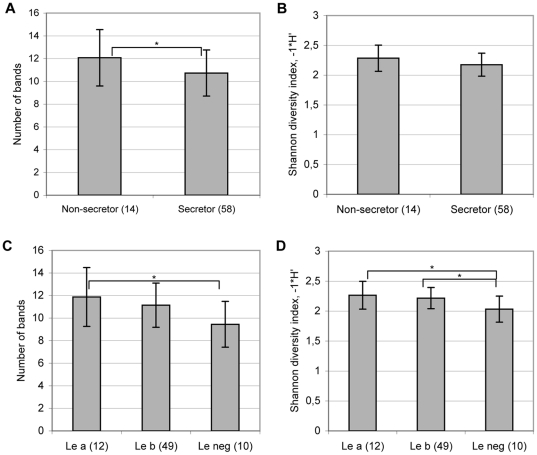
The richness (i.e. number of bands) of dominant bacteria obtained by PCR-DGGE with universal bacterial primers, was significantly higher in the non-secretor than in the secretor individuals (ANOVA; p = 0.03)(Fig. 1), showing that the non-secretor individuals had on average more bacterial genotypes over the detection limit. The bacterial diversity in the samples was measured by Shannon diversity index using the DGGE band intensity values. The Shannon diversity index showed a trend towards an increased bacterial diversity in the non-secretor individuals (p = 0.07). In addition, the individuals with the Lewis negative blood group had lower richness (ANOVA, p = 0.02) and diversity (ANOVA, p = 0.02) of dominant bacteria than the Lewis a individuals and lower diversity (ANOVA, p = 0.02) than the Lewis b individuals (Fig. 1).
Figure 1. Richness and diversity of dominant bacteria in faecal samples of the non-secretor and secretor individuals (A, B) and of the Lewis a, b and negative individuals (C, D) based on the DGGE profiles.
A significant difference between the groups by ANOVA: * p<0.05. In addition, a trend (p = 0.07) towards higher diversity in the non-secretor than in secretor individuals and towards higher richness in Le b individuals than in Le negative individuals was detected.
Eight DGGE band positions, or “DGGE genotypes”, were statistically (Fisher's exact test, p-value between 0.0005 and 0.05) more commonly detected in the non-secretor than in the secretor individuals (Table 3). However, no clustering between the secretor and non-secretor samples was detected in the PCA of DGGE profiles of the dominant microbiota suggesting that other factors dominate the variation in microbiota.
Table 3. The significantly differing band positions and the incidence of bands in secretor (14) and non-secretor samples (57) by PCR-DGGE with universal bacterial primers.
| Band positionA | Detected bands | % of non-secretors | % of secretors | ANOVA | Fisher's exact test |
| 25.20 % | 22 | 43 | 28 | 0.02 | 0.05 |
| 60.20 % | 19 | 36 | 25 | 0.01 | nsB |
| 56.60 % | 17 | 64 | 14 | ns | 0.0005 |
| 39.00 % | 11 | 36 | 11 | 0.004 | 0.01 |
| 42.40 % | 9 | 29 | 9 | 0.02 | 0.07 |
| 47.00 % | 7 | 21 | 7 | 0.05 | ns |
| 50.50 % | 6 | 29 | 4 | 0.001 | 0.01 |
| 61.10 % | 4 | 21 | 1.8 | 0.0002 | 0.03 |
Only band positions, which were statistically significantly different between the groups by ANOVA or/and Fisher exact test are shown. Bns = non-significant.
PCR-DGGE analysis of intestinal bifidobacterial community
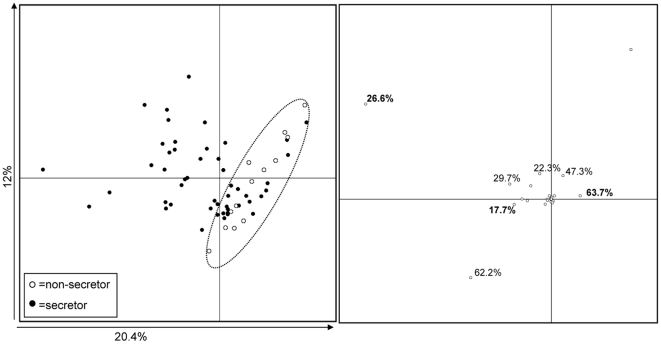
The non-secretor individuals formed a separate cluster within the secretor individuals in the PCA analysis of bifidobacterial DGGE profiles (Fig. 2), indicating that the bifidobacterial population was different in the non-secretor individuals in comparison with the secretor individuals. The Lewis negative individuals did not cluster separately from the Lewis a or Lewis b individuals.
Figure 2. PCA plot based on the bifidobacterial DGGE profiles of faecal samples from the non-secretor (open circles) and secretor (closed circles) individuals (A) and DGGE bands contributing to the principal components 1 and 2 (B).
In panel B, the numbers in bold indicate the band positions, which were significantly less commonly (Fisher's exact test, p<0.01) detected in the non-secretor individuals than in the secretor individuals (See Table 4).
