Abstract
Stem cells in the shoot apical meristem (SAM) of plants are the self-renewable reservoir for leaf, stem and flower organogenesis1,2. In nature, disease-free plants can be regenerated from SAM despite infections elsewhere, which underlies a horticultural practice for decades3. However, the molecular basis of the SAM immunity remains enigmatic. Here we show a surprising discovery that the CLAVATA3 peptide (CLV3p), expressed and secreted from the stem cells and functioning as a key regulator of stem cell homeostasis in the Arabidopsis SAM1,2,4, can trigger immune signalling and pathogen resistance via the flagellin receptor kinase FLS25,6. CLV3p-FLS2 signalling acts independently from the stem cell signalling pathway mediated through CLV1 and CLV2 receptors 1,2,4, and is uncoupled from the FLS2-mediated growth suppression5,6. Endogenous CLV3p perception in the SAM by a pattern recognition receptor FLS2 for bacterial flagellin breaks the previously defined self and nonself discrimination in innate immunity 6,7. The dual CLV3p perceptions illustrate co-evolution of plant peptide and receptor kinase signalling for both development and immunity. The enhanced immunity in SAM or germ lines may represent a common strategy toward immortal fate in plants and animals1,2,8.
In both plants and animals, innate immunity is triggered through pattern recognition receptors (PRRs) in response to microbe-associated molecular patterns (MAMPs)6,7 to provide the first line of inducible defense. Plant receptor kinases represent the main functions of known plasma membrane PRRs for MAMP recognition to distinguish nonself from self6. FLS2 is the first characterized plant leucine rich repeat (LRR) receptor kinase that perceives the bacterial flagellin and launches convergent downstream signalling and defense pathways for potentially broad-spectrum pathogen resistance5,6,9–11. The perception of bacterial flagellin is conserved in seed plants and functional FLS2 orthologs are found from A. thaliana to rice5,6. As FLS2 is expressed throughout the whole plant including the SAM (Supplementary Fig. 2)5,12, flagellin-FLS2 signalling could provide immune protection in different parts of the plant body after infections.
While developing a plant expression system to screen for peptide-mediated receptor-like kinase (RLK) signalling, we observed that the endogenously modified 12-amino acid CLV3p (Supplementary Table 1)13 triggered similar responses as flg22 (the conserved 22-amino acid peptide of bacterial flagellin) in mesophyll protoplasts5,6,9,14–16. This finding was unexpected because CLV3p is normally expressed, secreted and processed by the stem cells to control SAM maintenance via CLV1 and CLV2 receptors1,2,4,13,17. Flg22 and CLV3p, but not ΔCLV3p lacking the last His residue, activated similar MAPK activities detected by the in-gel kinase assay (Fig. 1a and Supplementary Fig. 3)6,9,14–16. Highly purified CLV3p synthesized by different sources displayed the same activities, ruling out the contamination possibility. We sought to identify the CLV3p receptor in leaf cells by examining MAPK activation in various receptor mutants. Neither the dominant clv1-1 mutant (Fig. 1a) nor the clv2-1 mutant (Supplementary Fig. 4)1,2,4,17–19 affected CLV3p-triggered MAPK activation. Surprisingly, two independent Ler and Col-0 fls2 mutant alleles5,6, but not efr-1 (the bacterial elongation factor EF-Tu receptor EFR) mutant20, failed to support the activation of MAPKs by both flg22 and CLV3p (Fig. 1a and Supplementary Fig. 3). Complementation with the WT FLS2 gene (Fig. 1b) but not CLV1 (data not shown) in the fls2 mutant confirmed that FLS2 could recognize both flg22 and CLV3p to mediate MAPK signalling. Importantly, CLV3p and flg22 activated similar early marker genes, including FRK1, WRKY29 and WRKY30 (Fig. 1c)9,10,14.
Figure 1. CLV3p and flg22 activate similar downstream responses through FLS2.
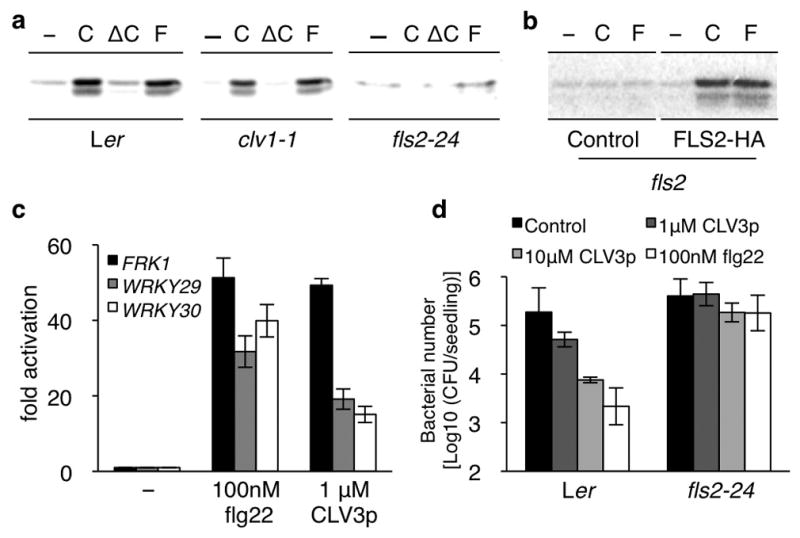
a, MAPK activity analysis. Ler, clv1-1 and fls2-24 protoplasts were treated with 1 μM CLV3p (C), 1 μM ΔCLV3p (ΔC) or 100 nM flg22 (F) for 10 min. b, FLS2-HA complements fls2. c, CLV3p triggers flg22 marker gene activation. Quantification by qRT-PCR. Peptides treatment for 1 h. Error bars, s.d. (n=3). d, CLV3p enhances resistance to Pst DC3000. Error bars, s.d. (n=3).
