Abstract
Precise 5′ splice-site recognition is essential for both constitutive and regulated pre-mRNA splicing. The U1 small nuclear ribonucleoprotein particle (snRNP)-specific protein U1C is involved in this first step of spliceosome assembly and important for stabilizing early splicing complexes. We used an embryonically lethal U1C mutant zebrafish, hi1371, to investigate the potential genomewide role of U1C for splicing regulation. U1C mutant embryos contain overall stable, but U1C-deficient U1 snRNPs. Surprisingly, genomewide RNA-Seq analysis of mutant versus wild-type embryos revealed a large set of specific target genes that changed their alternative splicing patterns in the absence of U1C. Injection of ZfU1C cRNA into mutant embryos and in vivo splicing experiments in HeLa cells after siRNA-mediated U1C knockdown confirmed the U1C dependency and specificity, as well as the functional conservation of the effects observed. In addition, sequence motif analysis of the U1C-dependent 5′ splice sites uncovered an association with downstream intronic U-rich elements. In sum, our findings provide evidence for a new role of a general snRNP protein, U1C, as a mediator of alternative splicing regulation.
Keywords: snRNP, splicing, zebrafish
Introduction
The majority of eukaryotic protein-coding genes contains intron sequences that have to be removed from messenger RNA precursors (pre-mRNA) to create a continuous and translatable open-reading frame. The cotranscriptional process of intron excision is catalysed by a dynamic macromolecular complex called spliceosome, which consists of small nuclear ribonucleoprotein particles (snRNPs) that assemble in a coordinated stepwise manner onto the nascent transcript. In the first step, the 5′ splice site is recognized by the U1 snRNP, and the branch point and polypyrimidine tract are bound by SF1/BBP and the U2AF heterodimer, respectively, to form the E complex, which commits the pre-mRNA to the splicing pathway. The recruitment of the U2 snRNP to the branch point region converts the E complex into the pre-spliceosomal A complex. Next, the pre-assembled U4/U6.U5 tri-snRNP joins to form the B complex, which undergoes several conformational rearrangements and compositional changes to become catalytically active, generating the C complex, in which both catalytic steps occur. Finally, the spliceosome disassembles and releases the mature mRNA (reviewed by Brow, 2002; Nilsen, 2003; Wahl et al, 2009).
Pre-mRNA splicing is a dynamic process and—particularly in higher eukaryotes—stringently regulated, based on the flexible recognition of splice sites (Black, 2003; Nilsen and Graveley, 2010). Most of the alternative splicing processes are regulated by trans-acting factors that belong to either the family of serine–arginine-rich (SR) proteins or the heterogeneous nuclear ribonucleoproteins (hnRNPs). Alternative splicing provides probably the most important mechanism to increase the functional complexity of higher eukaryotes by expanding the proteomic diversity. Recent studies indicate that most human protein-coding genes undergo alternative splicing (Wang and Burge, 2008) and additionally, defects in alternative splicing are relevant to numerous human disorders (reviewed by Buratti et al, 2006; Cooper et al, 2009; Tazi et al, 2009). In the past, researchers studied the function of various splicing regulators by focussing on a few model genes. More recently, genomewide approaches attempt to integrate the functions of different factors to describe global networks, giving insight into how multiple factors coordinately regulate alternative splicing through tissue- and development-specific mechanisms. These approaches aim at deciphering the splicing code and its underlying rules, and at correctly predicting alternative splicing patterns by computational analysis of several intrinsic features of any gene of interest (Blencowe, 2006; Ben-Dov et al, 2008; Barash et al, 2010).
