Abstract
The hyperparathyroidism-jaw tumor syndrome (HPT-JT) is a familial cancer syndrome that can result from germline inactivation of HRPT2/CDC73, a putative tumor suppressor gene that encodes parafibromin, a component of the transcriptional regulatory PAF1 complex with homology to the yeast protein Cdc73p. The vast majority of HRPT2/CDC73 germline mutations identified have been truncation or frameshift mutations, and loss-of-function due to missense mutation is rare. We report here a kindred with HPT-JT due to a germline L95P missense mutation in parafibromin. The mutant parafibromin was studied in vitro to understand the basis of its presumed loss-of-function. When transfected in cultured cells the L95P mutant was expressed to a lower level than wild-type parafibromin, a difference that was not overcome by inhibition of the proteasome degradation pathway. The L95P mutant parafibromin retained the ability to assemble with endogenous PAF1 complex components as evidenced by co-immunoprecipitation. Analysis of subcellular localization showed that the L95P mutant was markedly deficient in nucleolar localization compared to the wild-type, an impairment likely resulting from disruption of a putative nucleolar localization signal immediately upstream of the L95P mutation. Transfection of the L95P parafibromin mutant, but not the wild type, enhanced cell-cycle progression and increased cell survival in NIH-3T3 and HEK 293 cells, resulting apparently from dominant interference with endogenous parafibromin action. The simultaneous loss of nucleolar localization and acquisition of a growth stimulatory phenotype with the L95P mutation raise the possibility that parafibromin must interact with targets in the nucleolus to fully execute its tumor suppressor functions.
Keywords: CDC73, HRPT2, tumor suppressor, nucleolus, subcellular localization
INTRODUCTION
Germline mutation of the tumor suppressor gene HRPT2/CDC73 confers susceptibility to the hyperparathyroidism-jaw tumor syndrome (HPT-JT), an autosomal dominant syndrome whose major features are primary hyperparathyroidism (90%) including 15% of all affected by HPT-JT with parathyroid cancer, fibro-osseous tumors of the maxilla or mandible (30%), bilateral renal cysts (10%), and uterine tumors (Bradley, et al. 2005; Jackson, et al. 1990; Mallette, et al. 1987; Simonds, et al. 2002; Simonds, et al. 2004; Teh, et al. 1996; Teh, et al. 1998). The identification of HRPT2/CDC73 resulted from positional candidate cloning (Carpten, et al. 2002). Besides HPT-JT, germline inactivating HRPT2/CDC73 mutation has also been reported in a minority of kindreds with familial isolated hyperparathyroidism (FIHP) (Bradley, et al. 2006; Carpten et al. 2002; Guarnieri, et al. 2006; Howell, et al. 2003; Kelly, et al. 2006; Mizusawa, et al. 2006; Simonds et al. 2004; Villablanca, et al. 2004) and in up to 30% of patients with apparently sporadic parathyroid cancer (Cetani, et al. 2004; Shattuck, et al. 2003). HRPT2/CDC73 encodes parafibromin, a 531-amino acid putative tumor suppressor protein. The C-terminal region of parafibromin demonstrates sequence homology to Cdc73p, a budding yeast protein. Just as Cdc73p associates with the RNA polymerase II-associated Paf1 complex in yeast (Chang, et al. 1999; Shi, et al. 1997), mammalian parafibromin interacts with RNA polymerase II via a PAF1 complex, whose other protein components include Paf1, CTR9, Leo1 (Rozenblatt-Rosen, et al. 2005; Yart, et al. 2005; Zhu, et al. 2005), and the WD40-repeat protein Ski8 (Zhu et al. 2005).
The vast majority of clinically identified HRPT2/CDC73 germline loss-of-function mutations reported to date have been truncation or frameshift mutations. In the present study we describe a kindred with HPT-JT due to a germline L95P missense mutation in parafibromin. The mutant parafibromin was studied in vitro to better understand the basis of its impaired function. We report here that the L95P mutant retained the ability to interact with PAF1 complex components, but was expressed at a lower level than wild-type parafibromin and was deficient in nucleolar targeting. Furthermore transfection of the parafibromin L95P missense mutant promoted cell survival and enhanced cell cycle progression, properties presumed to reflect dominant interference with tumor suppressor functions of endogenous parafibromin. These results suggest nucleolar localization might be a sine qua non for the full anti-proliferative activity of parafibromin.
CASE REPORTS
Index case
The index patient (III-1, Fig. 1A) had incidental hypercalcemia discovered by routine blood testing at age 25 and primary hyperparathyroidism documented with an elevated intact PTH value of 135 pg/ml (normal 7 –82) with a concurrent serum calcium of 11.1 mg/dl (normal 8.6 –10.0) and hypercalciuria with 537 mg urinary calcium/ 24 hours (normal 100 –300). At cervical exploration an enlarged right inferior parathyroid adenoma (1.2 cm diameter) was excised that was hypercellular on pathologic examination, and right and left superior parathyroid glands were biopsied that were normocellular. Post-operatively the patient was rendered normocalcemic and has remained so for 3 years of follow-up. At age 18 and again at age 25 the patient had jaw tumors removed by an oral surgeon with no operative or pathological reports available.
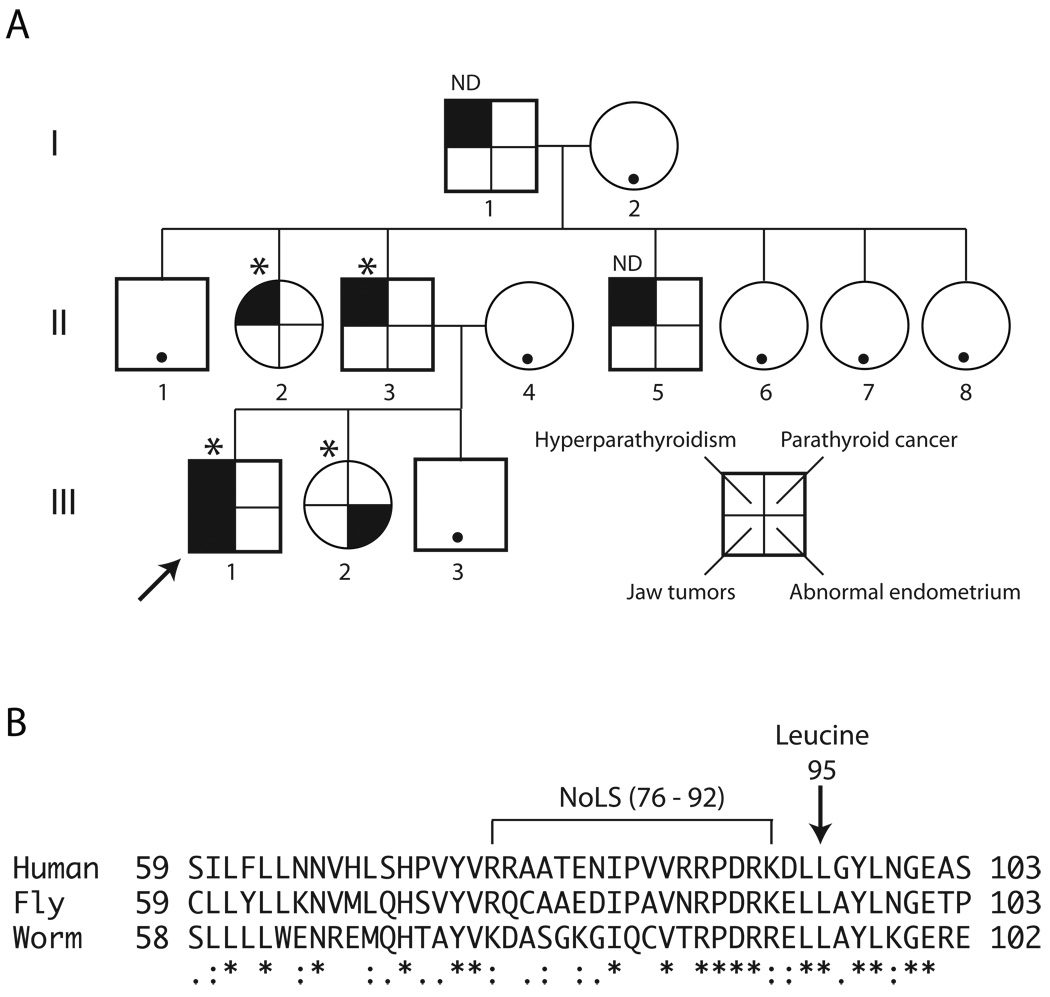
Figure 1. Kindred with the hyperparathyroidism-jaw tumor syndrome associated with germline HRPT2/CDC73 L95P missense mutation.