The bifidobacterial band positions that mainly contributed to the PCA clustering were 17.70%, 20.4%, 26.6%, 62.2% and 63.70% (Fig. 2). Three of these band positions (26.6%, 63.70% and 17.70%) were significantly more common in the secretor individuals than in the non-secretor individuals (Fisher's exact test, p<0.01) (Table 4). None of the 14 non-secretors had band in positions 17.70% and 63.70%. Furthermore, the band position 26.60% was clearly less frequent in the non-secretor individuals (N = 2/14; 14%) than in the secretor individuals (N = 38/57; 67%) (Table 4). These band positions were also among the most commonly detected genotypes in the secretor individuals (Table 4). Forty bands in the bifidobacterial DGGE gels, which represented 10 band positions, were excised from the DGGE gel and sequenced to identify which species/groups they represent (Table S1). With three exceptions, the band positions present in more than 10% of the samples, could be identified by sequencing (Table 4). The band position 26.60% was related to B. adolescentis and the position 63.70% to B. catenulatum/pseudocatenulatum, respectively (Table 4). Sequencing of the band position 17.70% was unsuccessful despite several attempts and was therefore not identified. The remaining band positions were related to B. longum, B. bifidum, uncultured bifidobacteria, and another B. adolescentis genotype (Table 4). In total, 26 band positions were detected in the Bifidobacterial-DGGE gels, 13 (50%) of these were detected at least in one non-secretor individual and all at least in one secretor individual.
Table 4. Identification of the bifidobacterial DGGE band positions by sequencing and the incidence of the bands in secretor (14) and non-secretor samples (57).
| Best Blast hit (best cultured hit, similarity) A | Band positionB | Detected bands | % of non-secretors | % of secretors |
| B. longum | 53.5% | 56 | 79 | 79 |
| Uncultured bifidob. (B. adolescentis, 99%) | 62.2% | 41 | 50 | 60 |
| B. adolescentis | 26.6% | 40 | 14 | 67** |
| not sequenced | 17.7% | 18 | 0 | 32** |
| B. catenulatum/pseudocatenulatum | 63.7% | 18 | 0 | 32** |
| B. bifidum | 29.7% | 17 | 7 | 28 |
| not sequenced | 20.4% | 16 | 7 | 26 |
| B. adolescentis | 22.3% | 13 | 14 | 19 |
| Uncultured bifidob. (B. adolescentis/ruminantium, 98–99%) | 43.8% | 13 | 14 | 19 |
| Uncultured bifidob. (B. catenulatum, 99%) | 47.3% | 9 | 7 | 14 |
| Uncultured bifidob. (B. adolescentis, 99%) | 55.0% | 9 | 7 | 14 |
| Uncultured bifidob. (B. ruminantium, 99%) | 44.5% | 8 | 7 | 12 |
| Not sequenced | 39.3% | 7 | 7 | 11 |
The similarity of the best Blast hit for a cultured strain is shown in parentheses, in cases where an uncultured bacterium was the best hit. Detailed data for the identification of each position is shown in table S1. B Only the band positions that were detected at least in 10 % of the samples are shown.** Significant differences: Band positions 26.6%, 17.70% and 63.7% were more frequently detected in secretor than non-secretor samples (Fisher's exact test, p<0.01), and in Lewis b than Lewis a samples (Fisher's exact test, p<0.01 for 17.70% and 26.60% ; p<0.02 for 63.70%).
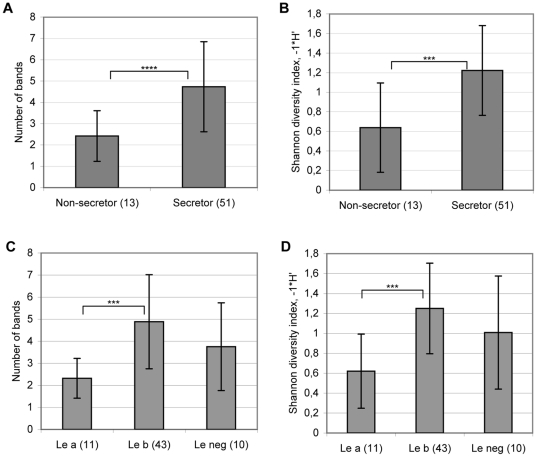
The lower incidence of bifidobacterial DGGE genotypes in the non-secretor individuals was also reflected in the bifidobacterial diversity in the samples. Bifidobacterial diversity and richness in the non-secretor individuals was significantly reduced in comparison with the secretor individuals (Fig. 3). On average, the number of bands per sample in the non-secretor individuals was almost twice (1.9) as low as that in the secretor individuals. The mean number of bands was 2.5 (range from 0 to 5) in the non-secretor samples and 4.7 (range from 0 to 11) in the secretor samples. The bifidobacterial diversity and richness did not significantly differ between the Lewis-negative individuals and the Lewis a or Lewis b individuals (Fig. 3).
Figure 3. Bifidobacterial richness and diversity in faecal samples of the non-secretor and secretor individuals (A, B) and Lewis a, b and negative individuals (C, D) based on the DGGE profiles.