For both constitutive and regulated alternative splicing, the initial 5′ splice-site recognition by the U1 snRNP is particularly important. The U1 snRNP contains, in addition to the U1 snRNA, a common set of seven Sm proteins and three specific proteins, U1-70K, U1A, and U1C. However, not all U1 snRNP-5′ splice-site interactions result in productive splicing, and RNA–RNA base pairing is certainly not sufficient for stable U1 snRNP binding and correct 5′ splice-site recognition (Zhuang and Weiner, 1986; Siliciano and Guthrie, 1988; Séraphin et al, 1988; Lund and Kjems, 2002; Lacadie and Rosbash, 2005; Hage et al, 2009). Specifically, both U1 snRNP constituents and U1 snRNP-interacting factors contribute to these early events in splice-site definition; excess of SR proteins, for example, can compensate for a lack of U1 snRNP-5′ splice-site interaction in vitro (Crispino et al, 1994; Tarn and Steitz, 1994). Du and Rosbash (2002) described that U1 snRNP particles lacking the 5′ end of the snRNA retain 5′ splice-site specificity and that recombinant yeast U1C is capable of selecting 5′ splice-site-like sequences independently of the snRNP. U1C was shown to stimulate E complex formation by stabilizing the base pairing between the 5′ end of the U1 snRNA and the 5′ splice-site region (Heinrichs et al, 1990; Will et al, 1996; Chen et al, 2001). U1C depletion in yeast affects pre-mRNA splicing in vivo, and extracts from U1C-deficient strains form low levels of commitment complexes and spliceosomes in vitro (Tang et al, 1997). A particularly important role of U1C in 5′ splice-site recognition is also supported by recent structural studies on the U1 snRNP: U1C was localized in close proximity to the 5′ end of the snRNA; moreover, certain amino-acid residues of U1C interact with the minor groove at the U1 snRNA-5′ splice-site duplex (Stark et al, 2001; Pomeranz Krummel et al, 2009). Taken together, these observations strongly suggest that interactions between U1C and the 5′ splice site may precede base pairing between the pre-mRNA and the U1 snRNA, providing a potential additional regulatory step in 5′ splice-site selection.
In the last decades, the zebrafish Danio rerio has become a very powerful model system to investigate vertebrate development and other complex biological processes, including human diseases (reviewed by Ackermann and Paw, 2003; Amsterdam and Hopkins, 2006). Our recent studies on the snRNP recycling factor p110 (Trede et al, 2007) have proven that genomewide studies on splicing defects are feasible in zebrafish and allow investigating the phenotypic consequences of specific splicing factor mutations on vertebrate development.
Here, we have focussed on the U1 snRNP-specific protein U1C, which is particularly interesting, since it may be directly involved in 5′ splice-site selection (see above). To investigate on a genomewide level, the potential role of U1C as a splicing regulator, we have made use of an embryonically lethal U1C mutant zebrafish, hi1371, which originates from a large insertional mutagenesis screen for genes essential in early zebrafish development (Golling et al, 2002; Amsterdam et al, 2004). We report here, that hi1371 mutant zebrafish embryos carry a stable, U1C-deficient U1 snRNP. To answer the question, whether U1C-deficiency results in aberrant splicing patterns, we performed high-throughput sequencing (RNA-Seq) of total RNA from wild-type versus mutant zebrafish embryos. As a result, we identified a specific set of target genes that display U1C-dependent splicing alterations, which appear to be associated with a U-rich intronic sequence motif. In sum, our study yielded new insights into the regulation of 5′ splice-site selection and evidence for a role of a general spliceosome component in alternative splicing.
Results
Hi1371 mutant zebrafish contain U1C-deficient U1 snRNPs
Based on a large insertional mutagenesis screen, >300 genes were identified that are essential in early zebrafish development (Golling et al, 2002; Amsterdam et al, 2004). In one mutant line, hi1371, a retroviral insertion was mapped within intron 1 of the U1C gene; the loss of U1C leads to severe phenotypic defects, including wide-range necrosis in the central nervous system and misdevelopment of several organs, resulting in an early embryonic lethality at about 5 days post-fertilization (dpf) (Golling et al, 2002).