A. Pedigree of study kindred with the hyperparathyroidism-jaw tumor syndrome including three generations of affecteds. Square symbols indicate males, and round symbols indicate females, with legend to phenotypes shown in lower right. The arrow indicates the proband (III-1). The asterisk above a symbol indicates positivity for germline HRPT2/CDC73 L95P missense mutation testing, and ND above a symbol indicates gene testing not yet performed. A black dot within an otherwise blank symbol indicates that the serum calcium was normal at the latest screening in an individual never known to have hypercalcemia. B. Sequence alignment of human parafibromin protein residues 59–103 with the corresponding regions of predicted homologs from Drosophila melanogaster (Hyrax, CG11990) and the nematode Brugia malayi (NCBI Access. No. XP_001895980). The position of the conserved leucine at residue 95 in human parafibromin is indicated (arrow), as is the position of a nucleolar localization signal (NoLS) encompassing residues 76 –92 previously described (Hahn and Marsh 2007). Multiple sequence alignment was generated with ClustalW2 software (v. 2.0.12) available at the European Bioinformatics Institute website (http://www.ebi.ac.uk/Tools/clustalw2/), using the default parameters (Larkin, et al. 2007). “*”, residues in the column are identical in all sequences in the alignment, “:”, conserved substitutions observed in the alignment; “.”, semiconserved substitutions observed in the alignment.
Other cases
The proband’s younger sister (III-2) is normocalcemic but with irregular menses associated with grossly abnormal thickened and irregular endometrium by vaginal ultrasound. She has not had jaw or parathyroid tumors. The proband’s father (II-3) presented at age 34 with an obstructing calcium oxalate stone in the distal ureter associated with hypercalcemia (14 mg/dl), hypophosphatemia (1.9 mg/dl), and elevated parathyroid hormone (204 pg/ml, normal 18–120). An enlarged cystic left inferior parathyroid adenoma (1.5 cm diameter) was excised at cervical exploration with no other parathyroid tissue identified. The patient’s calcium and phosphorus normalized postoperatively. The patient has remained normocalcemic during 16 years of follow-up although he has experienced recurrent nephrolithiasis for the last three years. He has not had jaw tumors. The proband’s aunt (II-2) had hypercalcemia discovered on an annual screening blood test at age 42 and underwent cervical exploration for primary hyperparathyroidism at which time enlarged left and right inferior parathyroid adenomas (both 1.5 cm diameter) were excised. The left upper gland was identified and proven normal on biopsy. The patient has since remained eucalcemic for 11 years. She has not had jaw tumors. The proband’s uncle (II-5) and grandfather (I-1) have also required surgery for primary hyperparathyroidism but operative and pathology records are not currently available. None has reported jaw tumors.
MATERIALS AND METHODS
HRPT2 mutational analysis
Patient genomic DNA was extracted from whole blood samples collected in EDTA tubes with informed consent approved by the NIDDK institutional review board and analyzed for HRPT2 mutation by GeneDx (Gaithersburg, MD).
Mammalian cDNA expression constructs
Expression constructs containing complementary DNAs encoding AU5-epitope tagged human parafibromin in pcDNA3 (Invitrogen) (Woodard, et al. 2005), and enhanced green fluorescent protein fused in-frame at its C-terminus with human parafibromin in pEGFP-C1 (Clontech) (Lin, et al. 2007), were previously described. The corresponding L95P missense mutant cDNAs were prepared using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene). The coding region of the mutant cDNAs was confirmed by DNA sequencing.
Antibodies
Antibodies used for immunoblots and/or immunoprecipitation were rabbit anti-human parafibromin antibody GRAPE (Lin et al. 2007), mouse anti-AU5 monoclonal (MMS-135R, Covance Research Products, Denver, PA), mouse anti-β-actin monoclonal (A5316, Sigma, St. Louis, MO), rabbit polyclonal anti-Leo1 (A300-175A, Bethyl Laboratories, Inc., Montgomery, TX), rabbit polyclonal anti-Paf1 (A300-172A, Bethyl Labs), and mouse monoclonal anti-fibrillarin antibody (ab18380, Abcam). Secondary antibodies utilized in immunoblots or immunocytofluorescence were Cy3-conjugated donkey anti-mouse IgG (715-165-150, Jackson ImmunoResearch Labs, West Grove, PA) and IR secondary antibodies (anti-rabbit IR 800 and anti-mouse Red and Green) from LI-COR Bioscience.
Cell Culture
Human cervical cancer-derived HeLa cells (Cat. No. CCL-2, ATCC, Manassas, VA), human embryonic kidney HEK 293 cells (Cat. No. CRL-1573, ATCC) and mouse NIH-3T3 cells (Cat. No. CRL-1658, ATCC) were grown in 75 cm2 flasks in DMEM supplemented with 10% fetal bovine serum, 4mM L-glutamine and penicillin/streptomycin at 37° C and 5 % CO2. Empty vector or expression plasmids were transfected using Lipofectamine 2000 (Invitrogen) or PolyJet transfection reagent (SL 100688, SignaGen Laboratories). The expression of the transfected proteins was verified by immunoblotting and/ or immunofluorescence.
Immunoprecipitation
Immunoprecipitation with anti-AU5 antibody was performed according to the manufacturer’s instructions (Immunoprecipitation Starter Pack, cat. No. 309410, Amersham Biosciences). Briefly, after transfection and 24 h incubation, HeLa cells were lysed with buffer A (250 mM NaCl, 50 mM Tris, pH 8.0, 5 mM EDTA, 0.5 % NP-40 and 1 X complete protease inhibitor cocktail [cat. No. 1836170, Roche]). The lysate was centrifuged at 1500 X g for 10 min. The supernatant was incubated overnight with AU5 antibody at 4° C. The mixture was incubated with protein G-Sepharose previously equilibrated in buffer A for 2 hours at 4° C. The beads were then washed 3 times with buffer A by centrifugation at for 1 min, and the washes discarded. Proteins were eluted from the beads with SDS sample buffer and detected by immunoblotting.