FLS2 signalling requires the recruitment of RLK BAK1 and the interaction between FLS2 and BAK1 represents the earliest event (within a minute) triggered by flg22 binding to FLS26,14–16. Like flg22, CLV3p also induced the immediate interaction between FLS2 and BAK1 detected by reciprocal co-immunoprecipitation (co-IP) (Supplementary Fig. 5a, b)14–16. Consistently, CLV3p signalling monitored by MAPK activation was greatly diminished in the bak1-4 mutant (Supplementary Fig. 6)14–16. Significantly, CLV3p pretreatment could confer enhanced resistance to the pathogenic bacteria Pseudomonas syringae pv. tomato (Pst) DC3000 in an FLS2 dependent manner (Fig. 1d)10,14. These comprehensive analyses strongly support our new finding that the conserved flagellin receptor FLS2 can recognize the stem cell peptide CLV3p and trigger the convergent innate immune signalling6,9,14.
Peptide titration experiments using the FLS2 and BAK1 co-IP assay showed that 1 nM flg22 was as potent as 1 μM CLV3p (Fig. 2a and Supplementary Fig. 5c) required for SAM suppression13,21. Intriguingly, flg22 and CLV3p peptides supported similar primary gene activation but distinct long-term growth effects in seedling assays (Figs 3 and 4 and Supplementary Fig. 7). Flg22 is a MAMP from non-self invaders and detected by a very sensitive perception system22 to induce innate immunity in a timely manner after infection. CLV3p, on the other hand, is an endogenous plant signal naturally secreted in the stem cell zone to activate constitutive innate immunity via FLS2 in the SAM, which might provide a type of “vaccination” before any infections to elicit sufficient immune protection without severe growth penalty caused by MAMPs (Fig. 3a–e)5,6,15,20.
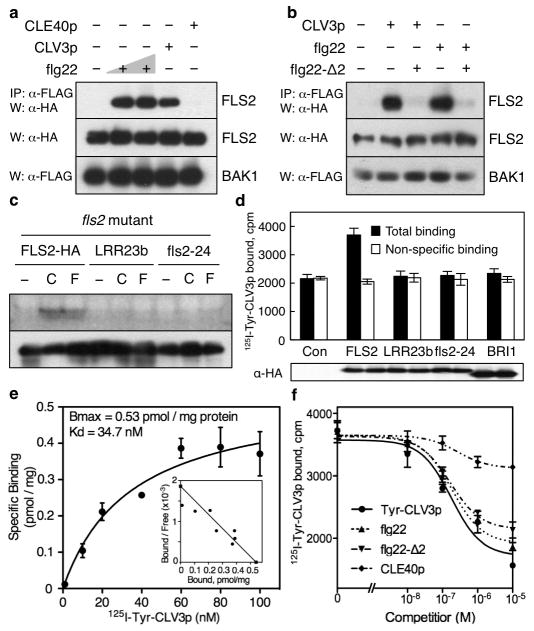
Figure 2. CLV3p and flg22 share similar perception through FLS2.
a, CLE40p fails to induce FLS2-BAK1 interaction. Treatments: flg22 (1, 10 nM), 1 μM CLV3p or 1 μM CLE40p for 10 min. b, Flg22-Δ2 blocks CLV3p and flg22 signalling. Flg22-Δ2 (50 μM). c, LRR mutations of FLS2 eliminate flg22 and CLV3p perceptions in fls2. Top, in-gel-kinase assay. Bottom, protein expression. d, 125I-Tyr-CLV3p specifically binds to FLS2. Non-specific binding (20 μM unlabeled Tyr-CLV3p). Error bars, s.d. (n=3). e, Saturation binding curve and Scatchard plot. Error bars, s.d. (n=3). f, Binding competition analysis with peptides. Error bars, s.d. (n=3). Non-specific binding (10 μM unlabeled Tyr-CLV3p).
Figure 3. CLV3p-mediated SAM arrest and immune signalling are uncoupled from flg22-triggered growth suppression.