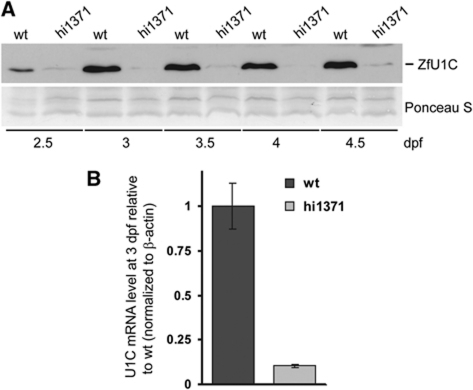
Western blot analysis of mutant embryos at early developmental stages from 2.5 to 4.5 dpf (Figure 1A) showed that endogenous U1C protein was almost undetectable in comparison to the respective wild-type individuals of the same age. Consistent with that, very low levels of U1C mRNA were measured by quantitative real-time PCR (Figure 1B). These very low residual levels of U1C protein and mRNA are most likely due to maternal contribution, supported by the microarray-based transcriptome analysis of zebrafish embryogenesis (Mathavan et al, 2005).
Figure 1.
U1C protein and mRNA levels in U1C mutant zebrafish embryos. (A) Wild-type (wt) and U1C mutant (hi1371) zebrafish embryos were collected every 12 h from 2.5 to 4.5 dpf and total embryo lysates were analysed by SDS–PAGE and western blot, using ZfU1C polyclonal antibody (top panel); Ponceau S staining of the western membrane is shown as loading control (bottom panel). (B) Total RNA from wild-type (dark grey) and mutant embryos (light grey) was isolated at 3 dpf, and endogenous U1C mRNA levels were measured by real-time RT–PCR.
For our studies, we chose 3-day-old embryos, because at that time the phenotype can be correctly distinguished between wild-type and mutant individuals; at the same time, the defects are mild enough to ensure that the effects observed are mainly primary and not due to the overall misdevelopment. Additionally, RT–PCR analysis revealed that several of our target genes were not expressed at 2 dpf even in the wild-type (data not shown), supporting the notion that our target genes are directly affected by the loss of U1C.
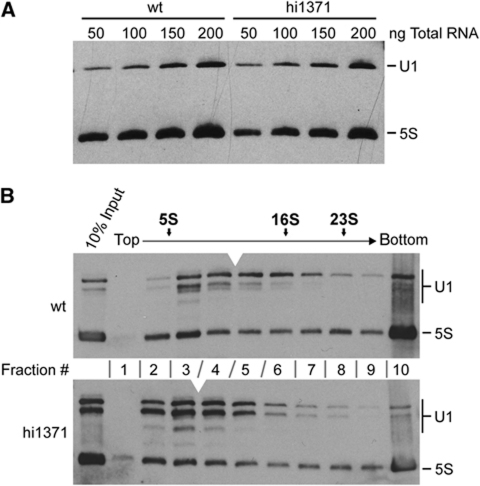
Despite the strong reduction of U1C protein levels, the U1 snRNA steady-state levels appear to be unaffected by the U1C knockout, as shown by northern blot analysis of total RNA (Figure 2A). However, glycerol gradient centrifugation revealed an aberrant sedimentation behaviour for the U1 snRNP in U1C mutant embryos (Figure 2B): While the U1 snRNP from wild-type embryo lysates sedimented in fractions #3–7, the U1 snRNP in mutant embryo lysates was detected in fractions #2–6. We conclude that the loss of the U1C protein results in a shift of the U1 snRNA peak by one fraction towards the top of the gradient, consistent with a stable, but U1C-deficient snRNP. In contrast to the U1 snRNP, none of the other snRNPs changed significantly in its sedimentation, comparing lysates from wild-type and U1C mutant embryos (Supplementary Figure S1).
Figure 2.
U1C deficiency does not change the U1 snRNA steady-state levels nor does it destabilize the U1 snRNP. (A) To assay the steady-state levels of U1 snRNA, 50–200 ng of total RNA (as indicated above the lanes) were prepared from either wild-type (wt) or U1C mutant embryos (hi1371) at 3 dpf and analysed by northern blotting with probes specific for U1 snRNA and, as a loading control, 5S rRNA (as marked on the right). (B) Lysates from either wild-type (upper panel) or hi1371 mutant embryos (lower panel) at 3 dpf were fractionated through a linear 10–30% glycerol gradient (#1–10 indicated between the panels, the positions of sedimentation markers above). RNA was isolated from each fraction and analysed by northern blot hybridization with probes specific for U1 snRNA and 5S rRNA (positions indicated on the right). The peak of the U1 snRNA is marked in each panel by the white arrowhead.