Gel electrophoresis, immunoblotting, chemiluminescence and infrared imaging
Cell lysates were boiled with equal volume of Laemmli’s 2X gel loading buffer and the hot solution was loaded onto 8% Tris-Glycine SDS-PAGE gels (Invitrogen) to separate the proteins, followed by transfer of the proteins on to 0.45-micron nitrocellulose membrane. Membranes were blocked with TBS or PBS (pH 7.4) containing 0.1% Tween 20 and 5% nonfat dry milk (blocking buffer) and incubated overnight with primary antibodies in the same buffer. The membranes were then washed seven times for 5 minutes each with the above buffer without milk, followed by a 1 hour incubation in blocking buffer including IR-labeled secondary antibodies (dilution 1: 20,000) protected from light. Then the membranes were washed 4 times and scanned for detecting the protein signals using the Odyssey infrared imaging system (LI-COR, Bioscience). For the quantification of the intensity of the protein bands membranes were dually probed, with the β-actin signal used as a loading control.
Protein decay rate analysis
HeLa cells were transfected with empty pcDNA3 vector, or cDNAs encoding AU5-tagged wild-type or L95P mutant parafibromin. At 6h after transfection, cells were treated with cycloheximide (Sigma) at a final concentration of 50µg/ml. At the indicated time points after adding cycloheximide, cells were collected, and whole cell lysates were subjected to immunoblotting using GRAPE anti-parafibromin or anti-β-actin antibodies, with quantification by Odyssey infrared imaging as described above. Linear regression analysis of the natural logarithm of the relative actin-normalized parafibromin protein band intensity versus time was performed using the Linear Regression analysis program of Prism software version 5.0b for Macintosh OSX (GraphPad Software, San Diego, CA).
Calculation of parafibromin half-life
Calculation of the parafibromin half-life was performed as previously described for c-myc (Lin, et al. 2008). In brief, according to the linear regression analysis described above, the y-value at time=0 for all plots was 4.61 (the natural log of 100[%] relative expression), and a y-value of 3.91 (indicating a relative expression of 50[%]) maps to the time corresponding to the parafibromin protein half-life. The Prism software program calculated the slope (m) ± SEM, y-intercept (b) ± SEM, r2 value, and x-value at y=3.91 (half-life) of the best-fit lines as part of the linear regression analysis. In order to calculate the standard error of the half-life, the relation y =mx+b was first re-ordered as x= (y−[b ±SEM])/m ±SEM, where x = the half-life, y=3.91 (the natural log of 50(%)), b= the mean y-intercept, and m= the mean slope of the best-fit line. The standard error of the half-life (SEMx) under each condition was then estimated using the following equation, assuming b and m to be Gaussian variables: SEMx= ([3.91−b]/m)*√([SEMb]2/b2 + [SEMm]2/m2) (Motulsky 1995). Student’s unpaired t test was used to evaluate for statistical significance.
Immunofluorescence
HeLa cells were transfected as described above and plated in chambers of poly-D-lysine-coated chamber slides and incubated for 6h. The culture medium was discarded and the cells were rinsed with PBS and fixed in 2% formaldehyde for 15 min. The slides were washed two times with PBS and incubated with PBS containing 8% FBS/ azide (buffer A) for 5 min at room temperature. Then primary antibody was diluted in buffer A containing 0.2% saponin (buffer B) and incubated at room temperature for 1h. The slides were rinsed 3 times with buffer A and incubated with the secondary antibody (fluorescein Anti-rabbit or Anti-mouse IgG (H+L), Vector Laboratories, Burlingame, CA) diluted in buffer B and incubated for 1h at room temperature protected from light. The slides were washed 3 times with buffer A and then 2 times with PBS and removed chamber from the slides. Then added mounting media containing DAPI (Vectashield H-1200) and incubated for 5 minutes in order to provide nuclear counterstaining. Finally the slides were covered with a cover slip, sealed and kept at 4° C until the time of confocal laser scanning microscopy.
Laser confocal imaging
Images were acquired on a confocal microscope (Leica TCS-SP2 -Leica Microsystems GmbH, Mannheim, Germany) using a 60× oil immersion objective. Fluorescein was excited with an argon laser (Enterprise model 651, Coherent Inc.) at 488 nm, DAPI with UV light at 364 nm, and Cy3 cyanine dye with a yellow helium neon laser at 594 nm. DIC images were acquired simultaneously using a transmitted light detector. Dyes were acquired in separate channels to minimize crosstalk. Images were processed using Leica TCS-SP2 software, Imaris v6.2 (Bitplane AG, Zurich Switzerland) and Adobe Photoshop CS3 (Adobe Systems).
Cell cycle analysis by flowcytometry and cell proliferation assay
For analyzing cell cycle distribution, NIH-3T3 and HEK 293 cells grown in 10 cm cell culture dishes were transfected with empty vector GFP-vector as control, and either with GFP-wild-type or -L95P parafibromin. After 48 hrs of transfection, cells were treated with trypsin and harvested in PBS using brief centrifugation. Cells were then treated with Vindelov’s PI [TBS containing propidium iodide (50µg/ml), RNase A (0.7 U/ml) (Sigma Chemical Co.) and NP-40 (0.1%)] and were stored at 4°C for at least 2h in dark prior to the acquisition. Samples were then run on a LSR-II flowcytometer (BD, San Jose, CA) and 50,000 cells were acquired. The intensity of propidium iodide was analyzed in GFP expressing cells using the cell-cycle platform (Dean/Jet/Fox mathematical model) of FlowJo software (Tree Star, Ashland, OR).
Cell viability was measured in vitro using the MultiTox-Fluor Multiplex Cytotoxicity Assay kit (Promega). Briefly, cells were plated into the wells of a 96-well plate at 5000 per well in octuplicate. Cells were allowed to grow for 24 h and then transfected with pcDNA3 vector alone, or with cDNA encoding AU5-tagged wild-type or L95P mutant parafibromin. 48 hours later, 20µl/well of combined substrate solution of glycyl-phenylalanyl-amino-fluorocoumarin and bis-alanyl-alanyl-phenylalanyl-rhodamine was added. After incubation of 1h at 37°C in a humidified 5% CO2 atmosphere, the absorbance at 405nM and 490nM was recorded by using a microplate reader.
RESULTS
Hyperparathyroidism-jaw tumor syndrome in a kindred associated with germline L95P missense mutation in parafibromin
An index case with primary hyperparathyroidism and a history of jaw tumors led to the identification of three generations of affected relatives that included four other individuals with primary hyperparathyroidism or menorrhagia associated with an abnormal endometrium as described in Case Reports (Fig. 1A). The proband and three other affected individuals harbored a heterozygous T →C germline transition in exon 3 of the HRPT2/CDC73 gene that resulted in a change in codon 95 of parafibromin from Leucine (CTT) to Proline (CCT) (Fig. 1A). An identical somatic HRPT2/CDC73 gene mutation was previously identified in an atypical parathyroid tumor taken from an individual with FIHP (Bradley et al. 2006). The leucine residue at position 95 in human parafibromin is highly conserved and is found in the corresponding position in parafibromin homologs from other species including fly and worm (Fig. 1B). The conserved leucine is immediately downstream of a putative nucleolar localization signal (NoLS) encompassing residues 76–92 that also serves as a secondary nuclear localization signal (NLS) (Hahn and Marsh 2007; Lin et al. 2007) (Fig. 1B).