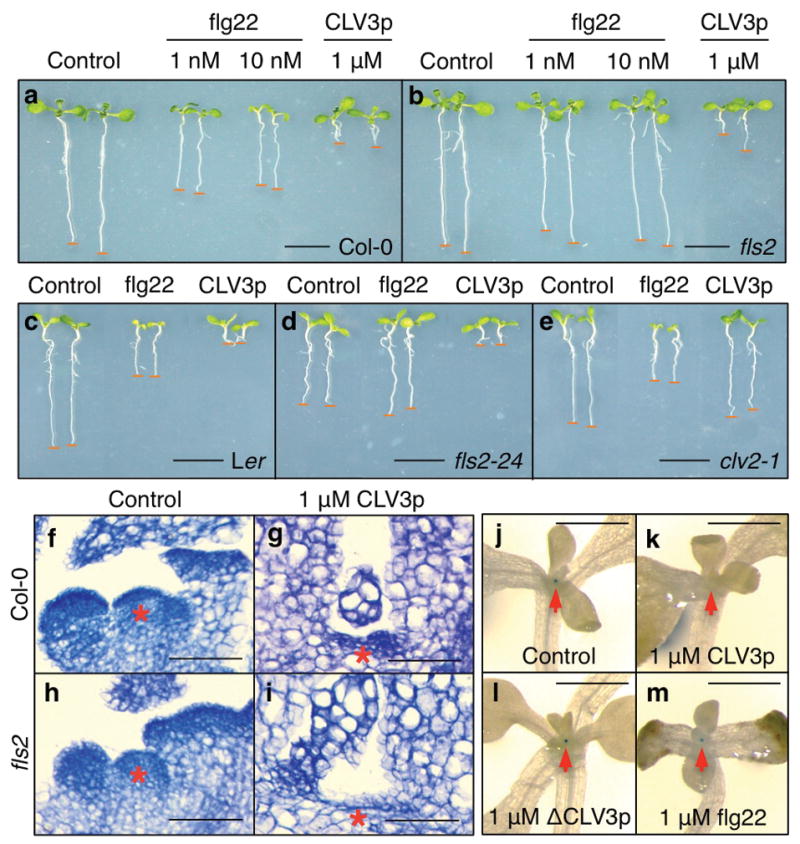
a-e, Growth inhibition analysis. Treatment with flg22 (1, 10 nM) or CLV3p (1 μM) in Col-0 (a) and fls2 (b), Ler (c), fls2-24 (d) and clv2-1 (e) seedlings. Scale bars, 1 cm. f–i, CLV3p suppresses the SAM in Col-0 and fls2 plants. Red asterisks, SAM region. Scale bars, 50 μm (f–i). j–m, CLV3p represses pWUS::GUS expression. Treatment was without (j) or with 1 μM CLV3p (k), 1 μM ΔCLV3p (l) or 1 μM flg22 (m). Red arrows, SAM region. Scale bars, 1 mm.
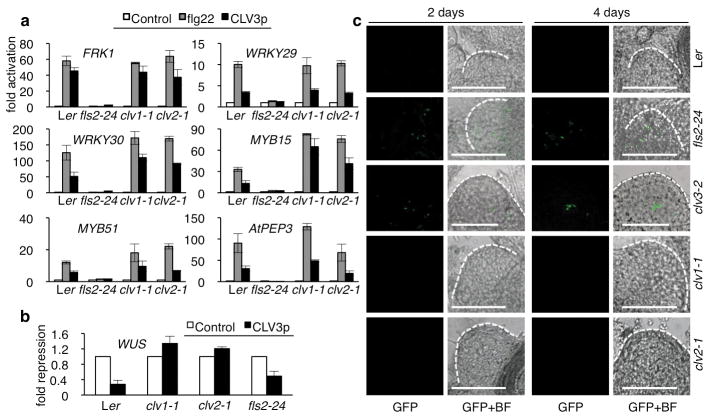
Figure 4. CLV3p-FLS2 signalling enhances innate immunity for SAM protection.
a, Flg22 marker gene activation by CLV3p through FLS2 in the SAM. b, WUS repression by CLV3p was mediated via CLV1 or CLV2 not FLS2. The SAM tissues were analyzed by qRT-PCR after treatment with 1 nM flg22 or 1 μM CLV3p for 1 h. Error bars, s.d. (n=3). c, Infection and proliferation of pathogenic bacteria in the SAM lacking CLV3p-FLS2 signalling. Pst DC3000-GFP was co-cultivated with seedlings for 2 or 4 days. Pst DC3000-GFP was visualized using a confocal microscope. Scale bars, 50 μm. BF, bright field.
CLV3 belongs to the CLV3/ERS-related (CLE) gene family, which is conserved in diverse plant species4,17–19. The Arabidopsis genome has 32 CLE genes4,17–19. CLV3p and many other CLE peptides share similar root growth inhibition activity through the CLV2-CRN (CORYNE)/SOL2 (SUPPRESSOR OF OVEREXPRESSION OF LLP1-2) receptor complex, revealing a high degree of redundancy in peptide signalling18,19. To assess the specificity of CLV3p-FLS2 signalling, we examined the activity of synthetic peptide CLE40p, which belongs to the same CLE subgroup as CLV3p4,17,18,21. CLE40p triggered the growth arrest of SAM, repression of WUSCHEL (WUS) detected by pWUS::GUS23, and root growth inhibition (Supplementary Fig. 8a, b) as previously reported for CLV3p and CLE19p4,17,18,21. WUS encodes a homeo-domain transcription factor playing a central role in Arabidopsis SAM maintenance 1,2. However, CLE40p did not promote FLS2-BAK1 interaction or flg22 early marker gene activation (Fig. 2a and Supplementary Fig. 8c). CLV3p-FLS2 signalling might be unique among CLE peptides.
