Abstract
Malignant melanoma is fatal in its metastatic stage. It is therefore essential to unravel the molecular mechanisms that govern disease progression to metastasis. MicroRNAs (miRs) are endogenous non-coding RNAs involved in tumourigenesis. Using a melanoma progression model, we identified a novel pathway controlled by miR-214 that coordinates metastatic capability. Pathway components include TFAP2C, homologue of a well-established melanoma tumour suppressor, the adhesion receptor ITGA3 and multiple surface molecules. Modulation of miR-214 influences in vitro tumour cell movement and survival to anoikis as well as extravasation from blood vessels and lung metastasis formation in vivo. Considering that miR-214 is known to be highly expressed in human melanomas, our data suggest a critical role for this miRNA in disease progression and the establishment of distant metastases.
Keywords: extravasation, melanoma, miR-214, TFAP2C, tumour progression
Introduction
The ability of tumours to acquire malignancy and spread in their host organism is one of the main issues in cancer treatment, as metastasis formation accounts for >90% of human cancer deaths. Nevertheless, the understanding of the molecular mechanisms that regulate metastatic dissemination remains fragmentary. The cascade of events that lead to metastasis is a complex multi-step process by which primary tumour cells acquire the ability to detach and invade adjacent tissues, intravasate, survive in the systemic circulation and translocate through the vasculature, adhere to the walls of capillaries of distant organs, extravasate in the parenchyma and finally proliferate in secondary tumours (Gupta and Massague, 2006). It is urgent to identify and characterize the genetic and epigenetic changes occurring during tumour progression. Several protein-coding genes involved in malignancy have been identified and characterized (Steeg, 2006; Nguyen et al, 2009). More recently, abnormalities in non-coding genes, such as microRNAs (miRs), have also been found to contribute to tumourigenesis (Croce, 2009; Valastyan and Weinberg, 2009). miRs are small endogenous non-coding RNAs able to post-transcriptionally downregulate expression of specific target genes by binding to the 3′UTRs of their mRNAs causing destabilization, degradation or translation inhibition (Filipowicz et al, 2008; Bartel, 2009). The ability of miRs to achieve simultaneous fine tuning of numerous different target genes makes them fundamental regulators of cellular signalling and implicates them in tumour progression (Inui et al, 2010). Several miRs, such as miR-21, miR-10b, miR-373 and 520c, miR-126, miR-335, miR-31, miR-200, miR-151 and miR-9, have already been reported to regulate tumour progression and metastasis (Valastyan and Weinberg, 2009; Ma et al, 2010).
Malignant melanoma is one of the most aggressive human cancers (Parkin et al, 2005) which progresses very rapidly via specific steps characterized by defined molecular alterations. Melanomas arise when the melanocytes of the epidermis become transformed and start to proliferate abnormally, leading to radial and vertical growth phases and subsequent spreading all over the body (Melnikova and Bar-Eli, 2008). The transition from the non-invasive to the invasive and metastatic stage is accompanied by gain of function of a number of transcription factors such as CREB/ATF-1, ATF-2, NFκB, SNAIL and STATs, while the loss of the AP-2 transcription factors (TFAP2) positively correlates with malignancy. At the same time, alterations in the repertoire of adhesion molecules, including MCAM-MUC18, E-cadherin, N-cadherin and several integrins, as well as changes in genes involved in angiogenesis, invasion and survival, such as VEGF, bFGF, IL-8, c-KIT, EGFR, MMP2 and PAR-1, are linked to the acquirement of higher metastatic potential (Melnikova and Bar-Eli, 2008). Several miRs, including miR-137, miR-221/222, miR-182 and miR-34a, have already been found to be involved in melanoma progression by regulating key genes such as c-KIT, MITF, FOXO3, ITGB3, CCND1 and p27Kip1 (Mueller and Bosserhoff, 2009). It now becomes fundamental to unravel how miRs control melanoma aggressiveness.
We identified a new pathway, coordinated by miR-214 and including TFAP2C, ITGA3 as well as multiple surface molecules, which controls melanoma metastasis dissemination by increasing migration, invasion, extravasation and survival of melanoma cells.
Results
miR-214 is upregulated in a metastatic melanoma model
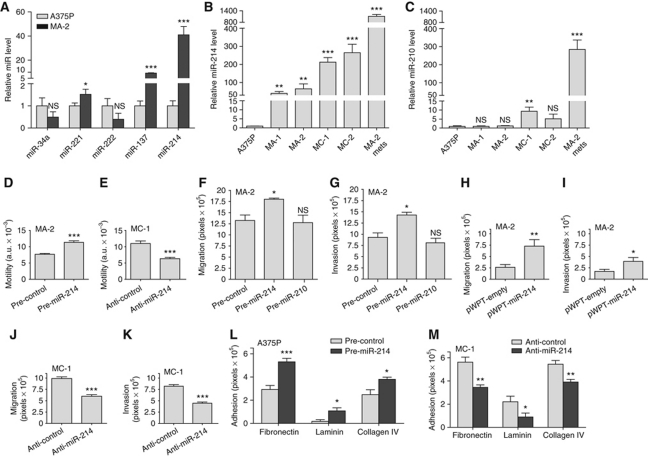
To assess a potential correlation between deregulation of miRs and melanoma malignancy, a miR profiling, which will be presented elsewhere (Cimino et al, unpublished), was performed in a melanoma progression model (Xu et al, 2008). The model consisted of a poorly metastatic A375 parental cell line (A375P) and its four highly metastatic variants, MA-1, MA-2, MC-1 and MC-2 derived by repeated passages in mice. Among the modulated miRs, miR-214 was found to be differentially expressed comparing metastatic (high) versus parental (low) cells in culture, as shown by qRT–PCR (Figure 1A and B). miR-214 showed a very strong enhancement of expression in samples derived from in vivo lung metastases following tail vein injections of MA-2 cells in immunodeficient mice (Figure 1B), suggesting an influence of the microenvironment for high expression. Induction of miR-214 expression in vivo was also observed in subcutaneous tumours derived from different melanoma cell lines expressing low miR-214 in culture (WK-Mel, GR4-Mel, 1300-Mel, Dett-Mel, SK-Mel-173, SK-Mel-197) (Supplementary Figure S1A). Other miRs, previously found to be involved in melanoma, such as miR-34a, miR-221, miR-222 and miR-137, also showed some differential expression in this system, however, not as pronounced as miR-214 changes (Figure 1A). When we extended expression analysis for miR-137 to other melanoma malignant cell lines it resulted to be overexpressed in some of them, such as WK-Mel, GR4-Mel, SK-Mel-173 and SK-Mel-197 compared with A375P. Instead no expression was detected in 1300-Mel, Dett-Mel and SK-Mel-187 cells (Supplementary Figure S1B). Some miRs were poorly expressed or did not show differential expression in our A375P isogenic model, including miR-210, which we used as a control (Figure 1C). miR-210 was expressed to some extent in most of the melanoma cells analysed although often at a low level (Supplementary Figure S1C). Importantly, miR-214 copy number gain was found in the genome of A375P, its MA-2 and MC-1 variants and in other melanoma cells, such as GR4-Mel, Dett-Mel, SK-Mel-103 and SK-Mel-187, as measured by genomic qRT–PCR (Supplementary Figure S1D) and SNP (not shown) analyses.
Figure 1.
miR-214 modulates cell migration and invasion. (A–C) Expression levels of the indicated miRs were evaluated in A375P cells or in its metastatic variants MA-1, MA-2, MC-1, MC-2 or in a pool of MA-2-derived lung metastases (MA-2 mets) by qRT–PCR. Results are shown as fold changes (mean±s.e.m.) relative to A375P cells, normalized on U44 RNA level. (D–M) Wound healing motility (D, E) or transwell migration or matrigel invasion (F–K) or adhesion on fibronectin, laminin or collagen (L, M) assays for cells either transfected with the indicated miR precursors or inhibitors or their negative controls (pre- and anti-miR or control) or stably transduced with pWPT-empty or miR-214 overexpression vectors. Results are shown as mean±s.e.m. of the reciprocal of the wound size (motility assay) or of the area covered by migrated or adherent cells (migration, invasion and adhesion assays). Two or three independent experiments were performed in triplicate and results were either shown as representative ones (A–C) or pooled together (D–M). *P<0.05; **P<0.01; ***P<0.001.
miR-214 expression enhances cell movement
The more pronounced expression of miR-214 in metastatic cells prompted us to investigate the potential pro-metastatic role of miR-214 by analysing cell movement following miR-214 expression modulations.
We stably or transiently overexpressed miR-214 in the miR-214-empty, poorly motile A375P cells and in the MA-2 metastatic variant, expressing an intermediate endogenous level of miR-214 (see Figure 1B), as well as in other melanoma cells, such as 1300-Mel, SK-Mel-187, WK-Mel and GR4-Mel, expressing low to undetectable miR-214 in culture (see Supplementary Figure S1A), by lentiviral infections with miR-214 expression or empty vectors (pWPT-miR-214 or pLemiR-214 or empty) or by transfections with miR-214 precursors or negative controls (pre-miR-214 or control). Conversely, we silenced miR-214 in the highly motile and invasive MC-1 and MC-2 variants following transfection with specific miR-214 antisense inhibitors or negative controls (anti-miR-214 or control). The efficacy of miR-214 modulations was tested by qRT–PCRs; miR-214 expression was increased up to 200 000- or 500-fold, respectively, in pre-miR-214 transiently transfected cells and stable infected cells (Supplementary Figure S2A–C) and almost completely silenced following miR-214 inhibition (Supplementary Figure S2D).
Significantly, miR-214 overexpression by pre-miR-214 transfection in MA-2 cells enhanced cell motility, migration and invasion as evaluated by wound healing assays (Figure 1D) or transwell assays in presence or absence of matrigel (Figure 1F and G) compared with negative controls. Migration and invasion were also increased 2–3-fold in stable miR-214-overexpressing MA-2 cells compared with cells containing the empty vector (Figure 1H and I). Moreover, miR-214 overexpression was sufficient to promote a significant increase in migration and/or invasion in poorly motile A375P cells (pWPT-miR-214, Supplementary Figure S3A) and in other unrelated melanoma cell lines, such as 1300-Mel, SK-Mel-187, WK-Mel and GR4-Mel (pre-miR-214, Supplementary Figure S3E–J). The effects observed on cell movement in the A375P isogenic model were specific for miR-214, since no significant variation in migration or invasion was observed following transient miR-210 overexpression (Figure 1F and G). In line with these results, transient miR-214 downregulation in MC-1 or MC-2 cells, following transfection with anti-miR-214, led to a 50% reduction in motility in a wound healing assay (Figure 1E, MC-1) and a 40–80% decrease in migration and matrigel invasion in transwell assays (Figures 1J and K, MC-1; Supplementary Figure S3B and C, MC-2). miR-214 functions were also evaluated by stable silencing in MC-1 cells using miR-214-specific sponges cloned in the 3′UTR of the green fluorescent protein (GFP) gene (see Materials and methods). MC-1 cells were transduced with pLenti-CMV-GFP-Puro-sponge1 (pLenti-sponge1) or pLenti-CMV-GFP-Puro-sponge3 (pLenti-sponge3) or pLenti-CMV-GFP-Puro (pLenti-empty) vectors and the efficacy of each sponge was evaluated by measuring the expression of the GFP in a western blot (WB) analysis (Supplementary Figure S2E). In presence of pLenti-sponge1 or pLenti-sponge3 GFP expression was highly decreased, indicating the efficient binding of miR-214 on the complementary sequences in the 3′UTR of the GFP. As shown in Supplementary Figure S3D, stable miR-214 silencing by sponge1 or sponge3 resulted in impairment of MC-1 cell migration in vitro (transwell assays), compared with control cells. Interestingly, miR-214 overexpression (pre-miR-214) was also able to significantly increase cell migration and matrigel invasion in other tumour cells, including human MDA-MB-231 or murine 4T1 mammary epithelial cancer cell lines (Supplementary Figures S2A and S3K–N). When we looked for the involvement of miR-214 in cell adhesion we observed significant adhesion alterations on different extracellular matrices (ECMs). In particular, transient miR-214 overexpression in A375P cells improved adhesion on fibronectin, laminin and collagen (1.5–3-fold increase), while miR-214 silencing in MC-1 cells consistently resulted in comparable adhesion defects on these matrices (Figure 1L and M).