Expression and stability of the L95P parafibromin mutant
Since the vast majority of germline and somatic HRPT2/CDC73 mutations identified in patients with HPT-JT, FIHP, or parathyroid cancer have been truncation or frameshift mutations, we presumed that the missense mutation identified in the HPT-JT kindred described here would impair the function of parafibromin. To better understand the molecular basis of its presumed loss of function, AU5 epitope tagged and green fluorescent proteins (GFP) fused versions of human L95P parafibromin mutant were made and compared to the wild type in cDNA transfection experiments in mammalian cell lines. When transfected into HeLa cells, both the GFP- and AU5-tagged versions of the L95P parafibromin mutant were expressed and had electrophoretic mobility similar to the corresponding wild-type protein as detected with anti-parafibromin antibodies (Fig. 2A). Quantitative immunoblotting by infrared imaging with normalization to the actin level showed that the L95P mutant parafibromin was expressed only to a level of about 50% of the corresponding wild-type GFP fusion, and to a level of about 30% of the corresponding wild-type AU5-tagged protein (Fig. 2A). Measurement of transcript levels of the transfected tagged parafibromin cDNAs by quantitative RT-PCR showed that levels of transcripts containing the L95P mutant were higher, not lower, than wild-type transcripts excluding missense or nonsense-mediated mRNA decay as a mechanism for reduced expression of the tagged parafibromin mutants (Supplementary Data Fig. 1). Because steady-state levels of both transfected L95P parafibromin mutants were lower than the corresponding wild-type proteins and could not be explained by lower transcript levels, we hypothesized that the missense mutation might destabilize the protein and confer accelerated degradation kinetics.
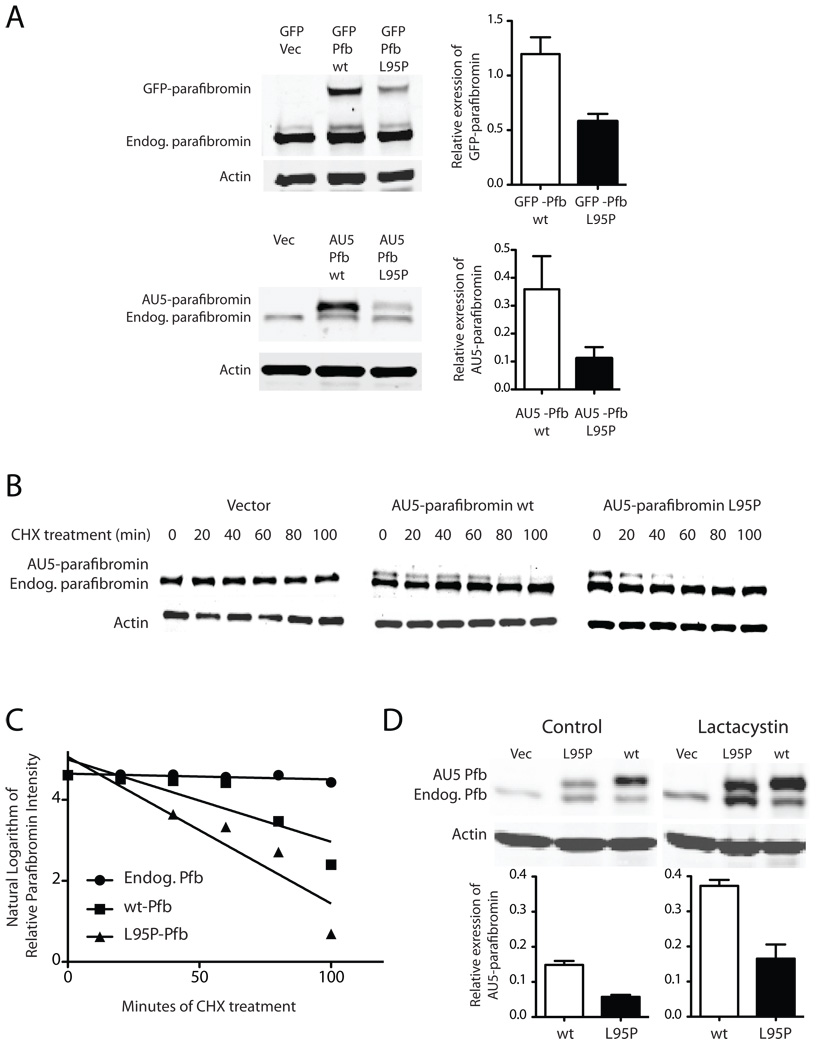
Figure 2. Diminished expression level of the L95P parafibromin mutant not due to accelerated proteasomal degradation.
A (Left) Immunoblots of GFP control or GFP fusions with wild-type (wt) or L95P mutant parafibromin (Pfb) (upper panels) and vector control (Vec), or AU5 epitope-tagged wild-type or L95P mutant parafibromin (lower panels) and endogenous parafibromin in cultured Hela cells after 24 hours of transfection. (Right) Histograms quantifying the relative expression level of wild type and L95P mutant parafibromin constructs compared to β-actin in the same sample. B. Time course of protein expression of endogenous parafibromin, AU5-parafibromin and AU5 L95P-parafibromin following cycloheximide treatment to block de novo protein synthesis. Cultured HeLa cells were transfected with the indicated constructs, then 6 hours following transfection cycloheximide was added to the culture medium and cells were collected at 0, 20, 40, 60, 80 and 100 min. Cell lysates were analyzed by immunoblotting at the indicated time points using anti-parafibromin antibody (upper) or anti-actin (lower) antibodies. The expression of β-actin is shown as a loading control. C. Linear regression analysis of the relative expression of transfected and endogenous parafibromin from (B) as described in Experimental Procedures. D. Immunoblots (above) and histograms (below) showing the expression, relative to β-actin, of wild type and L95P mutant AU5-tagged parafibromin without and with treatment with the proteasome inhibitor lactacystin for two hours.
We therefore studied the kinetics of protein disappearance in cycloheximide-treated HeLa cells previously transfected with vector alone, wild-type or L95P mutant AU5-tagged parafibromin by quantitative immunoblotting (Fig. 2B). The expression of parafibromin was examined at multiple time points between 0 and 100 minutes by quantitative analysis of the immunoblots by infrared imaging, and normalized by reference to the level of actin in each sample. The use of anti-parafibromin antibody in these experiments allowed the simultaneous monitoring of endogenous and transfected parafibromin (the latter migrating slightly slower on gel electrophoresis because of its epitope tag) (Fig. 2B). The natural logarithm of the actin-normalized parafibromin protein expression level (relative to t= 0) was plotted over time, and linear regression analysis used to estimate the parafibromin protein half-life assuming first-order kinetics (Fig. 2C). The estimated half-life of endogenous parafibromin was greater than that of the AU5 epitope-tagged wild-type parafibromin construct which was in turn greater than that of the L95P mutant parafibromin construct (Fig. 2C; endogenous parafibromin half-life, 520 ± 256 min; AU5-wt parafibromin half-life, 53 ± 17 min; AU5-L95P parafibromin half-life, 32 ± 7 min), but the differences among the estimated half-lives did not achieve statistical significance (endog. vs wt, p= 0.098; endog. vs. L95P, p= 0.086; wt vs. L95P, p= 0.28) (Fig. 2C).