Significant differences by ANOVA: **** p<0.0001, *** p<0.001.
The inter-individual variation in bifidobacterial profiles was high in both the non-secretor and the secretor samples, as indicated by relatively low similarity values for the DGGE profiles (on average 41% between the non-secretor individuals and on average 48% between the secretor individuals).
qPCR
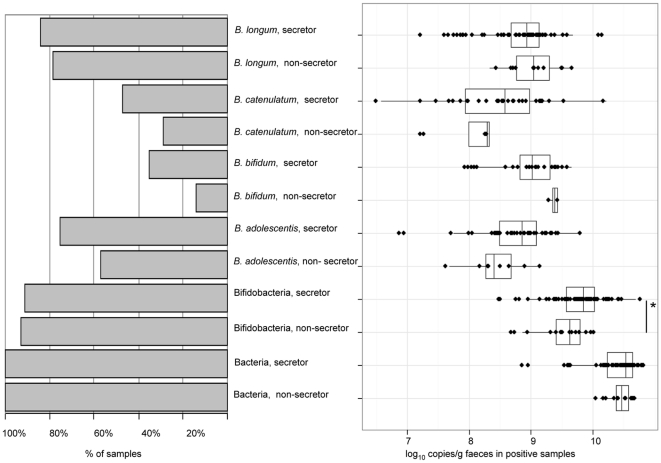
Using a qPCR approach, bifidobacterial 16S rRNA genes were detectable in over 90% of the faecal samples. The total number of bifidobacterial 16S rRNA gene copies was lower (Wilcoxon test, p = 0.05) and fewer bifidobacterial groups were present in the non-secretor individuals in comparison with the secretor individuals (Fig. 4). All the bifidobacterial groups, B. bifidum, B. longum group, B. catenulatum/pseudocatenulatum and B. adolescentis were detected less frequently in faecal samples of the non-secretor than in samples of the secretor individuals, confirming the PCR-DGGE results. In faecal samples with detectable amounts of B. adolescentis group, a trend towards lower abundance of B. adolescentis in the non-secretor than in secretor individuals (p = 0.06) was found. The abundances of other bifidobacterial groups in samples were not significantly different. The total bacterial numbers did not differ between the secretor and non-secretor individuals (Fig. 4).
Figure 4. Incidence (% of samples) (left) and Box-and-Whisker plots (right) (based on log10 16S rRNA gene copies per g faeces) of total bacteria, bifidobacteria and bifidobacterial groups in the faecal samples of the non-secretor and secretor individuals by qPCR.
A significant difference by Wilcoxon test: * p<0.05. In addition, a trend (p = 0.06) towards higher number of the 16S rRNA gene copies of B. adolescentis in the secretor individuals than in the non-secretor individuals was detected.
Effect of GG homozygocity versus GA heterozygocity in FUT2 gene 428G>A (W143X, se428, rs601338) SNP on the microbiota diversity
We also assessed whether homozygocity (GG) or heterozygocity (GA) of the FUT2 gene 428G>A SNP is associated with the composition of bifidobacteria or dominant bacteria. Neither the composition nor the diversity of dominant bacteria or bifidobacteria was significantly different between the secretor individuals homozygous or heterozygous for the position 428 in the codon 143.
Discussion
To study the association of the intestinal microbiota with the histo-blood group secretor status (defined by the FUT2 gene), we analysed the faecal microbiota in 71 individuals, of which 14 were non-secretors. We observed that the diversity and amount of faecal bifidobacteria was considerably reduced in the non-secretor individuals. In addition to bifidobacteria, indications that the composition of dominant bacteria differed between the non-secretor and secretor individuals were discovered. Altogether these results suggest that the FUT2 gene, which determines the presence of ABH histo-blood group glycans in mucus lining of the intestine, is a host genotypic feature significantly affecting the bacterial composition, particularly the bifidobacterial composition, in the intestine.
The secretor status determined by the FUT2 gene was strongly associated with the bifidobacterial diversity and composition. The non-secretor individuals only had about half of the bifidobacterial diversity and richness present in the secretor individuals based on the PCR-DGGE analysis. In addition, the non-secretor individuals had significantly reduced bifidobacterial abundance in comparison with the secretor individuals as measured by qPCR. Moreover, the non-secretor individuals lacked, or were rarely colonised by, several bifidobacterial DGGE genotypes, which were related to species B. adolescentis, P. catenulatum/pseudocatenulatum and B. bifidum, and were common in the secretor individuals. We applied the PCR-DGGE to compare the bacterial diversity and community structure between the secretor and non-secretor individuals. The PCR-DGGE method is known to detect only the predominant part of the bacteria present in a complex sample. Bifidobacterial population is usually composed of limited number (0–4) of species [10] and thus, could be captured by the PCR-DGGE analysis with bifidobacterial specific primers. We also showed that bifidobacterial-DGGE profiles were highly reproducible. Moreover, we isolated bifidobacterial strains from the non-secretor and secretor individuals and analysed their 16S rRNA gene fragments in a DGGE gel along with faecal samples. The isolated strains corresponding to the most common (present in >10% samples) band positions in bifidobacterial DGGE gels were found (data not shown), reducing the likelihood that the detected DGGE bands originate from PCR artefacts sometimes occurring in the PCR-DGGE. In contrast to the bifidobacteria-specific DGGE, it is likely that methodological limitations hinder the interpretation of the PCR-DGGE targeted at dominant bacteria (universal PCR-DGGE). It is probable that several secretor-status associated genotypes, which are present at low levels, are missed in the PCR-DGGE using universal primers and their associations with the secretor status could, thus, not be detected. Nevertheless, our finding on the differences in the dominant microbiota between the non-secretor and secretor individuals suggests that the association between microbiota and secretor status is not limited to bifidobacteria only. It remains to be seen which other bacterial groups and species are associated with the secretor status.