Taken together, these results show that miR-214 significantly enhances in vitro cell movement and modulates adhesion, suggesting that it might facilitate metastasis formation by favouring tumour cell invasion and adhesion to the surrounding ECM.
miR-214 expression enhances metastasis formation in vivo
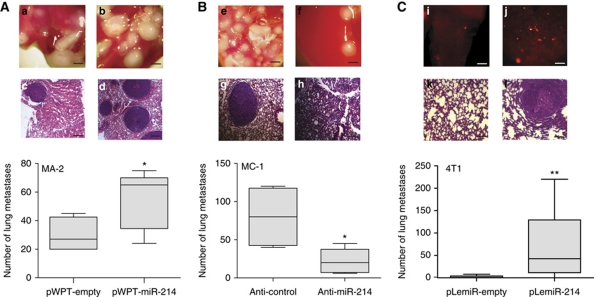
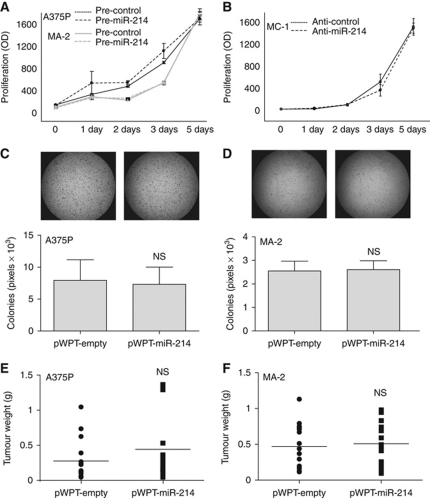
Because of its ability to induce motility and invasion in vitro, we asked whether miR-214 could influence cell movement and metastasis formation in vivo. Thus, miR-214-overexpressing MA-2 cells or miR-214-silenced MC-1 cells were injected into the tail vein of immunodeficient SCID mice and the number of lung metastases was evaluated 7 weeks later. A significantly higher number of macroscopic lung metastases could be observed for miR-214-overexpressing (pWPT-miR-214) MA-2 cells when compared with control (pWPT-empty) cells (Figure 2A). To investigate whether miR-214 was able to promote or regulate metastasis formation in the poorly metastatic parental cells, miR-214-overexpressing (pWPT-miR-214) or control (pWPT-empty) A375P cells were injected into SCID mice. Metastasis formation was analysed 9 weeks later and no macroscopic metastases were found on the lung surface for the two groups of mice. However, histological analyses revealed that 3 out of 10 mice injected with miR-214-overexpressing cells contained small metastatic formations in their lungs, while no micrometastases were found in control mice (Supplementary Figure S4). In contrast, miR-214-silenced MC-1 cells (by anti-miR-214 transfection) were significantly impaired in their ability to seed lung metastases and formed fewer lesions than the control (anti-control) cells (Figure 2B). High miR-214 expression levels were also able to enhance lung metastasis formation from a primary tumour. In fact, when the 4T1 mammary epithelial cancer cells, transduced with a lentiviral vector encoding for the turbo red-fluorescent protein (tRFP) alone (pLemiR-empty) or for miR-214 plus tRFP (pLemiR-214), were injected in mammary fat pad of female BALB/c mice, a significantly higher distribution of red-fluorescent micrometastasis in the lungs was observed for miR-214-overexpressing cells compared with controls, 20 days after injection (Supplementary Figure S2C; Figure 2C). However, miR-214-overexpressing cells gave rise to similar sized primary tumours as control cells (not shown). miR-214 enhancement effects on in vitro cell movement and in vivo metastasis formation are directly ascribed to the metastatic ability of the cells and not to proliferation effects. In fact, miR-214 overexpression (pre-miR-214) in A375P and MA-2 cells or miR-214 silencing (anti-miR-214) in MC-1 cells did not influence in vitro proliferation compared with controls (Figure 3A and B). Moreover, anchorage-independent growth in soft agar was not affected, since the number and the size of colonies for stable miR-214-overexpressing (pWPT-miR-214) A375P and MA-2 cells were comparable with controls (pWPT-empty) (Figure 3C and D). Furthermore, miR-214 did not influence primary tumour growth. In fact, when miR-214-overexpressing (pWPT-miR-214) or control (pWPT-empty) A375P or MA-2 cells were subcutaneously injected in the flanks of nude mice, the final tumour weight was similar for the different samples (Figure 3E and F).
Figure 2.
miR-214 enhances metastasis formation in vivo. (A, B) Metastasis formation in the lungs of SCID mice, 7 weeks after tail vein injection of MA-2 or MC-1 cells, stably transduced with pWPT-empty or miR-214 overexpression vectors (A) or transfected with miR-214 inhibitors or negative controls (anti-miR-214 or control) (B). Total number of macroscopic metastases per lung is shown as box and whiskers plots with median and minimum/maximum (n=5 mice per group). (C) Lung metastasis formation 3 weeks after injection of 4T1 cells stably transduced with pLemiR-tRFP (pLemiR-empty) or pLemiR-miR-214 overexpression (pLemiR-214) vectors in the mammary fat pad of BALB/c mice. Number of red-fluorescent micrometastasis per lung is shown as mean±s.e.m. for n=10 mice per group. Representative whole lung (a, b, e, f, i and j) and H&E staining (c, d, g, h, k and l) pictures are shown; bar=1 mm and 100 μm, respectively. Two independent experiments were performed and representative ones are shown (A, B) or results were pooled together (C). *P<0.05; **P<0.01.
Figure 3.
miR-214 does not affect cell and tumour growth. Proliferation (A, B) or anchorage-independent growth (C, D) or primary tumour growth 4 weeks after subcutaneous injection in the two flanks of nude mice (E, F) of cells transfected with miR-214 precursors or inhibitors or their negative controls (pre- and anti-miR-214 or control) or stably transduced with pWPT-empty or miR-214 overexpression vectors. Results are shown as mean±s.e.m. of the optical density (OD) (A, B) or of the area covered by colonies (C, D) or as tumour weight (dots or squares) and mean for n=15 (E) or 17 (F) mice per group. Two to four independent experiments were performed (in triplicate for A–D) and results were either shown as representative ones (A–D) or pooled together (E, F).
miR-214 modulates extravasation and survival
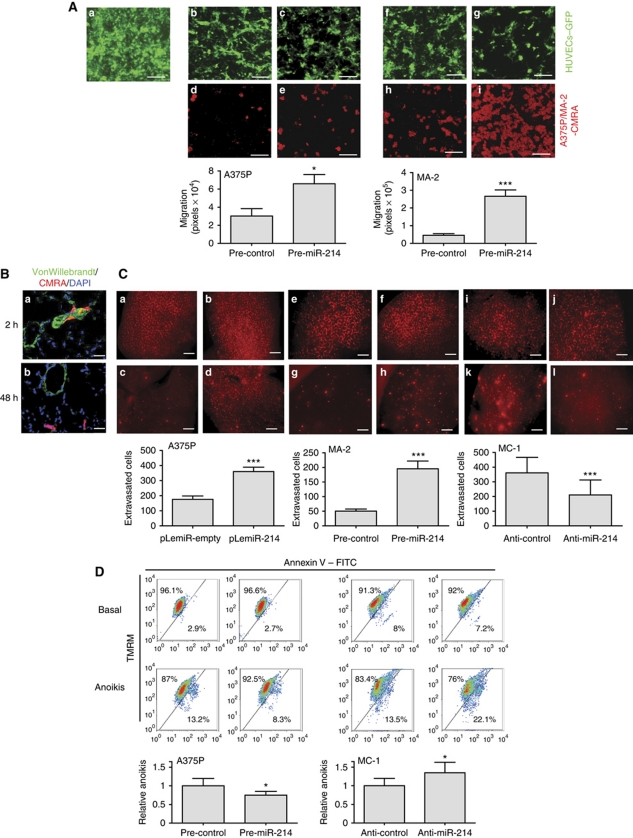
To assess the involvement of miR-214 in extravasation and survival, we combined in vitro and in vivo experiments. We first simulated transendothelial migration in vitro by seeding CMRA-labelled (red) miR-214-overexpressing (pre-miR-214) or control (pre-control) A375P or MA-2 cells in the upper chambers of fibronectin-coated transwells, covered by a confluent GFP-transduced human umbilical vein endothelial cell (HUVEC) monolayer. The remodelling of the endothelium and the consequent migration of melanoma cells in the lower chamber of the transwell were then evaluated (Figure 4A). The interaction of melanoma and endothelial cells induced the formation of fenestrations in the HUVEC monolayer. In the absence of melanoma cells (panel a) or in the presence of melanoma cell-conditioned medium (not shown), the HUVEC monolayer remains intact, suggesting the requirement of a direct cell–cell contact. Interestingly, the spaces formed in the endothelium were larger in the presence of miR-214-overexpressing A375P or MA-2 cells, compared with those formed with control cells (panels b, c, f and g) and, consequently, an increased number of cells migrated through the endothelial cells (panels d, e, h and i). In conclusion, miR-214 overexpression in A375P and MA-2 cells resulted in a 2–3-fold more efficient transendothelial migration. To evaluate the ability of miR-214 to influence extravasation in vivo, we first demonstrated that cell extravasation in the mouse lungs occurs within 48 h following injection of any metastatic variant in the blood circulation. For this, CMRA-labelled (red) negative control-transfected MC-1 (Figure 4B) or MA-2 (not shown) cells were injected in the tail vein of nude mice and their localization in the lungs was evaluated at different times. Two hours after injection, a high percentage of cells was located inside or associated with the lung blood vessels (Figure 4B, panel a), although some cells were already visible in the parenchyma (not shown). Forty-eight hours after injection, most of the cells present in the lungs appeared to be dispersed in the parenchyma (Figure 4B, panel b). To evaluate the role of miR-214 in the regulation of extravasation, CMRA-labelled miR-214-overexpressing A375P or MA-2 cells (pLemiR-214 or pre-miR-214, respectively) or silenced (anti-miR-214) MC-1 cells were injected via tail vein in nude mice and their ability to persist in the lungs was quantitated 48 h later. Two- or four-fold increased extravasation was observed, respectively, for miR-214-overexpressing A375P and MA-2 cells (Figure 4C, panels c, d, g and h), while about a 50% reduction in extravasation ability was observed for miR-214-silenced MC-1 cells (Figure 4C, panels k and l), compared with controls. This was not the consequence of a different lodging in the lung microvasculature, evaluated 2 h post-injection, since no difference was observed between miR-214-modulated cells and negative controls (Figure 4C, panels a, b, e, f, i and j). In vivo extravasation experiments were also performed with CMRA-labelled MC-1 cells stably transduced with specific miR-214 sponge (pLenti-sponge3) or empty (pLenti-empty) vectors as in Supplementary Figure S2E. In line with transient anti-miR-214 experiments, MC-1 cells expressing pLenti-sponge3 showed a highly reduced (80–90%) ability to extravasate compared with control cells, 48 h post-injection, as measured in the red (CMRA) or green (GFP) channel (Supplementary Figure S5A and B). No difference in lodging was observed at 2 h. In addition to transendothelial migration and extravasation ability, resistance to apoptosis was evaluated following miR-214 expression modulation in cells kept in the absence of adhesion (anoikis) and serum for 72 h, by AnnexinV–FITC and TMRM staining and cytofluorimetric analyses. miR-214-overexpressing (pre-miR-214) A375P cells showed a 20–30% reduction in cell death versus controls (Figure 4D, left); consistently, miR-214 silencing (anti-miR-214) in MC-1 cells led to 20–30% increased cell death (Figure 4D, right) compared with controls, suggesting a miR-214-driven cooperative role between extravasation ability and resistance to anoikis.