To pursue the question of possible mutant protein destabilization, we examined whether inhibition of the endogenous proteasomal degradation pathway might increase steady state levels of the missense parafibromin preferentially compared to the wild-type. This experiment was suggested by previous evidence that missense mutants of tumor suppressor proteins undergo accelerated degradation by the proteasomal pathway (Xu and Attisano 2000; Yaguchi, et al. 2004). Treatment of transfected HeLa cells with the proteasome inhibitor lactacystin increased the expression of both endogenous and transfected AU5 epitope-tagged parafibromin constructs compared to control (Fig. 2D). Analysis of the actin-normalized expression levels by quantitative immunoblotting revealed no preferential increase in the relative expression of the L95P parafibromin missense mutant however compared to the wild-type (Fig. 2D). Similar results were seen with the proteasome inhibitor MG132 (not shown). Interestingly, in both control and lactacystin treated cells, transfection of wild-type or L95P mutant parafibromin increased the steady-state level of endogenous parafibromin (Fig. 2D). The mechanism behind this phenomenon is not clear but might reflect competition for limited degradation capacity or else the existence of positive feedback circuits leading to increased HRPT2 promoter activity and/or enhanced stabilization of endogenous parafibromin protein. This effect may be subtle because it was not always readily demonstrable (cf. immunoblots in Fig. 2A).
Interaction of the L95P parafibromin mutant with Paf1 complex components
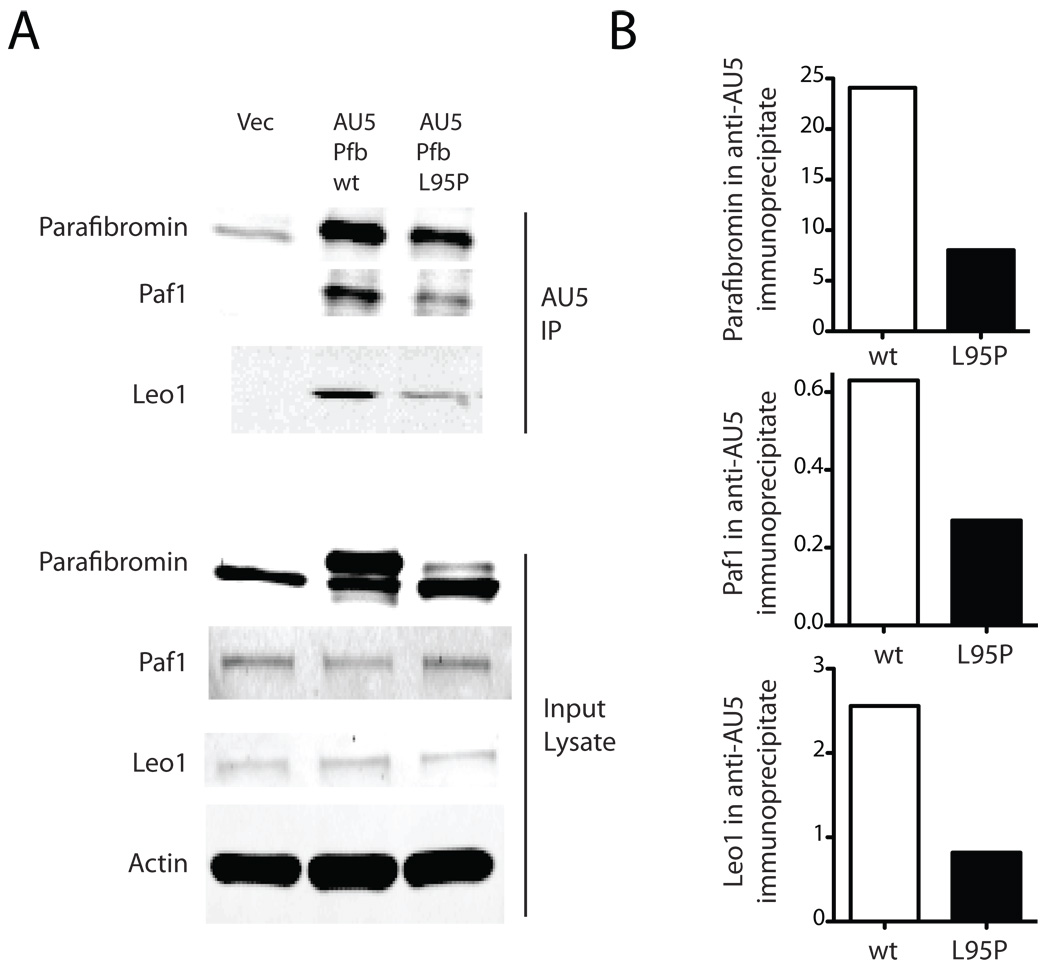
Mammalian parafibromin interacts with RNA polymerase II via a PAF1 complex (Rozenblatt-Rosen et al. 2005; Yart et al. 2005; Zhu et al. 2005), a function initially reported for the parafibromin homolog Cdc73p in yeast (Shi et al. 1997). Because impaired ability to interact with the PAF1 complex might account for the presumed loss-of-function of the L95P missense mutant, we examined the ability of wild-type and mutant parafibromin proteins to interact with PAF1 complex components by immunoprecipitation (Fig. 3). Both transfected wild-type and L95P mutant parafibromin could be immunoprecipitated with AU5 monoclonal antibody to their epitope tag, and both proteins could co-immunoprecipitate the PAF1 complex components Paf1 and Leo1 (Fig. 3A). Analysis of the precipitates by quantitative immunoblotting however revealed no preferential deficiency of the L95P parafibromin missense mutant in this assay, and for both the wild-type and mutant proteins the amount of Paf1 and Leo1 precipitated simply reflected the amount of epitope-tagged parafibromin protein in the AU5 immunoprecipitates (Fig. 3B).
Figure 3. The L95P parafibromin mutant retains interaction with PAF1 complex components.
A. 24 hours after transfection with vector only (Vec), or AU5 epitope-tagged wild-type (wt) or L95P mutant parafibromin (Pfb) HeLa cells were immunoprecipitated using AU5 monoclonal antibody and both the starting cell lysate (lower) and washed immunoprecipitates (upper) were analyzed by immunoblotting using anti-parafibromin, anti-Paf1 and anti-Leo1 antibodies as indicated. B. Histograms comparing the amount of immunoreactive parafibromin, Paf1 and Leo1 in the anti-AU5 immunoprecipitates.