The altered microbiota composition in the non-secretor individuals shown in this study may be an important factor contributing to the non-secretor disease susceptibility. Polymorphism of the FUT2 gene, determining the secretor status, has been suggested to modulate innate immune responses and even have an evolutionary role in humans' survival during different pathogen outbreaks [16],[17]. The non-secretor phenotype has been genetically associated with increased risk for Crohn's disease [18], [19], and necrotizing enterocolitis [20]. Secretor status is also associated with the susceptibility to several infectious diseases. Non-secretors have an increased risk for urinary tract infections [21], [22] and vaginal candidiasis [23], [24], but a reduced risk for diarrhoea caused by certain genotypes of Norovirus [25]. Interestingly, many of these secretor status associated diseases, such as Crohn's disease [26], [27], urinary tract infection [28], and NEC [29] have also been connected to changes in the intestinal microbiota composition or activity. Bifidobacteria have been shown to have health promoting effects on humans [13]. Bifidobacteria or bifidobacteria-containing strain mixtures have shown promising results e.g. in the alleviation of the symptoms of irritable bowel syndrome (IBS) [30], inflammatory bowel disease [31], and diarrhoea [32], although the mechanism of action is largely unknown. Reduced bifidobacterial abundance has been connected to intestinal disorders such as irritable bowel syndrome [33] and inflammatory bowel disease [26]. Taken together, the properties of bifidobacteria and the results of this study suggest that the secretor status, by effecting bifidobacterial diversity, may also play a role in susceptibility to the diseases associated with the decreased bifidobacterial abundance in the intestine.
Metagenomics studies indicate that a considerable number of intestinal microbiota genes are involved in carbohydrate metabolism. Kurokawa et al. [34] reported that the carbohydrate metabolism genes of microbiota are enriched in the intestine (24% of genes in adults and 34% in children) in comparison with the microbiota originating from other environments, such as soil and sea. Both plant polysaccharides and host derived glycans are important energy sources for intestinal bacteria. Fucosylated histo-blood group antigens, such as the ABH and Lewis b histo-blood group antigens, are terminal epitopes of glycan chains in glycoproteins and glycolipids mediating the interaction between host and both commensal and pathogenic intestinal bacteria [35], [36]. Non-secretor individuals have null-allele of FUT2 gene and do not express such α1,2-fucose containing glycan structures in their intestinal mucosa. Bacteria that can interact with these epitopes and compete for adhesion sites or to use them as energy sources have a better colonisation ability in secretor individuals than in non-secretor individuals (e.g. bifidobacteria in this study). Intestinal bifidobacteria, whose abundance and diversity were higher in the intestine of secretor individuals than non-secretor individuals in this study, are adapted to utilise glycans present in mucins and human milk [37]. It is known that some microorganisms secrete glycosidases capable of degrading histo-blood group antigens [14]. Comparatively small populations of human faecal bacteria produce α-glycosidases capable of degrading terminal ABH and/or Lewis groups in glycans [14]. Among them are bifidobacteria with 1,2-α-fucosidase to hydrolyse α-1,2-fucosyl linkages present in various glycans, such as the above-mentioned histo-blood group antigens [38]. Recently, Turroni et al. [39] showed, using genomic, proteomic and transcriptomic analysis of B. bifidum PRL2010, the existence of enzymes allowing further degradation of many core glycan chains and they concluded that the property is important for intestinal colonisation of B. bifidum. Such degradation of glycan cores may require the initial removal of terminal α-fucose, enabling subsequent processing of glycan chain by β-galactosidase and/or β-N-acetylhexosaminidase and endo-α-N-acetylgalactosaminidase, which catalyses the release of GalNAc from serine/threonine residues of various mucin-type glycoproteins, all of these enzymes being encoded in B. bifidum PRL2010 genome [39]. In addition, bifidobacteria but typically not the other common commensals, have lacto-N-biosidase degrading type 1 glycan chains, which are precursors of fucosylated histo-blood group antigens in the human intestine [40]. Therefore, it can be concluded that bifidobacteria have very specific strategies for the utilization of host glycans.