To determine whether flg22 and CLV3p triggered FLS2 signalling through the same or different extracellular sites, we carried out competition experiments using a well-established antagonist peptide lacking only the last 2 amino acids at the C-terminus of flg22 (flg22-Δ2)24. Based on the CLV3p- or flg22-triggered FLS2-BAK1 interaction assay by co-IP, we showed that flg22-Δ2 effectively competed with and blocked flg22 and CLV3p signalling (Fig. 2b). Moreover, two LRR mutants, one in the 10th LRR domain (fls2-24) and the other in the 23rd LRR domain (the LRR23b mutant) of FLS2, failed to activate flg22 and CLV3p signalling detected by MAPK activation despite normal FLS2 protein levels (Fig. 2c)9,14–16,24–26. These results suggest that flg22 and CLV3p likely share binding and activation sites in the extracellular LRR domain of FLS2.
Based on the established studies on the flg22-FLS2 and CLV3p-CLV1 interactions27,28, we developed a cell-based assay to show that 125I-Tyr-CLV3p interacted directly with FLS2 expressed in the null fls2 mutant protoplasts. Importantly, only the wild-type FLS2 protein but not the fls2-24 and LRR23b mutant protein or the brassinosteroid receptor kinase (BRI1) showed specific binding to 125I-Tyr-CLV3p (Fig. 2d). The saturation binding curve and Scatchard plot were generated using the specific binding assay. The estimated dissociation constant (Kd) for FLS2 and 125I-Tyr-CLV3p interaction was 34.7 nM (Fig. 2e), which was close to the Kd for CLV1 and CLV3p interaction28. The specific binding could be competed by the unlabeled Tyr-CLV3p, CLV3p, flg22, flg22-Δ2, but not CLE40 (Fig. 2f and Supplementary Fig. 9). Tyr-CLV3p and CLV3p displayed identical effectiveness in competing with 125I-Tyr-CLV3p for binding to FLS2 (Supplementary Fig. 9), immune marker gene activation and root inhibition (data not shown). 125I-Tyr-CLV3p binding to FLS2 shared characteristic of CLV3p binding to CLV128. Although the estimated Kd for 125I-Tyr-flg22 binding to FLS2 is lower27 and FLS2 responses to flg22 are more sensitive (Figs 1c, d and 2a and Supplementary Fig. 5a, c), flg22 did not compete more effectively than Tyr-CLV3p and CLV3p for 125I-Tyr-CLV3p binding to FLS2. Because the sequences of CLV3p and flg22 do not share any overt similarity and ΔCLV3p was ineffective in activating or blocking FLS2 signalling (Fig. 1a, data not shown), it was surprising to discover that they could still compete for some potentially shared binding sites on the LRR of FLS2 (Fig. 2b, c, f). The precise location of flg22 and CLV3p binding to FLS2 awaits future co-crystallographic analysis or mass spectrometry studies on cross-linked ligand-receptor complexes.
In previous studies, innate immune responses and growth inhibition are always tightly linked5,6,15,20. We investigated the effects of flg22 and CLV3p on seedling growth 5,6,15,20. Surprisingly, CLV3p did not inhibit shoot growth but caused stronger root growth arrest in Col-0 and Ler WT seedlings (Fig. 3a, c and Supplementary Figs 7a and 8b). While seedling growth inhibition in both shoots and roots by flg22 was eliminated in two independent fls2 mutants, the stronger root growth arrest by CLV3p was retained (Fig. 3b, d). Although CLV3p activated, via FLS2, a spectrum of innate immune responses similar to those via flg22-FLS2 signalling in mesophyll cells, seedlings and the SAM (Figs 1, 2 and 4 and Supplementary Figs 3, 5 and 8), CLV3p did not stimulate the typical flg22-FLS2-mediated growth suppression in whole seedlings (Fig. 3a–e). Similar to other synthetic CLE peptides, CLV3p triggered root growth arrest was abolished in the clv2-1 mutant (Fig. 3c–e and Supplementary Fig. 7)18,19. Thus, CLV3p-FLS2 signalling activated only immune responses but not the general growth inhibition. Significantly, CLV3p was very active in triggering the typical SAM arrest in null fls2 seedlings (Fig. 3f–i)1,2,4,13,17–19,23,28. The SAM growth arrest was correlated with the repression of a sensitive pWUS::GUS reporter in the organizing centre by the exogenous CLV3p but not ΔCLV3p or flg22 (Fig. 3j–m)23.