Subcellular localization of the L95P parafibromin mutant
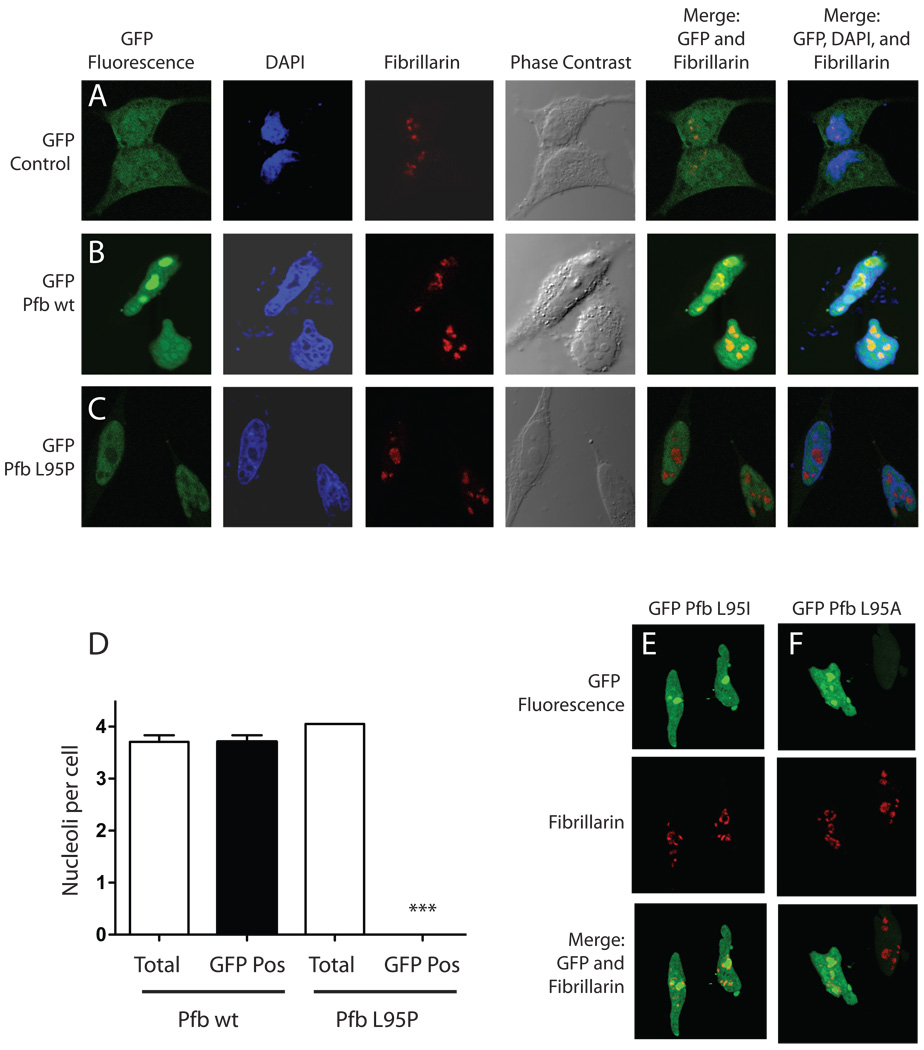
As noted above, the highly conserved leucine that is mutated to proline in affected individuals from the HPT-JT family described here is immediately downstream of a putative NoLS and secondary NLS in parafibromin (Hahn and Marsh 2007; Lin et al. 2007) (Fig. 1B). To see if the mutation might have an impact on parafibromin targeting, the subcellular localization of the GFP-tagged L95P parafibromin mutant was compared with that of the corresponding wild-type GFP fusion protein and GFP alone in transfected HeLa cells by laser confocal microscopy (Fig. 4). The cells were also treated with DAPI and anti-fibrillarin antibodies to delineate the nucleus and nucleolus respectively. Whereas GFP alone was distributed throughout the cytoplasm and nucleus (Fig. 4A), the wild-type parafibromin GFP fusion protein was localized to the cell nucleus with strong nucleolar concentration, as evidenced by co-localization with the anti-fibrillarin antibodies, as previously reported (Hahn and Marsh 2007) (Fig. 4B). In contrast the L95P parafibromin GFP fusion showed a striking lack of nucleolar localization even though the protein was clearly targeted to the cell nucleus (Fig. 4C, D). The change in codon 95 of parafibromin from leucine to proline seemed to be particularly deleterious to nucleolar localization since GFP fusions with parafibromin mutants bearing more conservative amino acid substitutions, such as to isoleucine (L95I) or alanine (L95A), demonstrated nucleolar localization resembling the wild-type on laser confocal analysis (Fig. 4E, F). Co-transfection of an excess of L95P mutant parafibromin did not block the nucleolar localization of the wild-type parafibromin GFP fusion protein (Supplementary Data Fig. 2).
Figure 4. The L95P parafibromin mutation blocks nucleolar but not nuclear localization.
Hela cells were cultured in chamber slides and transfected with GFP vector control (A) or else GFP fusions with wild-type (Pfb wt) (B) or L95P mutant parafibromin (Pfb L95P) (C). Cells were then treated with the DAPI nuclear stain, immunostained using anti-fibrillarin antibody as a nucleolar marker, and analyzed by confocal laser fluorescence microscopy. Phase contrast and overlay (merge) of GFP fluorescent signal with the nucleolar marker without or with the nuclear stain are also shown. D. Quantitation of nucleoli per Hela cell transfected with GFP fusions with wild-type (Pfb wt) or L95 mutant parafibromin (Pfb L95P): total nucleoli counted if red on fibrillarin-only stained images; GFP-positive nucleoli counted if yellow on merging of GFP fluorescence and fibrillarin stained images. Means shown from pooling of 2 or 3 experiments per transfection, with 19 to 24 cells (nuclei) analyzed per experiment. ***, two-tailed P value <0.0001 compared to total, employing Student’s unpaired t test. E., F. Confocal laser fluorescence microscopic images of Hela cells transfected with GFP fusions with L95I mutant (Pfb L95I) (E) or L95A mutant parafibromin (Pfb L95A) (F), and analyzed as indicated, in A–C.
Effect of the L95P parafibromin mutant on cell cycle progression and cell viability
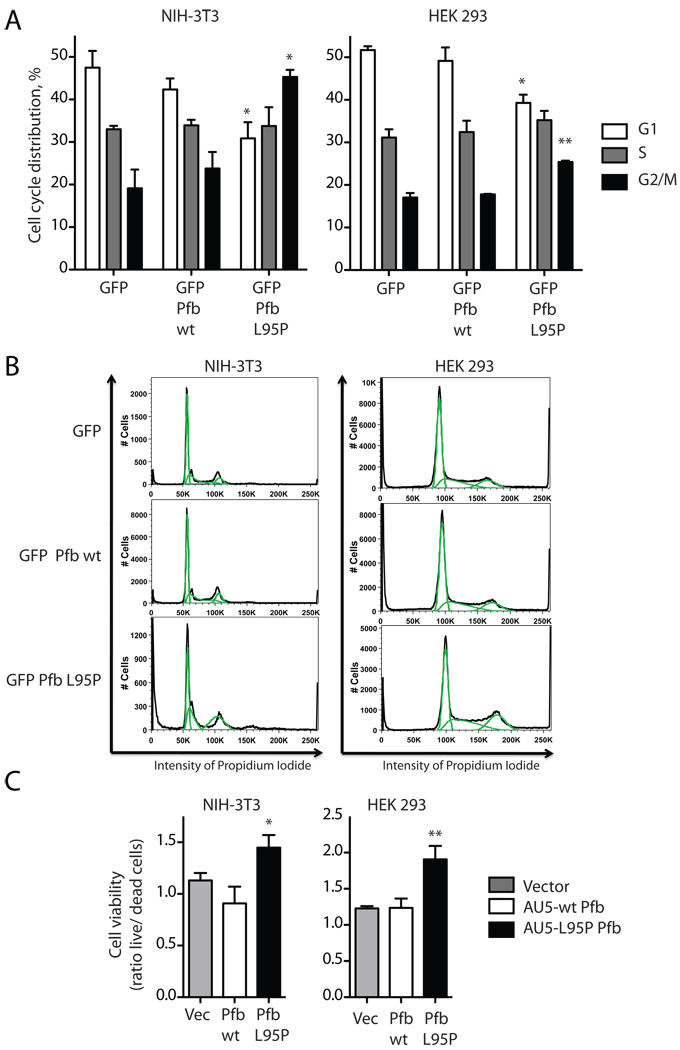
It has been previously shown that knockdown of endogenous parafibromin or transfection of patient-derived parafibromin mutants promotes cell cycle progression and/or cellular proliferation (Lin et al. 2008; Yart et al. 2005; Zhang, et al. 2006). We wondered if the presumed loss-of-function L95P parafibromin mutant might exhibit a phenotype in this regard and we therefore studied the cell cycle distribution of cells transfected with GFP alone or GFP-tagged wild-type or L95P parafibromin by flowcytometric analysis in GFP expressing cells (Fig. 5A, B). Compared to control, there was a significant loss of G1 phase and gain in G2/M phase cells in L95P parafibromin transfected NIH-3T3 and HEK 293 cells (Fig. 5A, B). No significant differences from control were seen in wild-type parafibromin transfected cells in either cell type.