In northern blot analysis of U1 snRNA prepared from embryo lysates we consistently detected an additional, heterogeneous band, which ran below the major, full-length U1 snRNA species and was much more prominent in mutant than in wild-type embryo lysates (Figure 2B, compare inputs). This contrasts with direct RNA isolation from embryos, where we observed only one discrete band for the U1 snRNA (Figure 2A). RACE experiments revealed that the shorter U1 snRNA species are truncated at their 5′ ends (data not shown). Since the single-stranded 5′-terminal sequence of U1 snRNA is likely bound by U1C within the U1 snRNA-pre-mRNA duplex (Pomeranz Krummel et al, 2009), the enhanced susceptibility of that region to RNA degradation suggests a protective function of U1C within the complex.
Global RNA-Seq analysis identifies a specific set of U1C-dependent splicing events in zebrafish
Since U1C has been implicated in 5′ splice-site selection (see Introduction), we next assayed whether the absence of U1C protein in the hi1371 mutant zebrafish embryos changes global alternative splicing patterns, using high-throughput sequencing (Solexa RNA-Seq). Total RNA from wild-type and mutant embryos at the age of 3 dpf was processed through standard protocols from Illumina, yielding a total of 67.3 millions 76 bp single-end sequence reads (31.8 millions from mutant, 35.5 millions from wild-type).
We looked for alternative splicing changes between these two samples in these six modes: single- and multiple-exon skipping, intron retention, alternative 5′ and 3′ splice-site usage, and mutually exclusive exons. A data analysis procedure was developed to predict U1C-dependent alternative splicing targets, consisting of the following five stages (for details, see Supplementary data):
alignment (both junction and non-junction) and mapping of sequence reads to the annotated zebrafish refSeq genes,
calculating the read-density (i.e. sequence-read coverage) of exonic and intronic regions as mRNA expression index,
measuring junction-count (number of sequence reads spanning a specific splice junction) to predict the alternative splicing mode,
calculating the ratio of the read-density of each exon or intron and the junction-count of each splice junction between the two samples,
defining two information groups for each of the alternative splicing modes (e.g. for exon inclusion and skipping information), and quantitating these values as index of expression changes between the alternative isoforms, thereby defining parameters of reciprocal effects for target prediction.
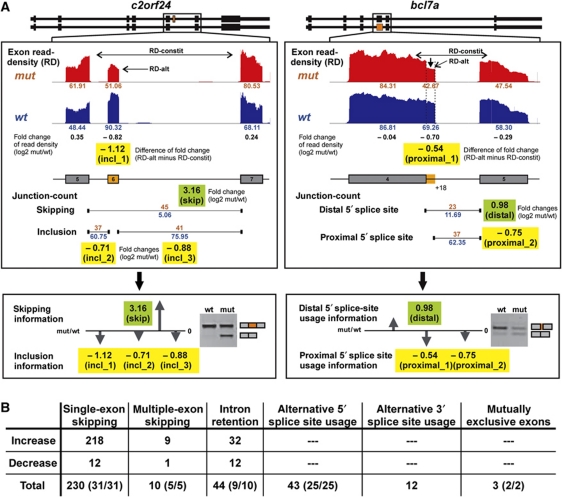
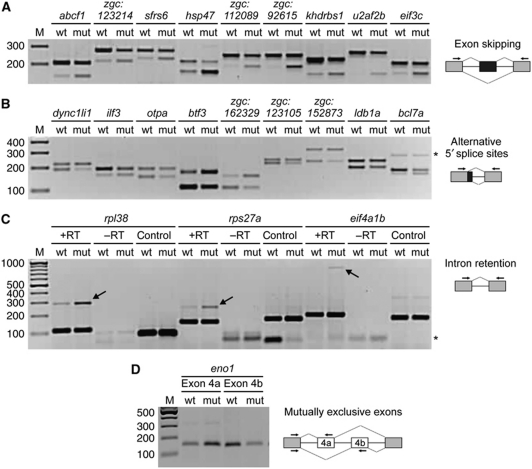
Figure 3A gives two representative examples of target prediction and RT–PCR validation: c2orf24 (exon skipping, left panel) and bcl7a (alternative 5′ splice sites, right panel). As a result, we predicted ∼350 U1C-dependent targets distributed to the six different alternative splicing modes mentioned above (Figure 3B; Supplementary Tables S3–S7). A total of 73 targets were selected for validation by semiquantitative RT–PCR. Figure 4 shows 22 RT–PCR validations with examples for four alternative splicing modes: single-exon skipping, alternative 5′ splice-site use, intron retention, and mutually exclusive exons (for the complete set of positive validation reactions, see Supplementary Figure S2). In sum, 72 of the 73 targets were positively validated.