Because CLV3p is specifically expressed and secreted from stem cells of the SAM1,2,4,13,17,23, it is critical to examine CLV3p-FLS2 signalling in the SAM to evaluate its physiological relevance. Using qRT-PCR analysis with isolated SAM tissues, CLV3p clearly triggered two parallel signalling pathways in the SAM (Fig. 4a, b and Supplementary Figs 10–13). The activation of important immune marker genes by CLV3p in WT, clv1-1, clv2-1 but not fls2-24 validated the action of innate immune signalling via FLS2 in the Arabidopsis SAM. These flg22 and CLV3p inducible genes, including FRK1 (a RLK), WRKY29, WRKY30, MYB15, MYB51 (transcription factors) and PEP3 (a peptide inhibiting Pst DC3000 growth via RLK signalling), play important roles in bacterial and fungal resistance6,9,10,14. Consistently, some of these marker genes showed reduced endogenous expression in the SAM of clv3-2, but were constitutively expressed at higher levels in the SAM but not other tissues of WT (Supplementary Figs 10, 11a and 12). Their endogenous expression levels were low in both the SAM and other tissues in clv3-2 and fls2-24 (Supplementary Fig. 12). Complementation of clv3-2 for immune marker gene expression in the SAM could be achieved with nM range of exogenous CLV3p, reflecting the physiological relevance of the Kd for CLV3p and FLS2 interaction and CLV3p-FLS2 signalling in the SAM (Fig. 2e and Supplementary Fig. 10). The flg22 induction of these immune marker genes was observed in the SAM of clv1-1, clv2-1 and clv3-2 mutants, supporting the potency of MAMP signalling via FLS2 (Fig. 4a and Supplementary Fig. 11b). At 10–100 pM flg22, the immune marker gene induction levels were reduced in the SAM of clv3-2 compared to those in the SAM of WT and clv1-1, suggesting the operation of both CLV3p-FLS2 and flg22-FLS2 signalling pathways in the SAM (Supplementary Fig. 13). The repression of WUS was triggered by CLV3p only in the SAM of WT and fls2-24, but not clv1-1 and clv2-1 (Fig. 4b)1,2,4,17,19,23.
Most strikingly, we have never detected the presence of a single live Pst DC3000-GFP bacterium in the SAM of WT seedlings in 11 infection experiments by visualizing bacteria proliferation using confocal microscopy for up to 4 days (Fig. 4c and Supplementary Fig. 14a). Because Pst DC3000-GFP could be easily visualized in the infected WT and clv3-2 cotyledons (Supplementary Fig. 14b, d), the WT SAM appeared to exhibit differential immunity (Fig. 4c and Supplementary Fig. 14a). Significantly, the SAM was no longer protected from Pst DC3000-GFP infection in fls2-24 and clv3-2 mutants (Fig. 4c and Supplementary Fig. 14c, e, f), supporting the important role of the endogenous CLV3p in the SAM protection through the FLS2-mediated innate immune signalling pathway (Fig. 4c and Supplementary Figs 1 and 14). To further demonstrate and quantify the proliferation and growth of Pst DC3000-GFP in the SAM of the fls2-24 and clv3-2 mutants observed after 2 to 4 days of infection, we counted the increasing bacteria numbers and carried out quantitative PCR analysis of the GFP DNA from the bacteria, both indicating the loss of the distinct SAM immunity (Fig. 4c and Supplementary Figs 14e and 15). There were higher numbers of Pst DC3000-GFP bacteria in the bigger clv3-2 SAM with more cells of similar size but not bigger cells (Supplementary Fig. 14c, e). Importantly, Pst DC3000-GFP was found to be completely excluded from the similarly enlarged clv1-1 and clv2-1 SAM, in which the CLV3p-FLS2 signalling remained active (Fig. 4c). Based on qPCR analysis of GFP DNA derived only from Pst DC3000-GFP, non-specific bacteria attachment background could be estimated from the SAM tissue samples 1 h after bacteria-seedling co-cultivation. Consistent with confocal microscopic observations, active bacteria growth and proliferation were exclusively detected only in the SAM of the clv3-2 and fls2-24 mutants (Supplementary Fig. 15).
We have uncovered a surprising mechanism underlying the stem cell-triggered immunity for pathogenic bacteria protection through CLV3p-FLS2 signalling (Supplementary Fig. 1). It will be interesting to examine the SAM protection from a variety of other pathogens. We propose that CLV3p is recognized by two distinct types of receptors involved in mostly non-overlapping functions in the SAM (Supplementary Fig. 1). The “constitutive” immunity in the SAM resembles flg22 pretreatment as a type of vaccination (before infection), which is more effective to confer protection against virulent pathogens such as Pst DC3000 possessing effectors to cripple MAMP signalling6,14. It is amazing that CLV3p-FLS2 signalling seems to have evolved to provide constitutive immune protection in the SAM but avoid the penalty from potent growth suppression associated MAMP signalling5,6,15,20. A future challenge is to elucidate the precise differential downstream signalling events via the same receptor in response to different peptide ligands. Lacking the genes for antibodies and immune cell receptors in humans29 to respond to new signals from diverse invaders, plant RLKs, displaying high polymorphism and fast evolution30, may provide an alternative means to recognize self or nonself in a beneficial manner through constant selections in evolution. It will also be important to explore the roles of other secreted plant peptides and known or orphan receptors in innate immunity4,6,17,30.
METHODS SUMMARY
Plasmid constructs
The constructs for expressing FLS2-HA and BAK1-FLAG for co-IP were reported previously14. LRR23b-HA and fls2-24-HA are mutant variants of the LRR domain of FLS2, which were generated by site-directed mutagenesis. The fls2-24 allele is a Gly to Arg mutation in the 10th LRR domain of FLS2 and insensitive to flg2225. The LRR23b is mutated in two amino acids (Gln to Leu and Phe to Leu) in the 23rd LRR domain of FLS2 and lacks flg22 signalling26.