Figure 5. The L95P parafibromin mutant enhances cell cycle progression and promotes cell survival in vitro.
A. Cell cycle analysis. NIH-3T3 cells and HEK 293 cells were transfected with GFP vector cDNA control or else cDNAs encoding fusion proteins of GFP with wild-type (wt) or L95 mutant parafibromin (Pfb). After 48 hours of transfection the cells were subjected to flowcytometric analysis in GFP expressing cells. Histograms show the percentage of cells in the indicated phases of the cell cycle. Values shown represent mean and standard errors of pooled results from of (HEK 293) or 3 (NIH-3T3) independent experiments; **, two-tailed P value <0.02; * <0.05 compared to GFP control, employing Student’s unpaired t test. B. Flowcytometric analysis plotting cell number versus intensity of propidium iodide signal for cells transfected under the indicated conditions, from a representative experiment described in A. C. Cell viability analysis. Cultured NIH-3T3 cells and HEK 293 cells were transfected with vector only (Vec), or AU5 epitope-tagged wild-type (wt) or L95P mutant parafibromin (Pfb) and after 48 hrs were analyzed for cell viability as described in Experimental Procedures. Ordinate values show the ratio of live to dead cells, representing the pooled data of six independent repeats. *, two-tailed P value <0.05; ** <0.01 compared to vector and wild-type controls, employing Fisher’s exact test.
The effect of transfected wild type and mutant parafibromin on cell viability was also studied in cultured cells (Fig. 5C). Transfection of the L95P parafibromin mutant but not the wild type significantly increased the survival of both NIH-3T3 and HEK 293 cells in this assay (Fig. 5C). In fact the cell viability of wild-type parafibromin-transfected NIH-3T3 cells was reduced, however this effect did not reach statistical significance (Fig. 5C, left panel). This outcome is somewhat at odds with previous results employing NIH-3T3 cells from our lab (Woodard et al. 2005) and others (Zhang et al. 2006) showing anti-proliferative effects in response to acute transfection of wild-type parafibromin. The basis for this apparent discrepancy is not known but may relate to the different proliferation assays employed with the NIH-3T3 cells (tetrazolium salt conversion colorimetric assay (Woodard et al. 2005) vs. focus formation and soft agar colony formation assays (Zhang et al. 2006) vs. dual fluorometric substrate cell viability assay (present study)).
DISCUSSION
The identification of a rare missense mutation in a family with HPT-JT provided an opportunity to study the mutant parafibromin function in vitro to understand the basis of its presumed loss of tumor suppressor function. We observed that the ability to associate with PAF1 complex components Paf1 and Leo1 was preserved in the L95P parafibromin mutant, perhaps unsurprising since the point mutation is remote from the core Cdc73p homology domain encompassing residues 233 to 525 (Pfam ID PF05179; (Finn, et al. 2008)) presumed to mediate the evolutionarily conserved interaction of the HRPT2/CDC73 gene product with the PAF1 complex (Chang et al. 1999; Shi et al. 1997). The key differences from wild type parafibromin we observed in the L95P mutant were its lower expression level, its impaired nucleolar localization, and its ability upon transfection to stimulate cell cycle progression and enhance survival of cells expressing a normal complement of endogenous parafibromin.
The lower expression level of the L95P parafibromin mutant might certainly account in part for its presumed loss-of-function. The difference in expression levels between wild type and L95P mutant parafibromin was not reduced or eliminated by treatment with proteasome inhibitors, however, suggesting that accelerated proteasomal destruction of the missense mutant was not occurring as has been described for other mutant tumor suppressor proteins (Xu and Attisano 2000; Yaguchi et al. 2004). Whether there is a causal relationship between the impaired nucleolar localization of the L95P parafibromin mutant, suspected to result from disruption of a nearby strong NoLS (Hahn and Marsh 2007), and its lower protein expression level and possibly decreased stability compared to the wild type remains an open question.
Whatever the mechanistic basis for its low steady state protein levels, the quantitative difference in expression from the wild type only underscores the significance of the qualitative effects observed for the L95P parafibromin mutant. Despite its documented lower expression level compared to wild type, the L95P missense mutant has a distinct pro-cell growth and survival phenotype in transfected cells: the mutant tumor suppressor protein enhanced cell cycle progression and increased survival of both NIH-3T3 and HEK 293 cells. The distinct phenotype shown by the parafibromin mutant argues strongly against the view that the L95P missense substitution is a null mutation that produces an inert and inactive protein. The demonstration of the growth and survival-promoting phenotype of the mutant parafibromin, qualities opposite those expected of a tumor suppressor protein, instead support the view that the L95P mutant is blocking or interfering with the functions of the endogenous parafibromin present in the transfected cells.
Dominant interfering mutants of cell regulatory molecules are those that retain some protein-protein interactions of the parent but lose others as a result of the mutation. In this formulation the retained interactions of the L95P parafibromin mutant with the PAF1 complex components might fall into the former category, while the loss of nucleolar targeting might reflect lost protein-protein interactions falling into the latter grouping. The combination of lost nucleolar localization with acquisition of a growth stimulatory phenotype following a single amino acid substitution in the L95P mutant raises the intriguing possibility that parafibromin must interact with targets in the nucleolus to execute its full repertoire of tumor suppressor functions. Examining how the loss of nucleolar targeting and associated protein-protein interactions as a result of the L95P missense mutation could be directly linked to its growth and survival-promoting phenotype may provide significant insight into the pathogenesis of sporadic and HPT-JT-associated parathyroid cancer.
Supplementary Material
ACKNOWLEDGEMENTS
The authors appreciate the support and encouragement of Metabolic Diseases Branch colleagues Steven J. Marx, Lee S. Weinstein, and Sunita K. Agarwal and the helpful suggestions of Dr. Ling Lin (Lombardi Cancer Center, Georgetown University).
FUNDING
This research was funded by the Division of Intramural Research of the National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland.
Footnotes
DECLARATION OF INTEREST
None of the authors has a conflict of interest that could be perceived as prejudicing the impartiality of the research reported herein.