In this study we present evidence that the FUT2 gene, which defines the secretor status and thus, the expression of the ABH and Lewis histo-blood group antigens in intestinal mucus, is one of the host genotypic features determining the composition of intestinal microbiota, particularly bifidobacterial population. We showed that bifidobacterial diversity and composition is strongly associated with the secretor status of the host. These results increase our understanding of the factors explaining inter-individual variations in intestinal microbiota composition and help us to evaluate the role of intestinal microbiota in health and disease.
Materials and Methods
Samples
Altogether 82 healthy adult individuals were recruited to the study from Helsinki metropolitan area, Finland. Individuals with clinically diagnosed intestinal diseases or regular intestinal disturbances were excluded from the study. The individuals had not received antibiotic therapy within two months of the faecal sampling time. Probiotic consumption was restricted one week before sampling and alcohol consumption was limited to one portion per day during three days before faecal sampling. All individuals consumed mixed diet. The study had the approval of the ethical committee of the Helsinki University Hospital and all subjects signed a written informed consent.
Both faecal and blood samples were collected from 71 subjects (7 males and 64 females). The distribution of blood groups was balanced towards the blood groups (Lewis a, Lewis negative or AB and B) rare in Finland by excluding 11 secretor individuals representing common blood groups A or O and Lewis b from faecal donation. The age of the volunteers who donated faecal samples ranged from 31 years to 61 years and was on average 44.7 years.
Faecal samples for the determination of the microbiota composition were frozen at −80°C within 5 hours of defecation. EDTA anticoagulated peripheral blood samples for blood group analysis were kept at +4°C and analysed within 24 hours. Buffy coats were extracted from citrate anticoagulated peripheral blood samples by centrifugation and stored at −80°C until DNA extraction.
Determination of ABH and Lewis blood group and secretor status
ABO blood groups were determined by hemagglutination assay with Olympus PK 7300 according to standard blood banking practise. Lewis a and Lewis b typings were performed in tubes by monoclonal antisera (Sanquin, the Netherlands). Determination of secretor status was based on Lewis antigens. Secretor status could not be determined by phenotyping for the samples of Lewis negative individuals and their secretor status assignments were based on genotyping of the FUT2 gene.
In addition to phenotyping, secretor status was genotyped by sequencing the coding exon of FUT2 as described in [41] and [42]. Briefly, the FUT2 exon was amplified with PCR and sequenced with ABI3100 in the Haartman Institute, Sequencing Core Facility (University of Helsinki, Finland) using primers described in Table 5. Individual's secretor genotype was defined as non-secretor, when the FUT2 428G>A SNP (se428, W143X, rs601338) was AA and as secretor when the FUT2 428G>A SNP was GA or GG. Based on the sequence analyses of the FUT2 exon in a separate Finnish cohort consisting of 184 secretor and non-secretor individuals, no other non-secretor alleles than the FUT2 428G>A (se428, W143X, rs601338) were found in Finnish population (data not shown).
Table 5. Primers used in sequencing of the FUT2 gene encoding fucosyltransferase 2.
| Primer | Sequence 5′→3′ | Reference |
| 1_FUT2forward | CCATCTCCCAGCTAACGTGTCC | [41] |
| 2_FUT2reverse | GGGAGGCAGAGAAGGAGAAAAGG | [41] |
| 3_FUT2forward | GGGGAGTACGTCCGCTTCAC | This study |
| 4_FUT2reverse | AGGATCTCCTGGCGGAGGTG | This study |
| 1_FUT2PCRforward | ACACACCCACACTATGCCTGCAC | [47] |
| 2_FUT2PCRreverse | ACTTGCAGCCCAACGCATCTT | [47] |
| 1_FUT2SEQforward | CCAGCTAACGTGTCCCGTTTTCC | [47] |
| 2_FUT2SEQreverse | GGCACTCATCTTGAGGGAGGCA | [47] |
DNA extraction
Total bacterial DNA was extracted from faecal samples using the FastDNA® SPIN Kit for Soil and the FastPrep® Instrument (MP Biomedicals, CA, USA) according the manufactures instruction with minor modifications. Shortly, 0.3 g faecal sample was mixed by vortexing with sodium phosphate buffer and MT buffer. Faecal slurry (1 ml) was homogenised and cells were lysed in lysing matrix E tubes with FastPrep® Instrument three times for 60 at speed setting 6.0 m/s. DNA was purified using silica based binding matrix, SPIN ™ filters, and SEWS-M wash solution. The DNA was eluted in 250 µl Dnase/pyrogen free water. Human DNA was extracted from buffy coat preparations using the QIAamp DNA Blood Mini Kit (QIAGEN Inc, CA, US). The DNA concentrations were determined with NanoDrop 1000 (Thermo Scientific, DE, USA). The extracted DNA samples were stored at − 20°C.