Mesophyll protoplas transient assays
Protoplast isolation and transient expression assays were performed as previously described9,14. For protein expression in co-IP and in-gel kinase assays, protoplasts were transfected with plasmid DNA and incubated for 6 h at room temperature. Then, peptides such as CLV3p and flg22 were added for 10 min to induce MAPK activation or FLS2-BAK1 interaction. For qRT-PCR analysis, protoplasts were treated with peptides for 1 h.
In-gel kinase assay and co-IP
Both experiments were performed according to previously described9,14. MBP (Invitrogen) was used as a substrate for endogenous MPK3 and MPK6 activation analysis9. For co-IP, FLS2-HA and BAK1-FLAG were co-immunoprecipitated by an anti-FLAG antibody (Sigma) and detected by an anti-HA antibody (Roche) in immunoblot analysis.
METHODS
Plant materials and growth conditions
Col-0 and Ler were used as wild type Arabidopsis plants in this study. The bak1-4, efr-1, and fls2 (Salk_141277) mutants are in Col-0 background14,15,20 and clv1-1, clv2-1, clv3-2, and fls2-24 are in Ler background5,31,32. The clv1-1 mutation is in the kinase domain and represents a dominant negative allele33. The mutation of clv2-1 causes the early stop codon at the 33rd residue and is a null allele34. The γ-ray-induced clv3-2 is a presumed null allele33,35. Wild type and mutant plants were grown on soil at 23°C, 65% humidity, and 75 μmol m−2 s−1 light intensity under 12 h light/12 h dark photoperiod condition for 4 weeks before mesophyll protoplast isolation36. For liquid culture of Arabidopsis seedlings, seeds were germinated and grew in 6-well plates containing 1 ml of liquid medium (0.5 × MS and 0.5% sucrose, pH 5.8 adjusted with KOH). For GUS assay in the SAM and qRT-PCR analysis using SAM tissues, seedlings were grown for 8 days. For growth inhibition analysis by flg22 and CLV3p, seedlings were grown at same condition for 8 days (Fig. 3a, b and Supplementary Fig. 8b) or 6 days (Fig. 3c–e and Supplementary Fig. 7b).
QRT-PCR analysis
Total RNA was isolated from protoplasts or SAM tissues with TRIzol reagent (Invitrogen). To harvest SAM tissues, seedlings were instantaneously frozen by liquid nitrogen in the mortar pre-chilled on the dry ice. Cotyledons, hypocotyls and roots were removed using fine forceps. The purity of the harvested SAM tissue was confirmed by SAM specific marker genes and the absence of marker genes not expressed in the SAM (Supplementary Fig. 2)12. First strand cDNA was synthesized from 1 μg of total RNA with M-MLV reverse transcriptase (Promega). All qRT-PCR analyses were performed by CFX96 real time PCR detection system with iQ SYBR green supermix (Bio-Rad). ACT2 (ACTIN2, At3g18780) was used as a control gene.
GUS staining
GUS staining was performed as described23. Plants were fixed in 90% cold acetone for 20 min and rinsed twice in staining buffer without X-Gluc (5-Bromo-4-chloro-3-indoxyl-beta-D-glucuronide). Samples were infiltrated with staining buffer [100 mM NaPO4 buffer, pH 7.0; 0.5 mM Ferrocyanide; 0.5 mM Ferricyanide; 0.1% Triton X-100; 10 mM EDTA, pH 8.0; 1 mM X-Gluc (Gold Biotechnology, Inc)] under vacuum for 10 min and incubated at 37°C for 3 h. After then, staining buffer was removed and dehydrated up to 70% ethanol. Microscopic analysis was carried out with Leica DFC 500 camera mounted on Leica MZ16F.
Bacterial infection assay in the SAM
Nine seeds were sowed in 1 ml liquid medium in 6-well plates and grown under constant light (50–65 μmol m−2 s−1) at 25–27°C without shaking. Pst DC3000-GFP culture was grown in KB liquid medium (50 μg/ml of rifampicin and 15 μg/ml of Tetracyclin) with shaking at 28°C. Overnight cultured Pst DC3000-GFP were washed twice with water and diluted to OD600=0.02. Diluted Pst DC3000-GFP (50 μl) was added into the liquid medium with 2 day-old seedlings. Plants and bacteria were co-cultivated with gentle shaking (50 rpm) for 2, 3 or 4 days under constant light. For observation of bacteria infected SAM, co-cultivated seedlings were washed with 70% ethanol twice and rinsed twice with water. All seedlings were placed on glasse slides with 1 ml of water, and squashed gently by cover slips for bacterial observation using a confocal laser-scanning microscope (Leica TCS-NT). The experiment was repeated at least five times with similar results in Ler, fls2-24, clv3-2, clv1-1 and clv2-1 (Fig. 4c). For the qPCR analysis of DC3000-GFP, diluted Pst Dc3000-GFP (OD600 = 0.5, 200 μl) was co-cultivated with WT or mutant seedlings for 3 or 4 days. After washing and rinsing seedlings twice, tissues from five SAMs were harvested and ground in 100 μl of water. Control experiments were conducted at 0 DAC (day after co-cultivation) to determine non-specific bacterial attachment 1 h after co-cultivation (black bar). The experiment was repeated three times with similar results. The GFP level determined by qPCR was correlated with specific bacterial growth, and normalized based on the Arabidopsis ACT2 gene in the SAM tissues.