Figure 4.
miR-214 promotes extravasation and cell survival. (A) Transendothelial migration assays of CMRA-labelled (red) cells, through a fibronectin-coated transwell membrane covered by a confluent monolayer of HUVECs–GFP. Upper panels: HUVECs–GFP monolayer on the upper side of the transwell at time 0 h (a) or 18 h following the seeding of A375P (b, c) or MA-2 (f, g) cells transfected with miR-214 precursors or negative controls (pre-miR-214 or control). Lower panels: transmigrated A375P (d, e) or MA-2 (h, i) cells transfected as indicated, on the lower side of the transwell. (B, C) In vivo extravasation assays following tail vein injections in nude mice of CMRA-labelled (red) A375P cells stably transduced with pLemiR-empty or pLemiR-214 vectors, or MA-2 cells transfected as in (A) or MC-1 cells transfected with miR-214 inhibitors or negative controls (anti-miR-214 or control). (B) Representative fields of murine lung sections 2 h (a) or 48 h (b) post-injections, stained for VonWillebrandt factor (green) and counterstained with DAPI (blue). (C) CMRA-labelled cells in whole lungs 2 h (a, b, e, f, i and j) or 48 h (c, d, g, h, k and l) post-injections. Results are shown as mean±s.e.m. of the area covered by migrated cells (A) or of the number of extravasated cells at 48 h for n=10 (A375P), 7 (MA-2) or 5 (MC-1) mice per group (C). Two independent experiments were performed (in triplicate for (A)) and representative ones are shown. Bar=30 μm (A), 10 μm (B) or 500 μm (C). (D) Anoikis assays for A375P or MC-1 cells transfected as in (C). Cell death percentage was evaluated by TMRM and AnnexinV–FITC stainings, displayed in bidimensional plots. HighTMRM–LowAnnexinV gate: healthy population; LowTMRM–HighAnnexinV gate: apoptotic population. Four independent experiments were performed in duplicate and pooled quantitations refer to fold increased or decreased cell death versus controls, shown as mean±s.e.m. *P<0.05; ***P<0.001.
miR-214 regulates the expression of a panel of target genes
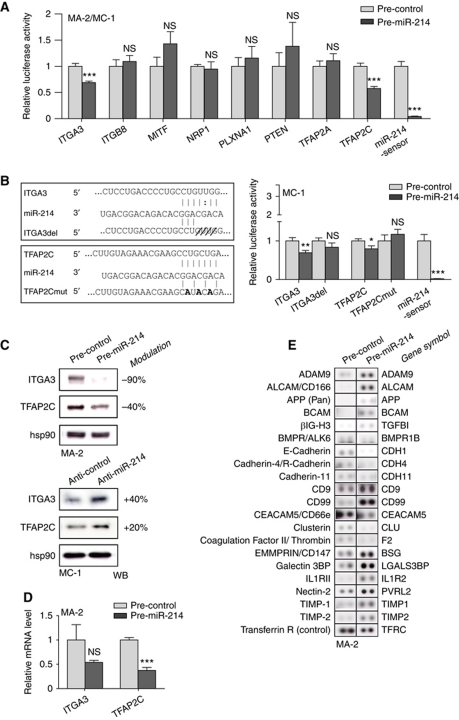
miR-214's ability to coordinate the various steps of metastasis formation can be attributed to multiple direct and indirect regulation of target genes. To identify direct targets of miR-214, we first used different algorithms such as miRecords (Xiao et al, 2009), miRanda (John et al, 2004) and TargetScan 5.1 (Friedman et al, 2009) and obtained >2000 predicted genes containing one or more putative binding sites for this miR. Then, genes were functionally classified using the Ingenuity Systems Analysis software and a large number of genes involved in Cancer (557 genes; P from 5.96E-13 to 9.7E-04) and Cellular Movement (302 genes; P from 7.82E-11 to 1.05E-03) were identified. A panel of 11 putative targets predicted by at least one algorithm and present in one or both of the functional categories were selected for experimental validation. For this purpose, the 3′UTRs of these 11 genes were cloned into a reporter vector and luciferase assays performed in MA-2 and/or MC-1 cells transfected with miR-214 precursors or negative controls (pre-miR-214 or control). As shown in Figure 5A, luciferase expression driven by the 3′UTRs of integrin α3 (ITGA3) or transcription factor AP-2γ (TFAP2C) was significantly repressed. No effect was revealed in the presence of the 3′UTRs of integrin β8 (ITGB8), microphthalmia-associated transcription factor (MITF), neuropilin 1 (NRP1), plexinA1 (PLXNA1), phosphatase and tensin homologue (PTEN) and transcription factor AP-2α (TFAP2A) (Figure 5A). A slight luciferase repression was also observed in the presence of the p21 protein (Cdc42/Rac)-activated kinase 2 (PAK2) 3′UTR while, surprisingly, a strong induction of luciferase activity occurred in the presence of met proto-oncogene (MET) and Rho GTPase activating protein 12 (ARHGAP12) 3′UTRs (Supplementary Figure S6A). To assess if the ITGA3 and TFAP2C luciferase expression regulation depended on the binding between miR-214 seed and the complementary sequence present on the 3′UTRs of ITGA3 or TFAP2C, a four nucleotide deletion or three point mutations were inserted in the ITGA3 or TFAP2C 3′UTRs, respectively, as indicated in Figure 5B, left. Both 3′UTR alterations completely abrogated the effect of miR-214 overexpression on luciferase expression in MC-1 (Figure 5B, right), A375P and MA-2 cells (not shown) indicating the specific and direct regulation of miR-214 on ITGA3 and TFAP2C 3′UTR binding sites in our A375P isogenic model. Similar results were obtained also in unrelated melanoma cell lines such as WK-Mel (Supplementary Figure S7A) or GR4-Mel (not shown) transfected with miR-214 precursors or negative controls. As a positive control, a miR-214-sensor construct, containing three perfect bindings for miR-214, was used for each experiment (Figure 5A and B; Supplementary Figures S6A and S7A). ITGA3 and TFAP2C modulations in luciferase activity corresponded to fluctuations of endogenous protein levels following transient miR-214 overexpression (pre-miR-214) in MA-2, WK-Mel, GR4-Mel (Figure 5C; Supplementary Figure S7C and D) or A375P and 1300-Mel (not shown) cells or miR-214 inhibition (anti-miR-214) in MC-1 cells (Figure 5C), as measured by WB analyses 48 h (not shown) or 72 h post-transfection. miR-214 overexpression led to a 30–90% and 40–80% protein decrease in ITGA3 and TFAP2C, respectively (Figure 5C; Supplementary Figure S7C and D). Reduction of protein expression was also observed in stable miR-214-overexpressing (pWPT-miR-214) MA-2 cells (10 and 15% decrease) compared with controls (Supplementary Figure S7B). Consistently, ITGA3 and TFAP2C proteins were upregulated (20 and 40%, respectively) in miR-214-silenced MC-1 cells (Figure 5C). Additionally, following transient miR-214 overexpression (pre-miR-214) in MA-2 cells, TFAP2C mRNA was significantly reduced, while ITGA3 mRNA showed a non-statistically significant decrease, as measured by qRT–PCR analyses 48 h post-transfection (Figure 5D).
Figure 5.
miR-214 downregulates TFAP2C and ITGA3 target genes and modulates multiple surface proteins. (A) Luciferase assays in MA-2 or MC-1 cells cotransfected with reporter constructs containing the 3′UTR of the indicated genes or a synthetic sequence including three perfect miR-214 binding sites (miR-214-sensor), cloned downstream of the luciferase coding sequence, together with miR-214 precursors or negative controls (pre-miR-214 or control). (B) Left panel: wild-type and miR-214 binding site mutant or deletant (ITGA3del, TFAP2Cmut) ITGA3 or TFAP2C 3′UTRs paired with miR-214 seed (respectively at positions 624 and 1071 of their 3′UTRs). Right panel: luciferase assays in cells cotransfected with wild-type or mutant 3′UTR reporter constructs, together with miR-214 precursors or negative controls (pre-miR-214 or control). Results are shown as mean±s.e.m. of Firefly luciferase activity relative to controls, normalized on Renilla luciferase activity. (C) Western blot (WB) analysis of ITGA3 and TFAP2C protein levels in MA-2 or MC-1 cells 72 h after transfections with miR-214 precursors or inhibitors or their negative controls (pre- and anti-miR-214 or control). Protein modulations were calculated relative to controls, normalized on the hsp90 loading control and expressed as percentages. (D) ITGA3 and TFAP2C mRNA levels measured by qRT–PCR in MA-2 cells 48 h after transfection as in (C, top). Results were calculated as fold changes (mean±s.e.m.) relative to controls, normalized on GAPDH. Three independent experiments were performed (in triplicate for (A, B); with independent protein or RNA preparations for (C, D)) and representative ones are shown. (E) Representative protein expression obtained for MA-2 cells 72 h after transfection with miR-214 precursors or negative controls (pre-miR-214 or control) by Proteome Profiler Human Soluble Receptor Antibody Arrays. Transferrin R was used as loading control. Doublets are shown. Two independent experiments were performed, a representative one is shown. Refer to Table I. *P<0.05; **P<0.01; ***P<0.001.
Consistent with the luciferase results, MET protein levels showed a 60 or 10% induction, respectively, following transient or stable miR-214 overexpression (Supplementary Figure S6B and D), which corresponded to mRNA fluctuations (Supplementary Figure S6E) and a 60% reduction following miR-214 silencing (Supplementary Figure S6C). Slight changes were also detected for PAK2 or PTEN protein expression following miR-214 overexpression or downmodulation, but no mRNA modulations were observed (Supplementary Figure S6B, C and E). Similarly, no mRNA changes were observed for ARHGAP12, ITGB8, MITF and NRP1, while a slight PLXNA1 mRNA decrease was found (Supplementary Figure S6E). On the contrary, TFAP2A was well modulated both at the protein and at the mRNA level (Supplementary Figure S6B and E); however, considering that TFAP2A is only slightly expressed in these melanoma cells.
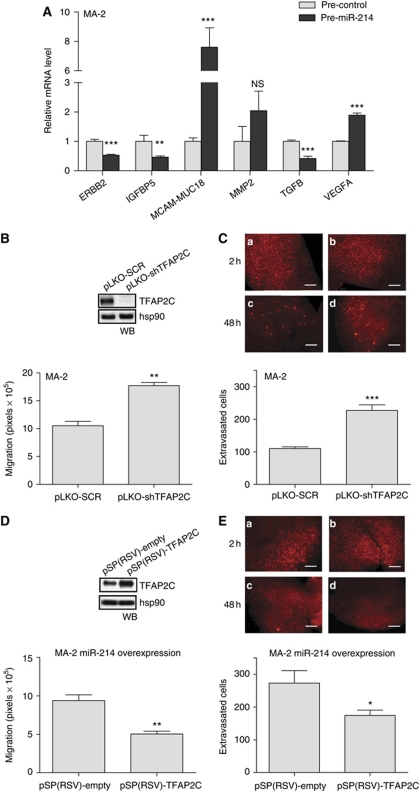
In parallel, we used a proteomic approach to identify miR-214-modulated surface proteins. The expression profile for 119 surface molecules involved in cell adhesion and movement was evaluated in MA-2 cells 72 h after transfection with miR-214 or control precursors; 65 differentially expressed molecules were found, 36 upregulated and 29 downregulated, following miR-214 overexpression (Table I; Figure 5E). Twenty-five modulated genes were predicted targets of miR-214 (Table I) and 17 genes, miR-214 predicted targets or not, were previously proven to be transcriptionally regulated by members of the AP-2 transcription factor family (TFAP2) or to contain TFAP2 binding sites in their promoters (Table I), suggesting a possible TFAP2-driven modulation in our melanoma model. Since it is well established that TFAP2 proteins contribute to melanoma progression via transcriptional regulation of key genes, we evaluated the expression of known TFAP2-activated or -repressed transcriptional targets by qRT–PCR, following miR-214 overexpression in MA-2 cells (pre-miR-214) compared with negative controls (pre-control). As shown in Figure 6A, the TFAP2-activated genes, ERBB2, IGFBP5 and TGFB, were found to be downregulated in the presence of miR-214, while the TFAP2-repressed genes, MCAM-MUC18 and VEGFA, but not MMP2, were upregulated (eight-fold and two-fold, respectively). Taken together, the direct or indirect TFAP2-dependent protein or mRNA modulations suggest that TFAP2 family members could have a relevant role in miR-214-dependent enhancement of melanoma malignancy.