PCR- DGGE
The similarity and diversity of microbiota in faecal samples of the study subjects was analysed by the PCR-DGGE. The partial 16S rRNA gene was amplified by PCR with universal bacterial primers and bifidobacterial specific primers. Amplification with universal primers U968F+GC (5′- CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGAACGCGAAGAACCTTA-3′) and U1401R (5′-CGGTGTGTACAAGACCC-3′) [43] was performed as described in Mättö et al. [44]. Bifidobacteria were amplified with primers Bif164F (5′- GGGTGGTAATGCCGGATG-3′) and Bif662R+GC (5′- CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCACCGTTACACCGGGAA-3′) as described in Satokari et al. [45], except elongation temperature of 72°C and Taq polymerase (Invitrogen) were used. Template DNAs were diluted to concentration 20 ng/µl for both PCRs. PCR products were obtained from all the samples with universal bacterial primers and from 64/71 samples with bifidobacterial primers. A volume of 20 µl PCR product was separated in 8% polyacrylamide gel with denaturing gradient of urea and formamide ranging from 38% to 60% (universal amplicons) or from 45% to 60% (bifidobacterial amplicons). The DGGE gels were run at 70 V for 960 mins using Dcode universal mutation detection system (Bio-Rad, CA, USA). The gels were stained with SYRB®Safe DNA gel stain (Invitrogen, Oregon, USA) for 30 mins and documented with SafeImager Bluelight table (Invitrogen) and AlphaImager HP (Alpha Innotech, South-Africa) imaging system. To estimate the gel-to-gel variation and reproducibility of the bifidobacterial PCR-DGGE method, 18 of the samples were run two or three times in different gels.
The bands were excised from the bifidobacterial DGGE gels by sterile Pasteur pipette and the DNA was eluted by incubating the gel slices in 50 µl of sterile H2O at +4°C overnight. The correct positions and purity of the DNA fragments eluted from the bands were checked by amplifying 1 µl of eluted DNA with primers Bif164F-Bif662R and re-running the amplified fragments along with the original samples in PCR-DGGE. Bands were sequenced in Eurofins MWG (Germany) using primers Bif164F and Bif66R without GC-rich clamp.
qPCR
The qPCR method was applied to detect and quantify the 16S rRNA gene copies of bacteria, bifidobacteria and 4 bifidobacterial species/groups, B. bifidum, B. longum group, B. catenulatum/pseudocatenulatum and B. adolescentis in faecal samples. The primers and annealing temperature for each primer pair are shown in Table 6. Reaction mixture (25 µl) was composed of 0.3 µM of each primer (Sigma-Aldrich, UK), 1 x Power SYBR Green PCR Master Mix (Applied Biosystems, CA, USA), 4 µl faecal DNA diluted to the concentration of 1 ng/µl for bifidobacterial group/species-specific primer pair and to the concentration of 0.1 ng/µl for universal primers and bifidobacterial primers. The amplification conditions in the ABI Prism 7000 instrument (Applied Biosystems, CA, USA) were one cycle of 95 °C for 10 mins, followed by 40 cycles of 95 °C for 15 s, and appropriate annealing temperature (see Table 6) for 60 s. Melting temperature curves from 60°C to 95°C were analysed to determine the specificity of the amplification. All the samples and standards were analysed in three replicates. Standard curves from bacterial strains were constructed for each bacterial group (Table 6) by 10-fold dilutions of known concentrations of the bacterial genomic DNA (from 10 ng/µl to 0.0001 ng/µl). The Genomic DNA from the standard strains was extracted by QIAmp®DNA mini kit (Qiagen) combined with cell lysis in the FastPrep® Instrument (MP Biomedicals, CA, USA). Bacterial cells were harvested from plates, transferred to lysis tubes containing 0.1 g zirconia beads (diameter of 0.1 mm) (BioSpec products, Inc., USA) and sterile solid glass beads (diameter of 3 mm) (Sigma) in 180 µl ATL-buffer from the QIAmp®DNA mini kit. Lysis tubes were treated with FastPrep® Instrument for 60 s at speed setting 5.5 m/s. DNA concentration measurements were performed similarly to the samples (see above). GenEx Enterprise v.5.2.6.34 (MultiD Analyses AB, Sweden) was used for the analysis of standard curves and reverse quantification of the samples. The amplification efficiencies were from 93% to 98% for all the other qPCR primer pairs except for B. bifidum specific primers, in which amplification efficiency varied from 80% to 92% and for B. catenulatum/pseudocatenulatum, in which efficiency varied from 87% to 91%.
Table 6. Primers targeting the 16S rRNA gene, annealing temperature and strains used as standards in qPCR analysis.