Seedling pathogen assay
The protocol was modified from previously described37,38. Nine seeds were sowed in 6-well plates containing 1 ml of liquid medium. Plants were grown under constant light at 25–27°C without shaking. Pst DC3000 culture were grown in KB liquid medium (50 μg/ml of rifampicin) with shaking at 28°C. Overnight cultured Pst DC3000 was washed twice with water and diluted to OD600=0.02, 1 × 107 cfu/ml. After 6 days of seedling growth, 50 μl of diluted Pst DC3000 was added into 1 ml of fresh 0.5 × MS liquid medium without sucrose. After adding Pst DC3000, plates containing plants and bacteria were co-cultivated with gentle shaking (<50 rpm) for 1 day under constant light (50–65 μmol m−2 s−1). For CLV3p- and flg22-induced immunity, seedlings were treated with 1 μM or 10 μM of CLV3p or 100 nM of flg22 peptide 1 day before adding Pst DC3000. For bacterial counting, co-cultivated seedlings were washed with 70% ethanol twice and rinsed twice with water. Then, three seedlings were put into each of three 1.5 ml tubes containing 100 μl of water and ground by a hand drill and blue pestles. After preparing serial dilutions, 10 μl of diluted bacteria (from 10−3 to 10−5) was spread on the KB plates and incubated for 2 days at 28°C before counting.
Assay for CLV3p-mediated SAM arrest
Twenty seeds of Col-0 and fls2 mutant were sowed in petri dishes (100 × 25 mm) containing 10 ml of liquid medium with or without 1 μM CLV3p. Plants were grown in a growth chamber at 23°C under short day condition (8 h light:16 h dark, 75 μmol m−2 s−1) for 4 weeks. The SAM tissues were cut and fixed using 4% (w/v) paraformaldehyde/4% (v/v) DMSO at 4°C for overnight. Collected samples were dehydrated through ethanol series (30%, 50%, 70%, 95% for 1 h in each step) at 4°C and stained with 0.1% Eosin Y (Sigma) in 100% ethanol at 4°C for overnight. Ethanol was changed through histoclear series (50% ethanol: 50% histoclear, 100% histoclear, 100% histoclear for 1 h in each step) (National Diagnostics). Histoclear was then gradually changed with melted paraffin (Fisher Scientific) in a 60°C chamber. Replacement of freshly melted paraffin was performed for 4 days. Paraffin embedded tissues were poured into the mold and adjusted in appropriated position. Section was carried out with a rotary microtome (Leica RM2255) at 8 μm thickness. Sectioned ribbons were placed on poly-prep glass slides (Sigma) with pre-warm water and incubated on slide warmer (Fisher Scientific) at 42°C for overnight. For meristem staining, paraffin of sections was removed by 100% histoclear twice for 10 min. Sectioned tissues were hydrated through reverse ethanol series (100%, 70%, 30% ethanol and water for 2 min in each step). Sections were stained with 0.1% Giemsa (Sigma) for 5 min and rinsed briefly with water. Stained sections were dehydrated through ethanol series (2 min in each step) and transferred to 100% histoclear for 2 min. For microscopic analysis, samples were dried and mounted with Cytoseal 60 (Richard-Allan Scientific) before taking pictures using Leica DM5000B.
Direct Binding Assay
The protocol was modified from previously described39–41. For reparation of receptor proteins, FLS2-HA, fls2-24-HA, LRR23b-HA and BRI1-HA were expressed in fls2 protoplasts (2.5 × 105 cells) for 6 h. Protoplasts were harvested and then resuspended in 500 μl of binding buffer (25 mM MES/KOH, pH 5.8, 3 mM MgCl2, 10 mM NaCl) with 2 mM DTT and protease inhibitor (Roche). Cells were vigorously mixed by vortex and kept on ice for 5 min, and centrifuged at 10,000 ×g for 20 min to yield pellet. The pellets were resuspended in 100 μl of binding buffer and the protein concentration from resuspended extract was measured by a NanoDrop 1000 spectrophotometer. This extract contained 50 μg of total protein. For the binding assay, resuspended extracts (100 μl) in binding buffer were mixed with 125I-Tyr-CLV3p (100 fmol in each sample) without or with unlabeled Tyr-CLV3p as a competitor for 30 min on ice. The average of specific radioactivity in five different batches of 125I-Tyr-CLV3p was 2,023.52 Ci/mmole. 125I-Tyr-CLV3p bound extracts were collected by a vacuum filtration system through glass fiber filters (Macherey-Nagel MN GF-2, 2.5-cm diameter), which were pre-incubated with 1% BSA, 1% bactotrypton, 1% bactopepton in binding buffer. Filters were washed with 15 ml of cold binding buffer and the retained radioactivity was determined by a gamma counter Beckman LS6500. Specific binding was measured by subtracting non-specific binding (with 10–20 μM unlabeled Tyr-CLV3p competitor) from total binding (without competitor). In the saturation binding assay, the non-specific binding in the presence of 20 μM unlabeled Tyr-CLV3p competitor showed linear increase with 1 to 100 nM 125I-Tyr-CLV3p and accounted for 40 ~ 60 % of total binding, which was similar to the range of non-specific binding observed with 125I-Tyr-flg22 binding to intact cells (Fig. 3A, Bauer et al., 2001). The dissociation constant (Kd), Scatchard plot and the non-linear regression analysis of competition assay were presented using the Prism 5 program (GraphPad). From the saturation data of Bmax = 0.53 pmol/mg protein and 50 ug protein from 2.5 × 105 cells, 6.36 × 104 125I-Tyr-CLV3p binding sites per cell could be estimated in total cell extracts. Specific CLV3p binding to FLS2 was similar to CLV3p binding to CLV1 and CLV2, the established CLV3p receptors40–42.