Table 1. miR-214 modulates multiple surface molecules involved in cell movement and adhesion.
| Protein | Gene symbol | Pre-miR-214/control FC | Up/down | miR-214 target | TFAP2-dependent modulations |
|---|---|---|---|---|---|
| ADAM15 | ADAM15 | 1.6 | Up | ||
| ADAM8 | ADAM8 | 1.6 | Up | Y | |
| ADAM9 | ADAM9 | 2.5 | Up | ||
| ALCAM/CD166 | ALCAM | 6.0 | Up | ||
| APP (pan) | APP | 1.8 | Up | Provenzano et al (2007) | |
| BCAM | BCAM | 1.9 | Up | Y | |
| βIG-H3 | TGFBI | 2.3 | Up | Orso et al (2008) | |
| BMPR-IB/ALK-6 | BMPR1B | −1.8 | Down | Y | |
| Cadherin-11 | CDH11 | −1.2 | Down | Y | Borchers et al (2001) |
| Cadherin-4/R-Cadherin | CDH4 | −1.8 | Down | Y | |
| Cathepsin D | CTSD | −1.2 | Down | Y | Redecker et al (1991) |
| CD155/PVR | PVR | 1.4 | Up | Solecki et al (1999) | |
| CD23/FcRII | FCER2 | 3.1 | Up | ||
| CD36/SR-B3 | CD36 | 1.7 | Up | ||
| CD40 Ligand/TNFSF5 | CD40LG | 1.3 | Up | ||
| CD40/TNFRSF5 | CD40 | −1.5 | Down | ||
| CD44H | CD44 | 1.7 | Up | ||
| CD9 | CD9 | 2.4 | Up | ||
| CD90/Thy1 | THY1 | 1.6 | Up | ||
| CD99 | CD99 | 1.8 | Up | ||
| CEACAM-1/CD66a | CEACAM1 | 1.4 | Up | ||
| CEACAM-5/CD66e | CEACAM5 | −2.0 | Down | ||
| Clusterin | CLU | −2.0 | Down | Bayon et al (2004) | |
| Coagulation factor II/Thrombin | F2 | −2.1 | Down | ||
| CX3CL1/Fractalkine | CX3CL1 | 1.7 | Up | Y | |
| CXCL8/IL-8 | IL8 | 1.4 | Up | Y | |
| E-Cadherin | CDH1 | −1.7 | Down | Y | Schwartz et al (2007) |
| EGF R/ErbB1 | EGFR | −1.6 | Down | Y | Wang et al (2006) |
| EMMPRIN/CD147 | BSG | 1.8 | Up | Y | |
| Endoglin/CD105 | ENG | 1.6 | Up | Y | Rìus et al (1998) |
| Endoglycan | PODXL2 | −1.4 | Down | ||
| EpCAM/TROP-1 | EPCAM | −1.5 | Down | ||
| Epiregulin | EREG | 1.2 | Up | Orso et al (2008) | |
| ErbB2/HER2 | ERBB2 | −1.4 | Down | Y | Bosher et al (1996) |
| ErbB3/HER3 | ERBB3 | −1.7 | Down | ||
| E-Selectin/CD62e | SELE | −1.9 | Down | ||
| Galectin-3BP/MAC-2BP | LGALS3BP | 2.7 | Up | ||
| HB-EGF | HBEGF | 1.7 | Up | ||
| HPRG | HRG | 1.2 | Up | ||
| ICAM-2/CD102 | ICAM2 | 1.6 | Up | Grether-Beck et al (1996) | |
| IL-1 RII | IL1R2 | 2.2 | Up | ||
| Integrin α6/CD49f | ITGA6 | −1.5 | Down | ||
| Integrin αV/CD51 | ITGAV | −1.4 | Down | Kambe et al (1998) | |
| Integrin β1/CD29 | ITGB1 | 1.7 | Up | ||
| Integrin β3/CD61 | ITGB3 | 1.6 | Up | ||
| JAM-A | JAM1 | −1.6 | Down | Y | |
| JAM-B | JAM2 | −1.5 | Down | ||
| JAM-C | JAM3 | −1.4 | Down | Y | |
| Jagged 1 | JAG1 | −1.2 | Down | Y | |
| Lipocalin-2/NGAL | LCN2 | 1.5 | Up | ||
| LRP-6 | LRP6 | −1.5 | Down | Y | |
| MMP-2 (total) | MMP2 | 1.5 | Up | Y | Nyormoi et al (2003) |
| MUCDHL | CDHR5 | 1.4 | Up | ||
| N-Cadherin | CDH2 | −1.5 | Down | Y | |
| NCAM-1/CD56 | NCAM1 | 1.6 | Up | Y | |
| Nectin-2/CD112 | PVRL2 | 2.1 | Up | Y | Lui et al (2007) |
| Periostin/OSF-2 | POSTN | −1.3 | Down | ||
| Podocalyxin | PODXL | −1.5 | Down | Y | |
| Semaphorin 3A | SEMA3A | −1.6 | Down | ||
| TIMP-1 | TIMP1 | 2.0 | Up | Clark et al (1997) | |
| TIMP-2 | TIMP2 | 2.2 | Up | Y | Hammani et al (1996) |
| TIMP-3 | TIMP3 | 1.4 | Up | Y | |
| VCAM-1 | VCAM1 | −1.2 | Down | ||
| VEGF R1/Flt1 | FLT1 | −1.3 | Down | ||
| VEGF R2/KDR2/Flk-1 | KDR | −1.5 | Down | Y | |
| Expression modulation of 65 out of 119 surface proteins in MA-2 cells 72 h after transfection with miR-214 precursors or negative controls (pre-miR-214 or control), obtained by Proteome Profiler Human Soluble Receptor Antibody Arrays (see also Figure 5E). Modulations are expressed as fold change (FC) referring to miR-214-overexpressing cells versus controls, normalized on Transferrin R expression levels. Y, predicted miR-214 target. Literature references indicate previously demonstrated TFAP2 family members-mediated gene regulation (see references in Supplementary data). | |||||
Figure 6.
miR-214-mediated downmodulation of TFAP2C accounts for miR-214 functions. (A) mRNA levels for the indicated genes measured by qRT–PCR in MA-2 cells 48 h after transfection of miR-214 precursors or negative controls (pre-miR-214 or control). Results were calculated as fold changes (mean±s.e.m.) relative to controls, normalized on GAPDH. (B, D) Transwell migration assays for cells stably transduced with TFAP2C-targeting or negative control scramble shRNA vectors (pLKO-shTFAP2C or SCR) (B) or first transduced with pLemiR-214 and then transfected with either pSP(RSV)-empty or TFAP2C expression vectors (D). In the boxes: western blot (WB) showing TFAP2C protein levels. (C, E) In vivo extravasation assays 2 h (a, b) or 48 h (c, d) following tail vein injection in nude mice of the cells described in (B, D), previously labelled with CMRA (red). Representative pictures of the lungs are shown; bar=500 μm. Results are presented as mean±s.e.m. of the area covered by migrated cells (B, D) or of the number of extravasated cells at 48 h for n=10 (C) or 5 (E) mice per group. Two to three independent experiments were performed (in triplicate for (A, B, D)) and results were either shown as representative ones or pooled together (C). *P<0.05; **P<0.01; ***P<0.001.
Connections between 70 protein-coding genes found to be modulated in a direct or indirect manner by miR-214 (65 surface molecules in addition to ITGA3, MET, PAK2, TFAP2A and TFAP2C; Figure 5; Supplementary Figure S6; Table I) were searched using the Ingenuity Systems Analysis. Two main networks were identified, such as Tissue Development, Cellular Movement, Cell Cycle (Supplementary Figure S8A) and Cell-To-Cell Signalling and Interaction, Tissue Development, Embryonic Development (Supplementary Figure S8B). A top pathway was also identified, Leukocyte Extravasation Signalling (not shown), including CD44, CD99, ITGB1, MMP2, JAM2, JAM3, VCAM1 and TIMPs, genes potentially involved in tumour cell movement and blood vessel extravasation.
miR-214-mediated downmodulation of TFAP2C and ITGA3 controls miR-214 functions
As presented above, miR-214 is able to modulate TFAP2C and genes known to be transcriptionally regulated by the TFAP2 family members. To investigate if the metastatic effects of miR-214 in this melanoma model could be explained, in part, via its TFAP2C target we first analysed how TFAP2C silencing affected in vitro and in vivo cell movement. MA-2 cells were transduced with either specific TFAP2C or scramble negative control (pLKO-shTFAP2C or SCR) shRNA lentiviral vectors, leading to almost complete silencing of TFAP2C expression as measured by WB analysis (Figure 6B, top). TFAP2C-silenced cells were used for in vitro cell migration and in vivo extravasation experiments leading to results that phenocopied miR-214 overexpression. Indeed, >1.5-fold increased in vitro cell migration (Figure 6B) and extravasation in the lungs 48 h following tail vein injections (Figure 6C, panels c and d) were observed. No effect on the initial lodging of silenced cells in the lung vasculature 2 h post-injection was observed (Figure 6C, panels a and b). Hence, we asked whether the pro-metastatic phenotype associated with miR-214 overexpression could be rescued by TFAP2C overexpression in miR-214-overexpressing cells. For this purpose, a TFAP2C expression construct lacking its 3′UTR (pSP(RSV)-TFAP2C) was transiently transfected in stable miR-214-overexpressing MA-2 cells (Supplementary Figure S2C), as shown by WB in Figure 6D, top. Remarkably, a 40% decrease in in vitro cell migration (Figure 6D) and a significant impairment of in vivo lung extravasation at 48 h (Figure 6E) were observed compared with pSP(RSV)-empty controls. In conclusion, these experiments proved that TFAP2C is a major functional player downstream of miR-214, in fact, its direct repression by miR-214 contributes to increased melanoma pro-metastatic traits.
The functional impact of ITGA3 modulation via miR-214 was evaluated with similar approaches, by in vitro migration and in vivo extravasation assays. For this purpose, ITGA3 was silenced in MA-2 cells following transfections with specific siRNAs (si-ITGA3), leading to strong protein downregulation compared with negative control cells (si-control), as evaluated by WB (Supplementary Figure S9A, top). In parallel, ITGA3 overexpression was obtained in miR-214-overexpressing MA-2 cells following transfection with a 3′UTR-deprived ITGA3 expression construct (pCMVzeo-ITGA3), compared with control (pCMVzeo-empty) cells (Supplementary Figure S9C, top). A significant increase in cell migration (Supplementary Figure S9A) was found in ITGA3-silenced cells, compared with controls, while decreased cell migration (Supplementary Figure S9C) was found for miR-214/ITGA3-overexpressing cells compared with controls. In contrast, lung extravasation of CMRA-labelled cells, 48 h post-injection as well as the lodging in the lung vasculature 2 h post-injection were not affected by ITGA3 silencing or overexpression (Supplementary Figures S9B and D). This observation is reinforced by the fact that long-term metastasis formation, 7 weeks following tail vein injection, was also not affected by ITGA3 overexpression in miR-214-overexpressing MA-2 cells (not shown).
Finally, the effect of TFAP2C on ITGA3 and MET protein expression was evaluated in MA-2 cells following TFAP2C silencing or overexpression. As shown in Supplementary Figure S10, only a slight protein modulation was observed for ITGA3 following TFAP2C silencing (10% reduction, left) or overexpression (10% increase, right). In contrast, MET protein expression strongly increased (over 100%) following stable TFAP2C silencing and decreased (20% reduction) when TFAP2C was transiently overexpressed (Supplementary Figure S10). These data suggest that the miR-214-dependent regulation of MET and ITGA3 is at least in part mediated by TFAP2C.
Expression of miR-214 and its modulated genes in human melanoma tumours
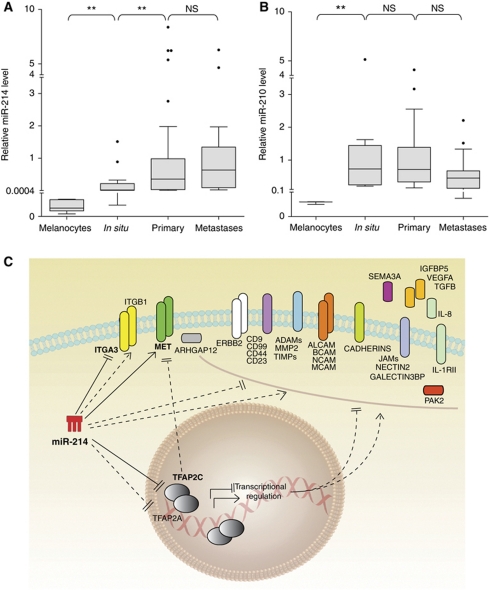
qRT–PCR analysis was used to assess miR-214 expression in human primary melanocytes (two preparations), in situ melanomas, confined into the basal membrane (n=13), primary invasive melanomas (n=57) and cutaneous metastases (n=18). As shown in Figure 7A, miR-214 was highly expressed in primary and metastatic melanomas, compared with in situ tumours and normal melanocytes. Instead, miR-210 expression, which we used as control, did not vary significantly among tumour samples; however, low miR-210 levels were found in primary melanocytes (Figure 7B). Considering the elevated levels of miR-214 in invasive melanomas and in metastases, we investigated the expression of over 70 miR-214-modulated genes (Figures 5 and 7C; Supplementary Figure S6; Table I) in a human protein-coding gene expression data set available in GEO, referring to mRNA expression in normal skin, nevi and primary melanomas (Talantov et al, 2005). Differential expression was observed for several of these genes relative to the three groups of samples analysed (Supplementary Figure S11), underlying the relevance of this gene group for melanoma progression. In agreement with our findings, TFAP2C mRNA was downregulated in melanoma but expressed in skin as well as nevus samples; in contrast, ITGA3 mRNA was upregulated in this cohort of melanomas, suggesting different or additional regulations.