| Group | PrimerA | Primer sequence 5′→3′ | Anneling-T, °C | Standard strain |
| Bacteria | p201, p1370 | GAGGAAGGNGNGGANGACGT, AGNCCCGNGAACGTATTCAC | 60 | L. rhamnosus E-96666 |
| Bifidobacteria | qBifF, qBifR | TCGCGTCYGGTGTGAAAG, CCACATCCAGCRTCCAC | 59 | B. bifidum E-97795 |
| B. longum group | BiLON-1, BiLON-2 | CAGTTGATCGCATGGTCTT, TACCCGTCGAAGCCAC | 62 | B. longum E-96664 |
| B. bifidum | BiBIF-1, BiBIF-2 | CCACATGATCGCATGTGATTG, CCGAAGGCTTGCTCCCAAA | 58 | B. bifidum E-97795 |
| B. catenulatum/pseudocatenulatum | BiCAT-1, BiCAT-2 | CGGATGCTCCGACTCCT, CGAAGGCTTGCTCCCGAT | 62 | B. catenulatum DSM 16992 |
| B. adolescentis group | BiADO-1B, BiADO-1b, BiADO-2 | CTCCAGTTGGATGCATGTC, TCCAGTTGACCGCATGGT, CGAAGGCTTGCTCCCAGT | 58 | B. adolescentis E-981074 |
Statistical analysis
Digitalised DGGE gel images were imported to the Bionumerics program version 5.0 (Applied Maths, Belgium) for normalisation and band detection. The bands were normalised in relation to a marker sample composed of 7 common intestinal bacterial strains (for the universal PCR-DGGE) or 5 bifidobacterial strains (for the bifidobacterial PCR-DGGE). Band search and band matching using band tolerance of 1% were performed as implemented in the Bionumerics. The bands and band matching were manually checked and corrected. Samples with no amplification in the bifidobacterial PCR-DGGE were excluded from the analysis. Similarity of the bifidobacterial profiles was calculated as implemented in Bionumerics, version 5. Matrices based on band intensities and presence /absence of bands were exported from Bionumerics and used for calculation of Shannon diversity indexes in Microsoft Excel. Shannon diversity index, H', was calculated using equation H' = Σpiln(pi), where pi was proportion of each species (i.e. DGGE band intensity) in the sample. The richness was calculated as a number of detected bands in the DGGE profile of the sample. Principal component analysis (PCA) based on the band intensities was calculated as implemented in Bionumerics, version 5.0. Other statistical analyses (ANOVA, Fisher exact test and two-sample Wilcoxon test) were computed with statistical programming language R, version 2.10.1. (http://www.r-project.org/). Gel-to-gel variation was measured by comparing the DGGE profiles of the samples run in different gels. The DGGE profiles based on either band intensities or presence/absence of bands were used for calculation of the Pearson correlation between replicate samples using statistical programming language R, version 2.12.0.
The sequences retrieved from the DGGE bands were trimmed and aligned by ClustalW [46]. The closest relatives of the sequences were searched using Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and NCBI nr database. Distance matrix of the aligned sequences was used to compare the similarity of the sequences. The accession numbers of the sequences are FR775384-FR775395.
Supporting Information
The best Blast hits of the sequences derived from bifidobacterial DGGE bands
(DOC)
Acknowledgments
The volunteers are thanked for the sample donations. Ms. Sisko Lehmonen, Ms. Elina Pusa, Ms. Paula Salmelainen and the technicians responsible for the blood group determinations, are thanked for skilful technical assistance. Dr. Pertti Sistonen is thanked for blood group expertise.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from europe and rural africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser, Arjan G. M J, et al. The host genotype affects the bacterial community in the human gastronintestinal tract. Microbial Ecology in Health and Disease. 2001;13(3):129–134. [Google Scholar]
- 7.Rajilic-Stojanovic M. Diversity of human gastrointestinal microbiota. novel perspestives from high throughput analyses. 2007. p. 214 p. PhD thesis, WageningenUniveristy and Research Center: Wageningen.
- 8.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lay C, Rigottier-Gois L, Holmstrom K, Rajilic M, Vaughan EE, et al. Colonic microbiota signatures across five northern european countries. Appl Environ Microbiol. 2005;71(7):4153–4155. doi: 10.1128/AEM.71.7.4153-4155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mättö J, Malinen E, Suihko ML, Alander M, Palva A, et al. Genetic heterogeneity and functional properties of intestinal bifidobacteria. J Appl Microbiol. 2004;97(3):459–470. doi: 10.1111/j.1365-2672.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 11.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65(10):4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, et al. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3(6):745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 13.Boesten RJ, de Vos WM. Interactomics in the human intestine: Lactobacilli and bifidobacteria make a difference. J Clin Gastroenterol 42 Suppl 3 Pt. 2008;2:S163–7. doi: 10.1097/MCG.0b013e31817dbd62. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, et al. Mucin degradation in human colon ecosystems. isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest. 1985;75(3):944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moulds JM, Nowicki S, Moulds JJ, Nowicki BJ. Human blood groups: Incidental receptors for viruses and bacteria. Transfusion. 1996;36(4):362–374. doi: 10.1046/j.1537-2995.1996.36496226154.x. [DOI] [PubMed] [Google Scholar]
- 16.Linden S, Mahdavi J, Semino-Mora C, Olsen C, Carlstedt I, et al. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4(1):e2. doi: 10.1371/journal.ppat.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anstee DJ. The relationship between blood groups and disease. Blood. 2010;115(23):4635–4643. doi: 10.1182/blood-2010-01-261859. [DOI] [PubMed] [Google Scholar]