Figure 3.
RNA-Seq-based selection of alternative splicing targets. (A) Our strategy to identify alternative splicing targets based on RNA-Seq data is summarized for two representative examples, c2orf24 (exon skipping) and bcl7a (alternative 5′ splice-site usage). For a detailed description, see Supplementary data. Exon–intron structures of both splicing isoforms for these two genes are given on top with the region boxed, which is analysed in detail below. c2orf24 (single-exon skipping; left panels): Read-density values of constitutive exons (RD-constit) and alternative exon (RD-alt) as well as junction-count values of skipping and inclusion junction reads are given in red (mut, mutant embryo) and blue (wt, wild-type embryo). The skipping information (green box) is the fold change (log2 mut/wt) of skipping junction-count. Shown in yellow boxes are three values of inclusion information: incl_1 (difference of fold change between RD-alt and RD-constit), incl_2 and incl_3 (fold changes of inclusion junction-count). The reciprocal effect of increased skipping information and decreased inclusion information predict an increase of the exon-skipping isoform in the mutant embryo. For direct comparison, RT–PCR validation is included on the right. bcl7a (alternative 5′ splice-site usage; right panels): Similarly as described above, the values of read-density and junction-count are shown. The distal 5′ splice-site usage information (green box) is the fold change (log2 mut/wt) of junction-count values for the distal splice sites. Shown in yellow boxes are two values of proximal 5′ splice-site usage information: proximal_1 (difference of fold change between RD-alt and RD-constit), proximal_2 (fold change of junction-count value for proximal 5′ splice site). The reciprocal effect of increased distal 5′ splice-site usage information and decreased proximal 5′ splice-site usage information predicts an increase of the shorter isoform with the distal 5′ splice site in the mutant embryo. For direct comparison, RT–PCR validation is included on the right. (B) Distribution of the 342 predicted targets in these six alternative splicing modes: single-exon skipping, multiple-exon skipping, intron retention, alternative 5′ or 3′ splice-site usage, and mutually exclusive exons, sorted by the effect (increase or decrease) observed in the U1C mutant zebrafish. In addition, the numbers of targets, which are positively validated among all targets selected for RT–PCR validation, are given in brackets (positive/total). For a complete list of all targets, see Supplementary Tables S3–S7.
Figure 4.
Genomewide effects of U1C deficiency on alternative splicing in the zebrafish. (A–D) Alternative splicing of selected target genes (names above the lanes) were analysed by RT–PCR, using total RNA from wild-type (wt) and U1C mutant (mut) embryos at 3 dpf and specific primer sets (arrows in the schematics on the right). M, DNA size markers (sizes in bp). (A) Increased exon skipping of nine target genes in the absence of U1C. Top and lower bands represent the exon inclusion and skipping products, respectively. (B) Alternative 5′ splice-site usage of nine target genes in the absence of U1C. Top and lower bands reflect usage of the proximal and distal 5′ splice sites, respectively. The asterisk marks an intron-retention product for bcl7a. (C) Three examples of increased intron retention in the absence of U1C. +RT refers to the validation reaction itself, and –RT represents the respective control reaction without reverse transcriptase. In addition, the intron specificity of the U1C effect was tested by amplifying another intron in the same gene (lanes control; see Results for the identities of the introns assayed). The arrows point to the intron-retention products, the asterisk to primer dimers. (D) Example for U1C-dependent mutually exclusive exons (indicated with 4a and 4b in the schematic and above the respective reaction).