Figure 7.
miR-214 is expressed in human melanomas and modulates multiple genes. (A, B) miR-214 (A) and miR-210 (B) expression levels were evaluated by qRT–PCR in human primary melanocytes (two independent preparations), in situ melanomas (n=13), primary melanomas (n=57) and cutaneous metastases (n=18). miR levels are shown as box and whisker plots; the bottom and the top indicate the 25th and 75th percentile (the lower and upper quartile, respectively), the band inside the box is the 50th percentile and the dots represent the outliers. Relative expression was calculated using the healthy skin miR median expression as reference and normalized on U44 RNA level. Statistical analysis was performed with the Mann–Whitney non-parametric statistical test. (C) Drawing summarizing the miR-214-modulated genes identified in this study and their connections. Continuous lines: miR-214 direct (luciferase proven) regulations; dashed lines: indirect regulations. *P<0.05; **P<0.01.
Discussion
Our work led to the identification of a novel functional pathway (Figure 7C), controlled by miR-214 and including its direct targets TFAP2C and ITGA3 as well as multiple surface proteins, that coordinates metastasis formation in a melanoma progression model, represented by the A375P poorly metastatic parental cell line and its derived metastatic variants. This pathway can be considered of general relevance for melanoma progression since it was also studied and validated in a panel of unrelated human melanoma cell lines. Importantly, overexpression of miR-214 or silencing of TFAP2C in melanoma cells enables them to acquire or increase invasive and metastatic behaviour. On the other hand, miR-214 silencing or concomitant miR-214 and TFAP2C (lacking its 3′UTR) overexpression impairs the establishment of a metastatic phenotype, suggesting a major role for miR-214 and TFAP2C in controlling melanoma tumour progression.
In agreement with our findings, also high-throughput screenings showed that miR-214 is strongly expressed in malignant cutaneous and ocular melanomas (Worley et al, 2008; Segura et al, 2010) as well as in various solid tumours, such as pancreas, prostate, gastric, breast and ovary cancers (Volinia et al, 2006; Blenkiron et al, 2007; Sempere et al, 2007; Yang et al, 2008; Ueda et al, 2010); thus, it will be fundamental to understand the mechanism of miR-214 overexpression in tumours. We showed that gene amplification for miR-214 locus occurs in A375P, MA-2 and MC-1 variants as well as in other melanoma cells. In addition, a main study of Comparative genomic hybridization demonstrated copy number gain in miR-214 locus for various melanomas, ovarian and breast cancer samples (Zhang et al, 2006). However, it was previously demonstrated that miR-214 is transcriptionally regulated by Twist-1 (Lee et al, 2009; Yin et al, 2010) and by Polycomb protein complex (Juan et al, 2009), suggesting important transcriptional controls for miR-214. The fact that miR-214 expression is generally low or undetectable in culture while it increases dramatically when cells are injected in vivo, i.e., tumours or lung metastases, suggests an essential regulation by the microenvironment, possibly involving secreted growth factors or cytokines or cell-to-cell contacts, currently under investigation.
So far, miR-214 was proposed to be mostly involved during differentiation and morphogenesis of tissues and organs (Flynt et al, 2007; Decembrini et al, 2009) and disruption of the mouse genomic locus including miR-214, Dnm3os, determined death within 1 month of birth with skeletal abnormalities (Watanabe et al, 2008). Considering that genetic programs controlling embryonic development are often reactivated during tumour progression, a role for miR-214 in tumourigenesis is expected. High levels of miR-214 in human tumours suggest an oncogene-like function, but we and others (Fei et al, 2008) did not find any positive regulation of cell growth for miR-214. Instead, increased levels of miR-214 could induce chemotherapy resistance in ovarian tumours via the direct downregulation of PTEN and consequent upregulation of the Akt pathway (Yang et al, 2008); however, we were not able to see any strong miR-214-dependent downmodulation of PTEN, suggesting a cell context role in PTEN targeting by miR-214.
The spread of melanoma cells from the primary lesion to the sites of metastasis formation involves multiple steps (Melnikova and Bar-Eli, 2008) that are coordinated by miR-214. Other miRs were previously found to be involved in some aspects of melanoma tumourigenesis: miR-221/222 and let7b mostly control proliferation and survival, while miR-182, miR-137, let7a and miR-34a affect migration and invasion as well as cell growth (Mueller and Bosserhoff, 2009). miR-214 exerts pleiotropic roles in our melanoma model due to its ability to directly or indirectly modulate the expression of many genes. We were able to identify over 70 miR-214-modulated genes that were previously found to be involved in cell movement and metastasis formation, some specifically in melanoma. Examples are integrins (Desgrosellier and Cheresh, 2010), cadherins (Jeanes et al, 2008), cell adhesion molecules (Kobayashi et al, 2007), the tyrosine kinase receptor MET (Benvenuti and Comoglio, 2007), MMP2 (Hornebeck et al, 2002), ADAMs (Duffy et al, 2009), TIMPs (Cruz-Munoz and Khokha, 2008), Semaphorin3A (Neufeld and Kessler, 2008) and PAK2 (Gadea et al, 2008). In addition to cell migration and invasion, we showed, for the first time, that a miR promotes transendothelial migration and extravasation of melanoma cells via direct tumour-endothelial cell interactions, as proven by the fact that melanoma cells but not conditioned medium are able to induce endothelial cell reorganization. Based on the extravasation experiments, we suggest that miR-214 pro-metastatic effect is mainly due to the earlier steps of the metastatic process, such as the passage through the endothelial barrier, more then post-colonization events. In fact, we observed a significant reduction in both extravasation (48 h) and long-term metastasis formation following transient silencing of miR-214 in cells, a condition in which we expected miR-214 levels to remain low only in the first 48 h post-transfection, as observed in in vitro experiments (not shown). Considering that extravasation data were confirmed by permanent knockdown of miR-214 via specific sponges and that miR-214 did not influence tumour cell growth, we can conclude that high levels of miR-214 favour extravasation and survival in the blood circulation but do not control later tumourigenesis steps. Heterotypic interactions are known to occur between endothelial and melanoma cells during extravasation involving, for instance, the CAM family molecules, such as ALCAM (Swart et al, 2005) and MCAM-MUC18 (Xie et al, 1997), or tetraspanin CD9 and ITGB1 complexes at the melanoma-endothelial cell contact regions (Longo et al, 2001), or IL-8 secretion, ICAM-1 expression and E-selectin reduction, as a consequence of B-Raf mutation in melanoma cells (Liang et al, 2007). Interestingly, these molecules are modulated by miR-214 in our cells (see Table I). Given that B-Raf is mutated in our melanoma model (Eskandarpour et al, 2005), a possible cooperation between B-Raf and miR-214 could occur during extravasation. Moreover, functional classification of miR-214-modulated molecules revealed an enrichment of genes involved in the leukocyte extravasation pathway, suggesting a common mechanism of migration through the endothelial walls for melanoma cells and leukocytes. Since miR-214 reduces anoikis in cell culture, we speculate that miR-214 also helps the tumour cells to survive in the blood circulation. Similar functions were shown for miR-31 in breast cancer (Valastyan et al, 2009). We found that miR-214 was able to confer various pro-metastatic traits also to the A375P parental cells; however, we observed only a slight effect on lung colonization in our experimental conditions. No macrometastases were found when miR-214-overexpressing or control A375P cells were injected. Nevertheless, micrometastases were detected exclusively for miR-214-overexpressing cells. These results suggest that miR-214 facilitates metastasis formation but it is not a strong metastasis inducer per se and that its role depends on the cellular context in which it operates. In any case, we cannot exclude that different experimental settings were needed for these investigations.
Based on bioinformatic predictions we looked for direct miR-214 targets and among various genes tested, we were able to prove that TFAP2C and ITGA3 are directly and specifically repressed via miR-214 binding on their 3′UTRs in melanoma cells. At the same time, we observed that miR-214 was able to induce MET overexpression. TFAP2C is a member of the AP-2 transcription factor family involved, together with TFAP2A, in the activation or repression of various genes implicated in tumourigenesis (Hilger-Eversheim et al, 2000; Eckert et al, 2005). Melanoma progression is clearly associated with loss of expression of total or nuclear TFAP2 proteins and this correlates with poor prognosis and advanced stages of the disease (Bar-Eli, 2001; Berger et al, 2005). Loss or deletions of the TFAP2 family member genomic loci were found in some, but not all, melanomas, suggesting other mechanisms for control of gene expression (Karjalainen et al, 2000). We were able to demonstrate that miR-214 downregulates expression of TFAP2C directly and TFAP2A indirectly in melanoma cells. We need to underline that only low TFAP2A protein expression was detected in the melanoma progression model we used, consistent with the fact that A375P cells were derived from a metastatic melanoma with low TFAP2A amount (Huang et al, 1998). Instead, TFAP2C is well expressed in our system, although at reduced levels in the most invasive variant MC-1 compared with the parental A375P cells due to miR-214 higher expression. Reduction of TFAP2A is, at least in part, due to direct control by TFAP2C, since TFAP2C silencing reduces TFAP2A protein levels partially (Orso et al, unpublished), which is in line with the fact that TFAP2 family members are able to regulate each other transcriptionally (Bauer et al, 1994). It is known that elimination of TFAP2A from non-metastatic primary melanoma cells increases malignancy (Gershenwald et al, 2001), while re-expression abrogates it (Huang et al, 1998), by controlling transcription of genes such as MCAM-MUC18, MMP2, PAR-1, VEGF, BCL2, CDKN1A/p21, E-cadherin and c-KIT (Melnikova and Bar-Eli, 2008). We were able to demonstrate that miR-214 overexpression increases the transcription of MCAM-MUC18 and VEGFA, two well-known TFAP2-repressed genes. In addition, it decreases expression of genes transcriptionally activated by TFAP2 family members, such as ERBB2 and IGFBP5, as well as TGFB. c-KIT, another well-known TFAP2-modulated gene, is expressed at very low levels in our system, consistently with the literature (Huang et al, 1998), so we were not able to evaluate its modulation. Other TFAP2-regulated genes including E-Cadherin/CDH1, CTSD, EREG, TGFBI, CDH11, CLU, PVR, EGRF, ERBB2, ICAM2, ITGAV, MMP2, PVRL2, TIMP1, TIMP2, all somehow connected with cell movement and/or malignancy, were found to be differentially expressed following miR-214 modulations. Considering that TFAP2C silencing in MA-2 cells phenocopied miR-214 overexpression biology, while its forced expression in miR-214-overexpressing cells rescued the migration and extravasation phenotypes, we conclude that TFAP2C is one of the main players through which miR-214 controls malignancy in melanoma. TFAP2C modulations were also proven to interfere, at least in part, with the expression of two other miR-214-modulated genes highly involved in cell movement and tumour progression, ITGA3 and MET, thus suggesting an additional relevance for TFAP2C in gene expression regulation in melanomas. The relevancy of TFAP2C is also underlined by its decreased expression in human melanoma samples from a public data set. On the contrary, we found that ITGA3 modulates cell migration in vitro, but it is not sufficient per se to control the extravasation process or metastasis formation in vivo, suggesting that the contribution of ITGA3 on miR-214-mediated functions is only partial and restricted to cell movement in culture. In line with our in vivo results, we did not find reduced expression of ITGA3 in the human melanoma public data set we analysed. Considering that ITGA3 is thought to promote invasion and metastasis in various human tumours (Mitchell et al, 2010), it is possible that, even if ITGA3 is a bona fide target of miR-214, a feedback loop that re-establishes high levels of this adhesion molecule occurs during melanoma progression. Unfortunately, only mRNA (no protein) data were available for the human data set we analysed. Since miR-214 was able to downregulate ITGA3 protein expression without affecting mRNA levels in cells, ITGA3 protein expression needs to be verified in a high number of human melanoma samples, before drawing any conclusions.