Supplementary Material
Acknowledgments
We thank T. Laux for the pWUS::GUS line, Y. Matsubayashi for the purified Ara-CLV3 peptide and generous and helpful advice, A. Collmer for the GFP-labeled Pst DC3000, L. Shan and P. He for constructs, G. Tena, F. Ausubel, J. Bush, J. Plotnikova and ABRC for mutant seeds and bacterial strains, and Y. Xiong, M. McCormack, Y. Niu, M. Ramon and J. Li for critically reading of the manuscript. Funding was provided by the NSF, NIH and the MGH CCIB fund to J.S.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.L. and J.S. initiated the project and designed the experiments; H.L. carried out the experiments and prepared the data with assistance from O.-K.C.; H.L and J.S. wrote the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Bäurle I, Laux T. Apical meristems: the plant’s fountain of youth. BioEssays. 2003;25:961–970. doi: 10.1002/bies.10341. [DOI] [PubMed] [Google Scholar]
- 2.Scheres B. Stem cells: a plant biology perspective. Cell. 2005;122:499–504. doi: 10.1016/j.cell.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Hollings M. Disease control through virus-free stock. Annu Rev Phytopath. 1965;3:367–396. [Google Scholar]
- 4.Jun JH, Fiume E, Fletcher JC. The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci. 2008;65:743–755. doi: 10.1007/s00018-007-7411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Gomez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 6.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 7.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: Making sense of microbial infections. Cell Host & Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asai T, et al. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 10.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 11.Yamamizo C, et al. Rewiring mitogen-activated protein kinase cascade by positive feedback confers potato blight resistance. Plant Physiol. 2006;140:681–692. doi: 10.1104/pp.105.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. PNAS. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo T, et al. A plant peptide encoded by CLV3 identified by in situ MALDI- TPF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 14.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell & Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defense. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 16.Heese A, et al. The receptor-like kinase SEERK3/BAK1 is a central regulator of innate immunity in plants. PNAS. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. Plant peptides in signaling: looking for new partners. TIPS. 2009;14:255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Miwa H, et al. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 2008;49:1752–1757. doi: 10.1093/pcp/pcn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;19:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 21.Fiers M, et al. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 2006;141:1284–1292. doi: 10.1104/pp.106.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 23.Bäurle I, Laux T. Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell. 2005;17:2271–2280. doi: 10.1105/tpc.105.032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinchila D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 ad determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Gómez L, Bauer Z, Boller T. Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell. 2001;13:1155–1163. [PMC free article] [PubMed] [Google Scholar]
- 26.Dunning FM, Sun W, Jansen KL, Helft L, Bent AF. Identification and mutational analysis of Arabidopsis FLS2 leucin-rich repeat domain residues that contribute to flagellin perception. Plant Cell. 2007;19:3297–3313. doi: 10.1105/tpc.106.048801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer Z, Gomez-Gomez L, Boller T, Felix G. Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. JBC. 2001;276:45669–45676. doi: 10.1074/jbc.M102390200. [DOI] [PubMed] [Google Scholar]
- 28.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 29.Goldsby RA, Kindt TJ, Osborne BA. KUBY IMMUNOLOGY. 6. W. H. FREEMAN & Co; New York: 2006. [Google Scholar]
- 30.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 31.Koornneef M, et al. Linkage map of Arabidopsis thaliana. J Hered. 1983;74:265–272. [Google Scholar]
- 32.Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- 33.Diévart A, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15:1198–1211. doi: 10.1105/tpc.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1933. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 36.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 37.Schreiber K, Ckurshumova W, Peek J, Desveaux D. A high-throughput chemical screen for resistance to Pseudomonas syringae in Arabidopsis. Plant J. 2008;54:522–531. doi: 10.1111/j.1365-313X.2008.03425.x. [DOI] [PubMed] [Google Scholar]
- 38.Boudsocq M, et al. Differential innate immune signaling via Ca2+ sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer Z, Gomez-Gomez L, Boller T, Felix G. Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. JBC. 2001;276:45669–45676. doi: 10.1074/jbc.M102390200. [DOI] [PubMed] [Google Scholar]
- 40.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 41.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.