The largest group of target genes exhibited increased exon skipping in the absence of U1C, which are 218 out of 230 cases for single-exon skipping and 9 out of 10 cases for multiple-exon skipping (Figure 3B). We show nine examples of RT–PCR validations (abcf1, zgc:123214, sfrs6, hsp47, zgc:112089, zgc:92615, khdrbs1, u2afb, and eif3c; Figure 4A); in each case, primers were designed to flank the target exon to amplify two different products, representing exon inclusion and skipping isoforms. The exon-skipping product was always more prominent for the mutant than for the wild-type, and in several cases, like zgc:92615 and u2af2b, exclusively detectable in the absence of U1C. This points to widespread effects on splice-site recognition in the U1C mutant.
Figure 4B summarizes nine examples of target genes that display an influence of U1C on alternative 5′ splice-site choice: dync1li1, ilf3, otpa, btf3, zgc:162329, zgc:123105, zgc:152873, ldb1a, and bcl7a. Here, the primers are located in the exon containing the alternative 5′ splice site and in the respective downstream exon, so that the two RT–PCR products reflect the usage of proximal and distal 5′ splice sites, respectively. In contrast to exon skipping, we found for alternative 5′ splice sites both cases, that is an increase in the use of the proximal or the distal alternative 5′ splice site in the U1C mutant (18 versus 8 cases, respectively). The 3.5-fold higher number of changes in 5′ rather than 3′ splice-site selection (Figure 3B) further confirmed that U1C directly participates in 5′ splice-site selection.
Next, a minor group of intron-retention cases was analysed, using primers in the two flanking exons (Figure 4C). In general, we see more cases of increased rather than decreased intron retention in the U1C mutant, which we could validate in most cases (see three examples, rpl38, rps27, and eif4a1b, lanes +RT). In each case, control reactions were included in the absence of reverse transcriptase (–RT) to exclude contaminations by genomic DNA. In another control reaction (control), we tested a different intron-spanning region in the same gene for its U1C dependency (intron 4 for rpl38, NM_00102486; intron 5 for rps27, NM_200502; intron 7 for eif4a1b, NM_201510): No differences between wild-type and mutant were found, demonstrating that these U1C-dependent intron-retention cases are intron specific.
Finally, we obtained evidence for U1C regulating mutually exclusive exons, of which Figure 4D shows one example (eno1). Using primer pairs specific for either one of the two mutually exclusive exons and the upstream exon, we amplified both isoforms separately and observed that the upstream exon 4a was preferred in the absence of U1C, while the downstream exon 4b was predominantly included in the wild-type.
Injection of ZfU1C cRNA rescues wild-type phenotype and restores splicing of target genes
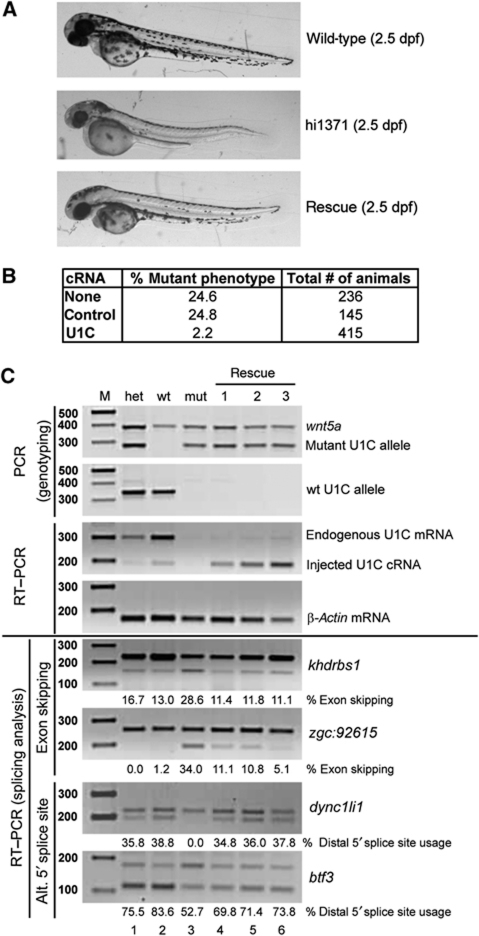