Considering that miR-214 is highly expressed in human melanomas and that the pool of miR-214-modulated genes correctly separates melanomas versus nevi or normal skin, we can conclude that the pathway including miR-214 and its modulated genes is able to coordinate melanoma progression and could be considered for therapeutic intervention.
Materials and methods
Cell culture
293T, MDA-MB-231 and 4T1 cells were from American Type Culture Collection and maintained in standard conditions. A375P and derived variants were provided by RO Hynes (Xu et al, 2008) and maintained in Dulbecco's Modified Eagle's Medium containing 10 mM Glutamax and 4.5 g/ml glucose (DMEM Glutamax™, GIBCO Invitrogen Life Technologies, Carlsbad, CA), supplemented with 10% heat-inactivated FCS (Seromed, GmbH), 1 mM sodium pyruvate, 25 mM HEPES pH 7.4, 1 × MEM vitamin solution, 1 × MEM non-essential amino acids and 100 μg/ml gentamicin (all from GIBCO Invitrogen Life Technologies). 1300-Mel, GR4-Mel, WK-Mel and Dett-Mel were provided by P Circosta and cultured according to Circosta et al (2009) in RPMI medium supplemented with 10% heat-inactivated FCS (Seromed, GmbH), 1 mM sodium pyruvate, 25 mM HEPES pH 7.4, 1 × MEM non-essential amino acids and 100 μg/ml gentamicin. HUVECs–GFP were provided by L Primo and grown as described (Primo et al, 2007). Human melanocytes were purchased from Lonza (Basel, CH). SK-MEL-103, -173, -187 and -197 melanoma cell lines were obtained and cultured as described in Segura et al (2009). Human epidermal melanocytes, derived from adult, lightly pigmented donor (HEMa-LP), were from Invitrogen Life Technologies (Cascade Biologics brand) and were maintained in Medium 254, supplemented with Human Melanocyte Growth Supplement-2, PMA-Free (Invitrogen Life Technologies, Carlsbad, CA), as indicated by manufacturer's guidelines.
Reagents and antibodies
pLKO.1-SCR (scramble) or pLKO.1-shTFAP2C lentiviral expression vectors were purchased from Open Biosystems (Huntsville, AL, cat. no. RHS3979-19745). pSP(RSV)-TFAP2C and pSP(RSV)-empty expression vectors were a gift from H Hurst (Bosher et al, 1995, 1996). siITGA3 (Hs_ITGA3_7 HP Validated siRNA) was purchased from Qiagen (Stanford, CA). pCMVzeo-ITGA3 and pCMVzeo-empty were a gift from M DiPersio (Iyer et al, 2005). miR precursors and inhibitors were pre-miR™ miRNA Precursor Molecules-Negative Control #1, pre-miR™ miRNA Precursor Hsa-miR-214 (PM12124), Hsa-miR-210 (PM10516), anti-miR™ miRNA Inhibitors-Negative Control #1, anti-miR™ miRNA Inhibitors Hsa-miR-214 (AM12124) (all from Ambion, Austin, TX). miRNA detection was TaqMan® MicroRNA Assays Hsa-miR-214 assay ID 000517 or Hsa-miR-214 assay ID 002306, Hsa-miR-210 assay ID 000512, Hsa-miR-221 assay ID 000524, Hsa-miR-222 assay ID 002276, Hsa-miR-137 assay ID 001129, Hsa-miR-34a assay ID 000426, Hsa-RNU44 assay ID 001094, U6 snRNA assay ID001973 (all from Applied Biosystems, Foster City, CA). Primary antibodies were anti-TFAP2A mAb 3B5, anti-TFAP2C mAb 6E4/4, anti-GAPDH pAb V-18, anti-hsp90 mAb F-8, anti-MET pAb C-12 (all from Santa Cruz Biotechnology, Santa Cruz, CA), anti-ITGA3 pAb 8-4 B7 gently provided by M DiPersio (DiPersio et al, 1995), anti-PAK2 pAb #2608, anti-PTEN pAb #9552 (Cell Signaling Technology, Danvers, MA), anti-Von Willebrand Factor pAb A0082 (DAKO Cytomation, Glostrup, Denmark), anti-GFP Ab (ab290, Abcam, Cambridge, UK). Secondary antibodies were goat anti-mouse IgG HRP conjugated, goat anti-rabbit IgG HRP conjugated, donkey anti-goat IgG HRP conjugated (all from Santa Cruz Biotechnology) and goat anti-rabbit IgG Alexa-Fluor-488 (Molecular Probes, Invitrogen Life Technologies). All antibodies were used at the producer's suggested concentrations.
Primers
Oligonucleotides employed in this study were cloning miR-214, TTTCATAGGCACCACTCACTTTAC and CCCTTTCCCCTTACTCTCCA; miR-214 synthetic binding site CTAGTCCACTGCCTGTCTGTGCCTGCTGTCGTAGGATCTACTGCCTGTCTGTGCCTGCTGTTGGACCTGACACTGCCTGTCTGTGCCTGCTGTCCCA and AGCTTGGGACAGCAGGCACAGACAGGCAGTGTCAGGTCCAACAGCAGGCACAGACAGGCAGTAGATCCTACGACAGCAGGCACAGACAGGCAGTGGA; cloning ARHGAP12 3′UTR, GCACTAGTGTTCCATCTTCGGACGTTGA and ATACGCGTCTTCAACAGACCATGCTCCC; cloning ITGA3 3′UTR, ATACTAGTAGGCTGACCGACGACTACTG and TAACGCGTTTTTGCATACAAACTGGGAGC; cloning ITGB8 3′UTR, ATGCCGGCGCTCACGGTCATGCCAGTTGCT and TAGTTTAAACCAGGGCTCATGGCCCGTGTA; cloning MET 3′UTR, ATACTAGTCCTCCTTCTGGGAGACATCA and TAACGCGTACAAGATGTTGCATCACTTTACTTT; cloning MITF 3′UTR, ATACTAGTCCTCCCTGCACTGCATTCGC and TAACGCGTCAGTGCTATGGACCATACAG; cloning NRP1 3′UTR, TAACTAGTGAGTACTTATTCGGAGGCATGAA and TAACGCGTAATTCAACTGTTTCTTTGGAATG, cloning PAK2 3′UTR, ATACTAGTCCTGTGCCTCTAACAAGCGATTC and TAACGCGTAAGTTTAGGAAAGGGAAAAATGCACA; cloning PLXNA1 3′UTR, ATACTAGTCCCCAGCTGTGATCATCC and TAACGCGTTGTTTAATCTCTCCTTAATGTGTGTG; cloning TFAP2A 3′UTR, ATACTAGTGCGGGTGACAGCTCCGGGAT and TAACGCGTTGATACCCTGCTCTGAACTCCAAGT; cloning TFAP2C 3′UTR, ATGAGCTCCTGGAGACCAGAGTCCAGCTGAT and TAACGCGTAGGGTTCAGCCCAACAGGAA; TFAP2C 3′UTR mutagenesis, TTCTGGTTTTAGGAAACTTGTAGAAACGAAGCATACAGATTGATTTTTTTCTCCTTTTTTTTTTTTTTT and AAAAAAAAAAAAAAAGGAGAAAAAAATCAATCTGTATGCTTCGTTTCTACAAGTTTCCTAAAACCAGAA; ITGA3 3′UTR deletions, CTCCTGACCCCTGCCTGCAGGCCCA and TGGGCCTGCAGGCAGGGGTCAGGAG, ARHGAP12 RT–PCR, TCAAAACCCTGCTTTCCTGA and CCATATTTCTCTTGATCCTTTGG and probe #7; IGFBP5, CTACCGCGAGCAAGTCAAG and GTCTCCTCGGCCATCTCA and probe #77; ITGA3 RT–PCR, GAGGACATGTGGCTTGGAGT and GTAGCGGTGGGCACAGAC and probe #13, MET RT–PCR AAATGTGCATGAAGCAGGAA and TCTCTGAATTAGAGCGATGTTGA and probe #59; NRP1 RT–PCR CACATTTCACAAGAAGATTGTGC and CATCAATTTTAATTTCTGGGTTCTTT and probe #85; PLXNA1 RT–PCR CAGTGATGTGGCTGTGTCG and CTCACGGGAGTTCCTCCA and probe #68 (all probes from Universal Probe Library, Roche, Mannheim, GmbH); ERBB2 RT–PCR QuantiTect Primer Assay QT00060746, GAPDH RT–PCR QuantiTect Primer Assay QT00079247, ITGB8 RT–PCR QuantiTect Primer Assay cat. no. QT00038507, MCAM-MUC18 RT–PCR QuantiTect Primer Assay cat. no. QT00079842, MITF RT–PCR QuantiTect Primer Assay cat. no. QT00037737, MMP2 RT–PCR QuantiTect Primer Assay cat. no. QT00088396, PAK2 RT–PCR QuantiTect Primer Assay cat. no. QT01677172, PTEN RT–PCR QuantiTect Primer Assay cat. no. QT01676969, RRN18S QuantiTect Primer Assay QT00199367 (all from Qiagen, Stanford, CA); TFAP2A RT–PCR, ACATTCCGATCCCAATGAGCA and TGTTTTGTAGCCAGGAGCATGTTT; TFAP2C RT–PCR, AACAGACCCACACACTTAGCCATT and GCGGACACAAAAACCAACCA; TGFB RT–PCR GCAACAATTCCTGGCGATACCT and AGCCCTCAATTTCCCCTCCA; VEGFA RT–PCRCTATGTCCTCACACCATTGAAACCA and GGAAGGTCAACCACTCACACACA. miR-214 genomic locus: TGGGCAAAACACAGTGAAAA and TGACGCGTGGAAGTATCAAA; UBE2E1 genomic locus: GGTGGGAAGTATTGCCACTCA and GTGAAACCCCAATTTATGTAGCGTAT.
Transient transfections of pre-miRs, anti-miRs, siRNAs or cDNAs
To obtain transient anti-miR, pre-miR or siRNA expression, cells were plated in 6-well plates at 30–50% confluency and transfected 24 h later using RNAiFect (Qiagen, Stanford, CA) reagent, according to the manufacturer's instructions, with 100 nM anti-miR, 75 nM pre-miR or 170 nM siRNA. For transient cDNA overexpression, cells were plated in 6-well plates at 90% confluency and transfected 24 h later using Lipofectamine™2000 (Invitrogen Life Technologies) reagent, according to the manufacturer's instruction. Cells were tested for miR or gene overexpression/knockdown 24 or 48 h later.
Plasmid construction and generation of stable cell lines
The human miR-214 gene was amplified from genomic DNA and cloned into pWPT (Addgene, Cambridge, MA) or pLemiR-tRFP (Open Biosystems) vectors to obtain pWPT-miR-214 and pLemiR-214 (still containing tRFP) vectors. For the preparation of the miR-214 sponges two different 292 nts long sequences, containing eight bulged target sites specific for miR-214 with linkers in-between, were designed as described in Krol et al (2010) synthesized by DNA2.0 (CA, USA) and cloned in the 3′UTR of the GFP gene in a pLenti-CMV-GFP-Puro vector (Addgene) generating the pLenti-sponge1 and -sponge3 vectors. The pLenti-CMV-GFP-Puro empty vector was used as control (pLenti-empty). All stable cell lines were generated via lentiviral infection. Lentiviruses were produced by calcium phosphate transfection of 20 μg vector plasmid together with 15 μg packaging (pCMVdR8.74) and 6 μg envelope (pMD2.G-VSVG) plasmids in 293T cells, according to Trono's lab protocol (http://tronolab.epfl.ch). Supernatant was harvested 48 h post-transfection, filtered with 0.45 μm filters, diluted and used to infect 3.5 × 105 cells in 6-well plates, in presence of 8 μg/ml Polybrene (Sigma-Aldrich, St Louis, MO). Luciferase reporter vectors containing the full-length or partial (PTEN, PAK-2, ITGB8) 3′UTR of the indicated miR-214 target genes were generated following PCR amplification of the 3′UTR from human genomic DNA and cloning into the Firefly Luciferase reporter pMIR-REPORT™ vector (Ambion). When indicated the 3′UTRs were mutagenized at the miR-214 recognition site using the QuickChange Site-Directed Mutagenesis kit (Stratagene, Cedar Creek, TX), according to the manufacturer's instructions. miR-214-sensor was obtained by annealing, purifying and cloning short oligonucleotides containing three perfect miR-214 binding sites into the SpeI and HindIII sites of the pMIR-REPORT vector.