REFERENCES
- Bradley KJ, Cavaco BM, Bowl MR, Harding B, Cranston T, Fratter C, Besser GM, Conceicao Pereira M, Davie MW, Dudley N, et al. Parafibromin mutations in hereditary hyperparathyroidism syndromes and parathyroid tumours. Clin Endocrinol (Oxf) 2006;64:299–306. doi: 10.1111/j.1365-2265.2006.02460.x. [DOI] [PubMed] [Google Scholar]
- Bradley KJ, Hobbs MR, Buley ID, Carpten JD, Cavaco BM, Fares JE, Laidler P, Manek S, Robbins CM, Salti IS, et al. Uterine tumours are a phenotypic manifestation of the hyperparathyroidism-jaw tumour syndrome. J Intern Med. 2005;257:18–26. doi: 10.1111/j.1365-2796.2004.01421.x. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, Simonds WF, Gillanders EM, Kennedy AM, Chen JD, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- Cetani F, Pardi E, Borsari S, Viacava P, Dipollina G, Cianferotti L, Ambrogini E, Gazzerro E, Colussi G, Berti P, et al. Genetic analyses of the HRPT2 gene in primary hyperparathyroidism: germline and somatic mutations in familial and sporadic parathyroid tumors. J Clin. Endocrinol. Metab. 2004;89:5583–5591. doi: 10.1210/jc.2004-0294. [DOI] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri V, Scillitani A, Muscarella LA, Battista C, Bonfitto N, Bisceglia M, Minisola S, Mascia ML, D'Agruma L, Cole DE. Diagnosis of parathyroid tumors in familial isolated hyperparathyroidism with HRPT2 mutation: implications for cancer surveillance. J Clin. Endocrinol. Metab. 2006;91:2827–2832. doi: 10.1210/jc.2005-1239. [DOI] [PubMed] [Google Scholar]
- Hahn MA, Marsh DJ. Nucleolar localization of parafibromin is mediated by three nucleolar localization signals. FEBS Lett. 2007;581:5070–5074. doi: 10.1016/j.febslet.2007.09.050. [DOI] [PubMed] [Google Scholar]
- Howell VM, Haven CJ, Kahnoski K, Khoo SK, Petillo D, Chen J, Fleuren GJ, Robinson BG, Delbridge LW, Philips J, et al. HRPT2 mutations are associated with malignancy in sporadic parathyroid tumours. J Med Genet. 2003;40:657–663. doi: 10.1136/jmg.40.9.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CE, Norum RA, Boyd SB, Talpos GB, Wilson SD, Taggart RT, Mallette LE. Hereditary hyperparathyroidism and multiple ossifying jaw fibromas: a clinically and genetically distinct syndrome. Surgery. 1990;108:1006–1012. [PubMed] [Google Scholar]
- Kelly TG, Shattuck TM, Reyes-Mugica M, Stewart AF, Simonds WF, Udelsman R, Arnold A, Carpenter TO. Surveillance for early detection of aggressive parathyroid disease: carcinoma and atypical adenoma in familial isolated hyperparathyroidism associated with a germline HRPT2 mutation. J Bone Miner Res. 2006;21:1666–1671. doi: 10.1359/jbmr.060702. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin L, Czapiga M, Nini L, Zhang JH, Simonds WF. Nuclear localization of the parafibromin tumor suppressor protein implicated in the hyperparathyroidism-jaw tumor syndrome enhances its proapoptotic function. Mol Cancer Res. 2007;5:183–193. doi: 10.1158/1541-7786.MCR-06-0129. [DOI] [PubMed] [Google Scholar]
- Lin L, Zhang JH, Panicker LM, Simonds WF. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc Natl Acad Sci U S A. 2008;105:17420–17425. doi: 10.1073/pnas.0710725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette LE, Malini S, Rappaport MP, Kirkland JL. Familial cystic parathyroid adenomatosis. Ann Intern Med. 1987;107:54–60. doi: 10.7326/0003-4819-107-1-54. [DOI] [PubMed] [Google Scholar]
- Mizusawa N, Uchino S, Iwata T, Tsuyuguchi M, Suzuki Y, Mizukoshi T, Yamashita Y, Sakurai A, Suzuki S, Beniko M, et al. Genetic analyses in patients with familial isolated hyperparathyroidism and hyperparathyroidism-jaw tumour syndrome. Clin Endocrinol (Oxf) 2006;65:9–16. doi: 10.1111/j.1365-2265.2006.02534.x. [DOI] [PubMed] [Google Scholar]
- Motulsky H. Intuitive Biostatistics. USA: Oxford University Press; 1995. Combining Probabilities; pp. 285–286. [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck TM, Valimaki S, Obara T, Gaz RD, Clark OH, Shoback D, Wierman ME, Tojo K, Robbins CM, Carpten JD, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003;349:1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonds WF, James-Newton LA, Agarwal SK, Yang B, Skarulis MC, Hendy GN, Marx SJ. Familial isolated hyperparathyroidism: Clinical and genetic characteristics of thirty-six kindreds. Medicine (Baltimore) 2002;81:1–26. doi: 10.1097/00005792-200201000-00001. [DOI] [PubMed] [Google Scholar]
- Simonds WF, Robbins CM, Agarwal SK, Hendy GN, Carpten JD, Marx SJ. Familial isolated hyperparathyroidism is rarely caused by germline mutation in HRPT2, the gene for the hyperparathyroidism-jaw tumor syndrome. J Clin. Endocrinol. Metab. 2004;89:96–102. doi: 10.1210/jc.2003-030675. [DOI] [PubMed] [Google Scholar]
- Teh BT, Farnebo F, Kristoffersson U, Sundelin B, Cardinal J, Axelson R, Yap A, Epstein M, Heath H, III, Cameron D, et al. Autosomal dominant primary hyperparathyroidism and jaw tumor syndrome associated with renal hamartomas and cystic kidney disease: linkage to 1q21-q32 and loss of the wild type allele in renal hamartomas. J Clin. Endocrinol. Metab. 1996;81:4204–4211. doi: 10.1210/jcem.81.12.8954016. [DOI] [PubMed] [Google Scholar]
- Teh BT, Farnebo F, Twigg S, Höög A, Kytölä S, Korpi-Hyövälti E, Wong FK, Nordenström J, Grimelius L, Sandelin K, et al. Familial isolated hyperparathyroidism maps to the hyperparathyroidism-jaw tumor locus in 1q21-q32 in a subset of families. J Clin. Endocrinol. Metab. 1998;83:2114–2120. doi: 10.1210/jcem.83.6.4896. [DOI] [PubMed] [Google Scholar]
- Villablanca A, Calender A, Forsberg L, Hoog A, Cheng JD, Petillo D, Bauters C, Kahnoski K, Ebeling T, Salmela P, et al. Germline and de novo mutations in the HRPT2 tumour suppressor gene in familial isolated hyperparathyroidism (FIHP) J Med Genet. 2004;41:e32. doi: 10.1136/jmg.2003.012369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- Xu J, Attisano L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor beta signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 2000;97:4820–4825. doi: 10.1073/pnas.97.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi H, Ohkura N, Takahashi M, Nagamura Y, Kitabayashi I, Tsukada T. Menin missense mutants associated with multiple endocrine neoplasia type 1 are rapidly degraded via the ubiquitin-proteasome pathway. Mol Cell Biol. 2004;24:6569–6580. doi: 10.1128/MCB.24.15.6569-6580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kong D, Tan MH, Pappas DL, Jr, Wang PF, Chen J, Farber L, Zhang N, Koo HM, Weinreich M, et al. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem Biophys Res Commun. 2006;350:17–24. doi: 10.1016/j.bbrc.2006.08.169. [DOI] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.