To address the question whether the phenotypic and splicing defects in the hi1371 mutant zebrafish really depend on the loss of U1C we performed rescue experiments. In vitro transcribed ZfU1C cRNA was injected into F1 embryos of heterozygous hi1371+/− individuals at the one-cell stage. 415 eggs were injected with the ZfU1C cRNA, 145 were treated with a control cRNA, and 236 were left untreated, serving as controls for the rescue and the injection procedure. At 2.5 dpf, the embryos were sorted according to their phenotypic appearance (Figure 5A). At this stage, the mutant phenotype is characterized by microphtalmia, a dorsally bent body axis, pericardic oedema, and reduced pigmentation, which we did not or only weakly observe in individuals after rescue (for a phenotypic description, see also Amsterdam et al, 2004). Control-cRNA-injected embryos showed about the same percentage (24.8%) of phenotypically mutant individuals as an uninjected clutch (24.6%); in contrast, injection of ZfU1C cRNA strongly reduced the number of phenotypic mutants (down to ∼2%), verifying that the phenotypic defects of the homozygous hi1371 mutants resulted from the loss of U1C protein (Figure 5B).
Figure 5.
Injection of ZfU1C cRNA rescues phenotypic defects of hi1371 mutant embryos and restores normal splicing patterns. (A) Phenotype at 2.5 dpf of wild-type (top panel) and hi1371 mutant embryos without (middle panel) or with cRNA injection (bottom panel). (B) Summary of cRNA injection experiments, comparing the score of mutant phenotype (in percentage) for uninjected embryos (none), or after control (control) and ZfU1C cRNA injection (U1C). (C) Rescue of splicing defects. (Upper four panels) To identify rescued individuals, DNA and RNA were isolated simultaneously from single zebrafish embryos (three examples rescue #1–3 shown); as controls, two wild-type embryos (het (heterozygous), lane 1; wt (homozygous), lane 2) and one mutant embryo (mut) were included in the analysis. DNA was used for genotyping, detecting by PCR separately the mutant and wild-type U1C alleles (first two panels; primers for wnt5a were included in the multiplex reaction as a PCR control). In addition, total RNA was subjected to RT–PCR analysis to detect the endogenous and the injected U1C RNA (third panel), using β-actin mRNA as a loading control (fourth panel). In the lower four panels, the rescue of splicing defects of four validated target genes was assayed by RT–PCR (gene names indicated on the right, alternative splicing types on the left); exon-skipping ratios and the use of the distal 5′ splice site are quantitated in percentage below each lane. M, DNA size markers (sizes in bp).
On the basis of the successful phenotypic rescue we asked the question whether the wild-type splicing pattern of U1C-dependent target genes was also restored. Therefore, all phenotypic wild-type individuals from one U1C cRNA injection were used to isolate DNA and RNA from single embryos. PCR on genomic DNA was carried out to detect the mutant allele (in the absence of the wild-type allele) to identify the U1C-knockout individuals among the rescued embryos (Figure 5C, panels PCR/genotyping). The injected U1C cRNA carries a shortened 3′ untranslated region (3′-UTR), allowing the detection of both endogenous and injected U1C mRNA by RT–PCR and two reverse primers specific for two 3′-UTR regions. In the semiquantitative RT–PCR on total RNA, the endogenous U1C mRNA levels differed a little between a heterzygous and a homozygous wild-type embryo, while an uninjected mutant showed no signal at all (Figure 5C, panels RT–PCR, lanes het, wt, and mut). The levels of injected U1C cRNA in the three rescued individuals ranged between the endogenous amount for the hetero- and the homozygous wild-type (Figure 5C, panels RT–PCR, lanes rescue 1–3).