- 18.McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with crohn's disease. Hum Mol Genet. 2010;19(17):3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, et al. Genome-wide meta-analysis increases to 71 the number of confirmed crohn's disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrow AL, Meinzen-Derr J, Huang P, Schibler KR, Cahill T, et al. Secretor phenotype and genotype are novel predictors of severe outcomes in premature infants. FASEB J 24(1_MeetingAbstracts) 2010:480.6. [Google Scholar]
- 21.Kinane DF, Blackwell CC, Brettle RP, Weir DM, Winstanley FP, et al. ABO blood group, secretor state, and susceptibility to recurrent urinary tract infection in women. Br Med J (Clin Res Ed) 1982;285(6334):7–9. doi: 10.1136/bmj.285.6334.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheinfeld J, Schaeffer AJ, Cordon-Cardo C, Rogatko A, Fair WR. Association of the lewis blood-group phenotype with recurrent urinary tract infections in women. N Engl J Med. 1989;320(12):773–777. doi: 10.1056/NEJM198903233201205. [DOI] [PubMed] [Google Scholar]
- 23.Thorn SM, Blackwell CC, MacCallum CJ, Weir DM, Brettle RP, et al. Nonsecretion of ABO blood group antigens and susceptibility to infection by candida species. FEMS Microbiology Immunology. 1989;47:401–406. doi: 10.1111/j.1574-6968.1989.tb02428.x. [DOI] [PubMed] [Google Scholar]
- 24.Hurd EA, Domino SE. Increased susceptibility of secretor factor gene Fut2-null mice to experimental vaginal candidiasis. Infect Immun. 2004;72(7):4279–4281. doi: 10.1128/IAI.72.7.4279-4281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson MM, Rydell GE, Grahn A, Rodriguez-Diaz J, Akerlind B, et al. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or lewis (FUT3) genotype. J Infect Dis. 2006;194(10):1422–1427. doi: 10.1086/508430. [DOI] [PubMed] [Google Scholar]
- 26.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, et al. Alterations of the dominant faecal bacterial groups in patients with crohn's disease of the colon. Gut. 2003;52(2):237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal crohn's disease. Inflamm Bowel Dis. 2009;15(5):653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 28.Kirjavainen PV, Pautler S, Baroja ML, Anukam K, Crowley K, et al. Abnormal immunological profile and vaginal microbiota in women prone to urinary tract infections. Clin Vaccine Immunol. 2009;16(1):29–36. doi: 10.1128/CVI.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3(8):944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner DM, Chey WD. Bifidobacterium infantis 35624: A novel probiotic for the treatment of irritable bowel syndrome. Rev Gastroenterol Disord. 2009;9(1):7–15. [PubMed] [Google Scholar]
- 31.Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. The gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15(13):1528–1536. doi: 10.2174/138161209788168146. [DOI] [PubMed] [Google Scholar]
- 32.Chouraqui JP, Van Egroo LD, Fichot MC. Acidified milk formula supplemented with bifidobacterium lactis: Impact on infant diarrhea in residential care settings. J Pediatr Gastroenterol Nutr. 2004;38(3):288–292. doi: 10.1097/00005176-200403000-00011. [DOI] [PubMed] [Google Scholar]
- 33.Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100(2):373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 34.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14(4):169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273(5280):1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 36.Hooper LV, Gordon JI. Glycans as legislators of host-microbial interactions: Spanning the spectrum from symbiosis to pathogenicity. Glycobiology. 2001;11(2):1R–10R. doi: 10.1093/glycob/11.2.1r. [DOI] [PubMed] [Google Scholar]
- 37.Turroni F, van Sinderen D, Ventura M. Bifidobacteria: From ecology to genomics. Front Biosci. 2009;14:4673–4684. doi: 10.2741/3559. [DOI] [PubMed] [Google Scholar]
- 38.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, et al. Molecular cloning and characterization of bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol. 2004;186(15):4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, et al. Genome analysis of bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, et al. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74(13):3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva LM, Carvalho AS, Guillon P, Seixas S, Azevedo M, et al. Infection-associated FUT2 (fucosyltransferase 2) genetic variation and impact on functionality assessed by in vivo studies. Glycoconj J. 2010;27(1):61–68. doi: 10.1007/s10719-009-9255-8. [DOI] [PubMed] [Google Scholar]
- 42.Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol. 2009;26(9):1993–2003. doi: 10.1093/molbev/msp108. [DOI] [PubMed] [Google Scholar]
- 43.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, et al. Sequence heterogeneities of genes encoding 16S rRNAs in paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178(19):5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mättö J, Maunuksela L, Kajander K, Palva A, Korpela R, et al. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome–a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43(2):213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67(2):504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit. 2002;2.3 doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 47.Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol. 2009;26(9):1993–2003. doi: 10.1093/molbev/msp108. [DOI] [PubMed] [Google Scholar]
- 48.Tseng CP, Cheng JC, Tseng CC, Wang C, Chen YL, et al. Broad-range ribosomal RNA real-time PCR after removal of DNA from reagents: Melting profiles for clinically important bacteria. Clin Chem. 2003;49(2):306–309. doi: 10.1373/49.2.306. [DOI] [PubMed] [Google Scholar]
- 49.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 50.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70(1):167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The best Blast hits of the sequences derived from bifidobacterial DGGE bands
(DOC)