RNA isolation and qRT–PCR for miRNA or mRNA detection
Total RNA was isolated from cells or tumours using TRIzol® Reagent (Invitrogen Life Technologies). qRT–PCRs for miR detection were performed with the indicated TaqMan MicroRNA Assays (Applied Biosystems) on 10 ng total RNA, according to the manufacturer's instructions. For mRNA detection, 1 μg of DNAse-treated RNA (DNA-free™ kit, Ambion) was retrotranscribed with RETROscript™ reagents (Ambion) and qRT–PCRs were carried out using gene-specific primers, using a 7900HT Fast Real Time PCR System. Quantitative normalization was performed on the expression of the RNU44 small nucleolar RNA or of GAPDH, for miR or mRNA detection, respectively. The relative expression levels between samples were calculated using the comparative delta CT (threshold cycle number) method (2−ΔΔCT) with a control sample as the reference point (Bookout and Mangelsdorf, 2003).
Motility assay
For the wound healing motility assay, cells were grown to confluency in 6-well plates, serum starved for 24 h, then a cross wound was made on the monolayer using a pipette tip. Cells were rinsed with PBS buffer and placed in complete growth medium. Photographs in different regions of the wound (at least 10 fields) were taken at t=0 h and at t=18 h, using Olympus IX70 microscope. The distance covered by the migrating cells is inversely proportional to the size of the wound; motility was quantitated by calculating the reciprocal of the wound size measured using the ImageJ software (http://rsbweb.nih.gov/ij/) (Orso et al, 2008).
Migration, invasion and transendothelial migration transwell assays
To measure migration 3.5 × 104 MC-1 or 7.5 × 104 MA-2 and WK-Mel or 1.2 × 105 1300-Mel, SK-Mel-187 and GR4-Mel or 2 × 105 A375P or 105 4T1 and MDA-MB-231 were seeded in serum-free media in the upper chambers of cell culture inserts (transwells) with 8.0 μm pore size membrane (24-well format, Becton Dickinson, NJ). To measure invasion of MC-1 and MA-2 cells and of 1300-Mel and SK-Mel-187, the previously mentioned inserts were pre-coated with 4 or 2 μg/well growth factor reduced Matrigel (Becton Dickinson), respectively, and the cells seeded as indicated above. For 4T1 and MDA-MB-231 cells, invasion assays were performed using BioCoat™ Matrigel Invasion Chambers with 8.0 μm pore size membrane (Becton Dickinson). For migration and invasion, the lower chambers were filled with complete growth media. After 18–20 h, the migrated cells present on the lower side of the membrane were fixed in 2.5% glutaraldehyde, stained with 0.1% crystal violet and photographed using an Olympus IX70 microscope (Orso et al, 2008). For transendothelial migration assay, 105 HUVECs–GFP were seeded in complete medium in the upper part of transwell inserts with 5.0 μm pore size membrane (24-well format, Costar, Corning Incorporated, NY) coated by fibronectin at 5 μg/cm2, and grown for 72 h, till confluency. Then, 5 × 104 cells were labelled with CellTracker™ Orange CMRA (Molecular Probes, Invitrogen Life Technologies), according to the manufacturer's instructions and seeded in HUVEC's complete medium onto the HUVEC–GFP monolayer on the upper side of the transwell. After 20 h, the HUVEC–GFP monolayer was photographed using Zeiss Axiovert200M microscope. Then, HUVECs and non-transmigrated cells were removed and the red-fluorescent (CMRA) cells that migrated on the lower side of the membrane were fixed in 4% paraformaldheyde and photographed using Zeiss AxioObserver microscope with ApoTome Module. Migration, invasion and transendothelial migration were evaluated by measuring the area occupied by migrated cells using the ImageJ software (http://rsbweb.nih.gov/ij/).
Adhesion assays
To test adhesion, 5 × 104 cells/well were seeded onto 5 μg/ml collagen IV or 10 μg/ml fibronectin or 5 μg/ml laminin (all from Sigma-Aldrich) pre-coated 96-well plates, for 1 h at 37°C. Cells were then washed thoroughly to remove non-adherent cells, fixed with methanol and stained with haematoxylin and eosin (Diff-Quik, Medion Diagnostics, Dudingen, CH). Wells were photographed using Olympus IX70 microscope and the area occupied by the adherent cell was measured using the ImageJ software (http://rsbweb.nih.gov/ij/) (Orso et al, 2008).
Proliferation assays
In all, 5 × 103 cells/well were plated in 96-well plates in complete medium and starved for 24 h. Complete medium was then added and cells were allowed to grow for 1, 2, 3 and 5 days, fixed with 2.5% glutaraldehyde and stained with 0.1% crystal violet. The dye was solubilized using 10% acetic acid and optical density measured directly in plates using a Microplate Reader Mithras LB940 (Berthold Technologies, GmbH) at 570 nm wavelength (Kueng et al, 1989).
Anchorage-independent growth assays
In all, 5 × 104 cells were resuspended in 8 ml of complete DMEM containing 0.45% Difco Noble Agar (Becton Dickinson) and plated in 6 cm bacterial dishes. Medium was changed every 3 days. After 20 days, the dishes were stained with nitroblue tetrazolium (Sigma-Aldrich), photographed with Nikon SMZ1000 stereomicroscope and colonies were counted using the ImageJ software (http://rsbweb.nih.gov/ij/) (Hynes et al, 1990).
Anoikis analysis
Cells were plated on a 2% agarose pad in serum-free medium for 72 h, collected, washed in PBS buffer, resuspended in 10 mM Hepes, 150 mM NaCl, 5 mM CaCl2 buffer containing FITC-conjugated Annexin-V (Bender MedSystems, GmbH) and 200 nM tetramethyl-rhodamine-methyl-ester (TMRM, Molecular Probes, Invitrogen, CA) and incubated at 37°C for 15 min. Flow cytometry analysis of apoptosis was carried out by using a FACSCalibur flow cytometer (Becton Dickinson). Data acquisition was performed using CellQuest software (Becton Dickinson) and data analysis with WinMDI software (version 2.8, Scripps Institute, CA). Results were displayed in bidimensional plots, with gates indicating the percentages of healthy and apoptotic populations (Rasola and Geuna, 2001).
In vivo tumour and metastasis assays
All experiments performed with live animals complied with ethical animal care. Seven-week-old female CD1 nude mice (Charles River Laboratories, Wilmington, MA) were subcutaneously injected in the two flanks with 8 × 106 A375P, MA-2, MC-1 cells or with 106 WK-Mel, GR4-Mel, 1300-Mel, SK-Mel-173, SK-Mel-197 (in PBS). Tumour growth was monitored every 3 days. Mice were dissected 4 weeks after injection and tumours were weighted. For experimental metastasis assays, 5 × 105 A375P, MA-2 or MC-1 cells (in PBS) were injected into the tail vein of 7-week-old female CB.17 SCID mice (Charles River Laboratories). Mice were dissected 7 (MA-2 and MC-1) or 9 (A375P) weeks later and lung macrometastases counted in fresh total lungs using a Nikon SMZ1000 stereomicroscope. Spontaneous metastases were evaluated in 14-week-old female immunocompetent BALB/c mice injected with 5 × 105 4T1 cells (in PBS) transduced with pLemiR-214 or pLemiR-empty vectors, expressing turbo red-fluorescent protein (tRFP), in the fourth left mammary gland fat pad and dissected 3 weeks later. Red-fluorescent metastatic foci were counted in the fresh total lungs using a Leica MZ16F fluorescence stereomicroscope. For all metastasis studies, lungs were formalin fixed, cut in small pieces and paraffin embedded, sectioned and haematoxylin and eosin (H&E) stained. Micrometastases were evaluated on specimens, with an Olympus BH2 microscope, on at least three different sections.
In vivo extravasation assay
In all, 1.6 × 106 A375P, MA-2 or MC-1 cells, previously labelled with CellTracker™ Orange CMRA (Molecular Probes, Invitrogen Life Technologies) and resuspended in PBS were injected into the tail vein of 7-week-old female CD1 nude mice (Charles River Laboratories). After 2 or 48 h, mice were sacrificed and 4% paraformaldehyde was injected into the trachea. Total lungs were dissected and photographed using a Leica MZ16F fluorescence stereomicroscope and red-fluorescent (CMRA) cells were counted 48 h following injections using the ImageJ software (http://rsbweb.nih.gov/ij/). Lungs were included in freezing resin (OCT Killik, Bio-Optica, IT) and cryostat-cut in 6 μm thick sections. For immunofluorescent stainings for blood vessels, samples were acetone fixed for 10 min, blocked in 5% bovine serum albumin (Sigma-Aldrich) for 1 h, incubated with anti-Von Willebrandt Factor (vWF) primary antibody (1:50 dilution) for 1 h and anti-rabbit Alexa-Fluor-488 secondary antibody for 30 min and counterstained with DAPI (Sigma-Aldrich) for 5 min. Specimens were examined and photographed using a Zeiss AxioObserver microscope with the ApoTome Module.
Luciferase assays
In all, 5 × 104 cells were cotransfected with 50 ng of the pMIR-REPORT™ (Ambion) Firefly luciferase constructs containing the 3′UTRs of the indicated miR-214 potential target genes and 20 ng of pRL-TK Renilla Luciferase normalization control (Promega, Madison, WI), using Lipofectamine2000 (Invitrogen Life Technologies). Lysates were collected 48–72 h after transfection and Firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter System (Promega).
Protein preparation and immunoblotting
Total protein extracts were obtained using a boiling buffer containing 0.125 M Tris/HCl, pH 6.8 and 2.5% sodium dodecyl sulphate (SDS). In all, 25 or 50 μg proteins were separated by SDS polyacrylamide gel electrophoresis (PAGE) and electroblotted onto polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA). Membranes were blocked in 5% non-fat milk Tris-buffered saline (TBS)–Tween buffer (137 mM NaCl, 20 mM Tris/HCl, pH 7.6, 0.1% Tween-20) for 1 h at 37°C, then incubated with appropriate primary and secondary antibodies in 1% milk TBS–Tween buffer, respectively, overnight at 4°C and for 1 h at room temperature and visualized by enhanced chemiluminescence (ECL®, Amersham Biosciences, Piscataway, NJ).
Proteomic analysis
The expression profile of a panel of 119 soluble receptors expressed by non-hematopoietic cells present in the Human Soluble Receptor Array Non-Hematopoietic Panel (R&D Systems, Minneapolis, MN) was evaluated in total protein extracts of MA-2 cells 72 h after transfection of pre-control or pre-miR-214, according to the manufacturer's instructions.
Ingenuity pathway analysis systems
The Ingenuity Pathways Knowledge Base (http://www.ingenuity.com) is currently the world's largest database of knowledge on biological networks, with annotations organized by experts. We exploited this database to look for functional network connections among the miR-214-modulated genes.
Collection and analyses of human melanoma samples
Archival melanoma tumour samples (13 in situ melanomas, 57 primary melanomas and 18 cutaneous metastases) were collected from the Ist. Dermatologic Clinic of the University of Torino and institutional approvals were obtained for all samples. As reference, seven healthy skin samples were included in the analysis. Tumour tissue harvested from patients was quickly frozen or OCT embedded and stored at −80°C, or it was formalin fixed and paraffin embedded (FFPE). Total RNA from frozen and/or OCT-embedded samples was isolated via TRIzol Reagent (Invitrogen Life Technologies), according to the manufacturer's guidelines. Briefly, each sample was homogenized in the denaturing lysis solution and an acid-phenol:chloroform extraction followed. RNA extraction from FFPE samples was performed using the miRNeasy FFPE Kit (Qiagen, Stanford, CA), according to the manufacturer's guidelines. Briefly, samples were treated with the lysis buffer with proteinase K, then incubated at 80°C to reverse formalin crosslinking. Genomic DNA was then removed using gDNA Eliminator spin columns, and the concentrated RNA was purified using RNeasy MinElute spin columns. miR-214 and miR-210 expression was evaluated by qRT–PCR as described above, starting from 10 ng of total RNA. Quantitative normalization was performed on the expression of the RNU44 small nucleolar RNA. Relative expression was calculated using as reference the median expression of miR-214 and miR-210 in the healthy skin samples. Two-tailed Mann–Whitney non-parametric statistical test was used to assess significant differences with a P-value <0.05 considered significant.
Analysis of human melanoma data sets
Expression data for human primary melanomas, nevi and normal skin were obtained from the GEO series associated with Talantov et al (2005), Accession GSE3189, log-transformed and variance stabilized by adding a constant equal to 16 before taking the logarithm. The samples were clustered using Pearson correlation-based hierarchical clustering. Clustering and heatmap were obtained using Bioconductor (Gentleman et al, 2004).
Analysis of miR-214 genomic locus
Genomic DNA was isolated from melanoma cell lines using the Qiagen QIAmp DNA Mini kit (Qiagen, Valencia, CA). PCRs were performed using MyiQ Single Color Real time PCR Detection System (Bio-Rad) in 96-well plates, with reaction mixture (25 μl) containing 20 ng of DNA and 12.5 μl of ABsolute Blue QPCR SYBR Green Mix (Thermo Scientific, Waltham, MA). Triplicate reactions were run with specific primers for all samples using the PCR protocol: 95°C for 15 min, 40 cycles of 95°C for 15 s, 55°C 30 s and 72°C for 30 s, followed by melting curve analysis to ensure amplification specificity. Under the selected conditions, only single peaks were detected in the melting curve. The products of the PCRs were also run on a 2% agarose gel to confirm the correct size of the amplified product. To determine the genetic load, the ΔΔCT method was used according to Pfaffl (2001) and assuming 100% efficiency in the amplification from normal human genomic DNA (Promega). UBE2E1 housekeeping gene was used as reference. This gene has not been found genetically altered in melanomas (Lazar et al, 2009).
Statistical analyses
Unless otherwise noted, data are presented as mean±s.e.m. and two-tailed Student's t-test was used for comparison, with *P<0.05; **P<0.01; ***P<0.001 considered to be statistically significant. NS indicates a not statistically significant P-value.
Supplementary Material
Acknowledgments
This work was supported by grants from the University of Torino (Local Research Funding 2007/DT, 2008/DT), Regione Piemonte Ricerca Scientifica Applicata (CIPE2004/DT), Compagnia di San Paolo (DT), Torino, PRIN 2008 (DT), AIRC 2010 (IG 10104 DT) and FIRB giovani 2008 (RBFR08F2FS-002 FO). FO and DC are fellows of the Regione Piemonte. We thank Lei Xu and Richard Hynes for giving us the A375P cells and its metastatic variants; Paola Circosta and Alessandro Cignetti for providing several melanoma cell lines; Helen Hurst for the pSP(RSV)-NN and pSP(RSV)-TFAP2C vectors; Mike DiPersio for the anti-ITGA3 antibody and the pCMVzeo-ITGA3 vector; Luca Primo for preparing HUVECs–GFP cells; Jacek Krol for advices with sponges; Roberto Chiarle for help with the tumour samples; Nancy Hynes, Pier Paolo Pandolfi and Richard Hynes for critical reading of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bar-Eli M (2001) Gene regulation in melanoma progression by the AP-2 transcription factor. Pigment Cell Res 14: 78–85 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R, Imhof A, Pscherer A, Kopp H, Moser M, Seegers S, Kerscher M, Tainsky MA, Hofstaedter F, Buettner R (1994) The genomic structure of the human AP-2 transcription factor. Nucleic Acids Res 22: 1413–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti S, Comoglio PM (2007) The MET receptor tyrosine kinase in invasion and metastasis. J Cell Physiol 213: 316–325 [DOI] [PubMed] [Google Scholar]
- Berger AJ, Davis DW, Tellez C, Prieto VG, Gershenwald JE, Johnson MM, Rimm DL, Bar-Eli M (2005) Automated quantitative analysis of activator protein-2alpha subcellular expression in melanoma tissue microarrays correlates with survival prediction. Cancer Res 65: 11185–11192 [DOI] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA (2007) MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 8: R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Mangelsdorf DJ (2003) Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 1: e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC (1996) A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene 13: 1701–1707 [PubMed] [Google Scholar]
- Bosher JM, Williams T, Hurst HC (1995) The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA 92: 744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circosta P, Granziero L, Follenzi A, Vigna E, Stella S, Vallario A, Elia AR, Gammaitoni L, Vitaggio K, Orso F, Geuna M, Sangiolo D, Todorovic M, Giachino C, Cignetti A (2009) T cell receptor (TCR) gene transfer with lentiviral vectors allows efficient redirection of tumor specificity in naive and memory T cells without prior stimulation of endogenous TCR. Hum Gene Ther 20: 1576–1588 [DOI] [PubMed] [Google Scholar]
- Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Munoz W, Khokha R (2008) The role of tissue inhibitors of metalloproteinases in tumorigenesis and metastasis. Crit Rev Clin Lab Sci 45: 291–338 [DOI] [PubMed] [Google Scholar]
- Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F (2009) MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci USA 106: 21179–21184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier JS, Cheresh DA (2010) Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 10: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPersio CM, Shah S, Hynes RO (1995) alpha 3A beta 1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J Cell Sci 108(Pt 6): 2321–2336 [DOI] [PubMed] [Google Scholar]
- Duffy MJ, McKiernan E, O’Donovan N, McGowan PM (2009) Role of ADAMs in cancer formation and progression. Clin Cancer Res 15: 1140–1144 [DOI] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H (2005) The AP-2 family of transcription factors. Genome Biol 6: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandarpour M, Kiaii S, Zhu C, Castro J, Sakko AJ, Hansson J (2005) Suppression of oncogenic NRAS by RNA interference induces apoptosis of human melanoma cells. Int J Cancer 115: 65–73 [DOI] [PubMed] [Google Scholar]
- Fei J, Lan F, Guo M, Li Y, Liu Y (2008) Inhibitory effects of anti-miRNA oligonucleotides (AMOs) on A549 cell growth. J Drug Target 16: 688–693 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG (2007) Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ (2008) DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol 18: 1456–1465 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenwald JE, Sumner W, Calderone T, Wang Z, Huang S, Bar-Eli M (2001) Dominant-negative transcription factor AP-2 augments SB-2 melanoma tumor growth in vivo. Oncogene 20: 3363–3375 [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J (2006) Cancer metastasis: building a framework. Cell 127: 679–695 [DOI] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, Buettner R (2000) Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene 260: 1–12 [DOI] [PubMed] [Google Scholar]
- Hornebeck W, Emonard H, Monboisse JC, Bellon G (2002) Matrix-directed regulation of pericellular proteolysis and tumor progression. Semin Cancer Biol 12: 231–241 [DOI] [PubMed] [Google Scholar]
- Huang S, Jean D, Luca M, Tainsky MA, Bar-Eli M (1998) Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. EMBO J 17: 4358–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Taverna D, Harwerth IM, Ciardiello F, Salomon DS, Yamamoto T, Groner B (1990) Epidermal growth factor receptor, but not c-erbB-2, activation prevents lactogenic hormone induction of the beta-casein gene in mouse mammary epithelial cells. Mol Cell Biol 10: 4027–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S (2010) MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Iyer V, Pumiglia K, DiPersio CM (2005) Alpha3beta1 integrin regulates MMP-9 mRNA stability in immortalized keratinocytes: a novel mechanism of integrin-mediated MMP gene expression. J Cell Sci 118(Pt 6): 1185–1195 [DOI] [PubMed] [Google Scholar]
- Jeanes A, Gottardi CJ, Yap AS (2008) Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27: 6920–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V (2009) Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 36: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karjalainen JM, Kellokoski JK, Mannermaa AJ, Kujala HE, Moisio KI, Mitchell PJ, Eskelinen MJ, Alhava EM, Kosma VM (2000) Failure in post-transcriptional processing is a possible inactivation mechanism of AP-2alpha in cutaneous melanoma. Br J Cancer 82: 2015–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Boelte KC, Lin PC (2007) Endothelial cell adhesion molecules and cancer progression. Curr Med Chem 14: 377–386 [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, Oertner TG, Schratt G, Bibel M, Roska B, Filipowicz W (2010) Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141: 618–631 [DOI] [PubMed] [Google Scholar]
- Kueng W, Silber E, Eppenberger U (1989) Quantification of cells cultured on 96-well plates. Anal Biochem 182: 16–19 [DOI] [PubMed] [Google Scholar]
- Lazar V, Ecsedi S, Szollosi AG, Toth R, Vizkeleti L, Rakosy Z, Begany A, Adany R, Balazs M (2009) Characterization of candidate gene copy number alterations in the 11q13 region along with BRAF and NRAS mutations in human melanoma. Mod Pathol 22: 1367–1378 [DOI] [PubMed] [Google Scholar]
- Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB (2009) Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res 37: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Sharma A, Peng HH, Robertson G, Dong C (2007) Targeting mutant (V600E) B-Raf in melanoma interrupts immunoediting of leukocyte functions and melanoma extravasation. Cancer Res 67: 5814–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo N, Yanez-Mo M, Mittelbrunn M, de la Rosa G, Munoz ML, Sanchez-Madrid F, Sanchez-Mateos P (2001) Regulatory role of tetraspanin CD9 in tumor-endothelial cell interaction during transendothelial invasion of melanoma cells. Blood 98: 3717–3726 [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova VO, Bar-Eli M (2008) Transcriptional control of the melanoma malignant phenotype. Cancer Biol Ther 7: 997–1003 [DOI] [PubMed] [Google Scholar]
- Mitchell K, Svenson KB, Longmate WM, Gkirtzimanaki K, Sadej R, Wang X, Zhao J, Eliopoulos AG, Berditchevski F, Dipersio CM (2010) Suppression of integrin alpha3beta1 in breast cancer cells reduces cyclooxygenase-2 gene expression and inhibits tumorigenesis, invasion, and cross-talk to endothelial cells. Cancer Res 70: 6359–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DW, Bosserhoff AK (2009) Role of miRNAs in the progression of malignant melanoma. Br J Cancer 101: 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld G, Kessler O (2008) The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat Rev Cancer 8: 632–645 [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massague J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9: 274–284 [DOI] [PubMed] [Google Scholar]
- Orso F, Penna E, Cimino D, Astanina E, Maione F, Valdembri D, Giraudo E, Serini G, Sismondi P, De Bortoli M, Taverna D (2008) AP-2alpha and AP-2gamma regulate tumor progression via specific genetic programs. FASEB J 22: 2702–2714 [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primo L, di Blasio L, Roca C, Droetto S, Piva R, Schaffhausen B, Bussolino F (2007) Essential role of PDK1 in regulating endothelial cell migration. J Cell Biol 176: 1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasola A, Geuna M (2001) A flow cytometry assay simultaneously detects independent apoptotic parameters. Cytometry 45: 151–157 [DOI] [PubMed] [Google Scholar]
- Segura MF, Belitskaya-Levy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, Osman I, Hernando E (2010) Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res 16: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E (2009) Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA 106: 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN (2007) Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67: 11612–11620 [DOI] [PubMed] [Google Scholar]
- Steeg PS (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 12: 895–904 [DOI] [PubMed] [Google Scholar]
- Swart GW, Lunter PC, Kilsdonk JW, Kempen LC (2005) Activated leukocyte cell adhesion molecule (ALCAM/CD166): signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev 24: 223–236 [DOI] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y (2005) Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res 11: 7234–7242 [DOI] [PubMed] [Google Scholar]
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM (2010) Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 11: 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA (2009) A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137: 1032–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Valastyan S, Weinberg RA (2009) MicroRNAs: crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle 8: 3506–3512 [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T (2008) Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn 237: 3738–3748 [DOI] [PubMed] [Google Scholar]
- Worley LA, Long MD, Onken MD, Harbour JW (2008) Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res 18: 184–190 [DOI] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37 (Database Issue): D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Luca M, Huang S, Gutman M, Reich R, Johnson JP, Bar-Eli M (1997) Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res 57: 2295–2303 [PubMed] [Google Scholar]
- Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, Wagner SN, Ramaswamy S, Mesirov JP, Hynes RO (2008) Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res 6: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ (2008) MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 68: 425–433 [DOI] [PubMed] [Google Scholar]
- Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, Rutherford T, Mor G (2010) TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene 29: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O’Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G (2006) microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 103: 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.