Finally, we investigated the changes in alternative splicing of several target genes after rescue (Figure 5C, bottom four panels). For some target genes the expression at 2.5 dpf was too low to be detected properly by RT–PCR, independently of the loss of U1C (data not shown). Four examples are presented here, which showed sufficient expression and a clear difference in alternative splicing between wild-type and mutant embryos: two for exon skipping (khdrbs1 and zgc:92615) and two for alternative 5′ splice-site usage (dync1li1 and btf3). Comparing the splicing patterns in the rescued embryos (lanes 4–6) with those for wild-type and mutant individuals (lanes 1–3) we clearly observed that the wild-type splicing patterns were completely or at least partially restored, demonstrating that injection of ZfU1C cRNA was sufficient to rescue U1C-dependent splicing regulation.
U1C-dependent 5′ splice sites are associated with intronic U-rich sequence motif
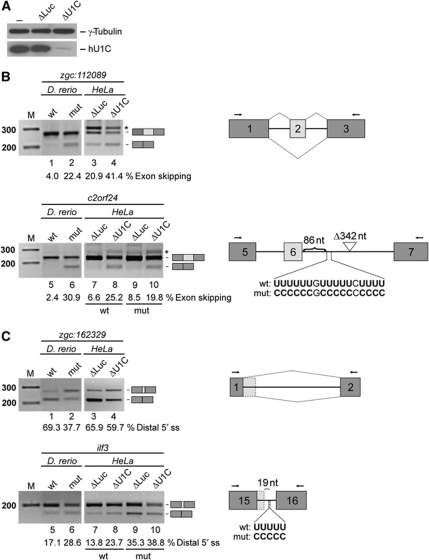
To further investigate the U1C dependency of a subclass of 5′ splice sites and the functional conservation of the effects described above, we used HeLa cells as a heterologous system for in vivo splicing assays. Minigene constructs of several validated zebrafish target genes were generated and—after siRNA-mediated U1C knockdown—transfected into HeLa cells (Figure 6; for biological replicates, see Supplementary Figure S3; for more details on the analysis of the U1C-deficient U1 snRNP after RNAi in HeLa cells, see Supplementary Figure S4). Figure 6A demonstrates that 3 days after siRNA transfection, the levels of U1C protein are reduced to <10%. At that time, the minigene constructs were transfected, and 24 h later total RNA was isolated for RT–PCR analysis of alternative splicing of the minigene (Figure 6B and C). We show two examples each for U1C-dependent exon skipping, zgc:112089 (Figure 6B, lanes 1–4) and c2orf24 (Figure 6B, lanes 5–10), and for alternative 5′ splice-site usage, zgc:162329 (Figure 6C, lanes 1–4) and ilf3 (Figure 6C, lanes 5–10). In all four cases, the RT–PCR analyses were directly compared with the validation RT–PCRs performed on zebrafish total RNA (compare panels labelled with D. rerio and HeLa). Regarding exon skipping, we found that, after transfection of these two zebrafish minigenes into HeLa cells, the skipping isoform was more pronounced in the U1C knockdown than in the control-siRNA-transfected cells, clearly reproducing the effects observed in U1C mutant and wild-type zebrafish, respectively (Figure 6B, compare lanes 1/2 with 3/4 and lanes 5/6 with 7/8). In addition, the splicing patterns of both minigenes with alternative 5′ splice sites responded to U1C knockdown in HeLa cells, resembling the effects observed for the zebrafish mutant (Figure 6C, compare lanes 1/2 with 3/4 and lanes 5/6 with 7/8).
Figure 6.
U1C knockdown in HeLa cells reproduces U1C-dependent alternative splicing changes in zebrafish and reveals functional role of associated U-rich elements. (A–C) Minigene constructs of four zebrafish target genes (zgc:112089, c2orf24, zgc:162329, and ilf3; as indicated above the panels) were transfected into HeLa cells after siRNA-mediated knockdown of U1C (ΔU1C) or luciferase (ΔLuc, as control). The splicing patterns were analysed by RT–PCR on total RNA, using specific primer sets (indicated with arrows in the schematics). RT–PCR reactions from HeLa cells (panels labelled HeLa) are compared side-by-side with the corresponding validation RT–PCRs from zebrafish embryos (panels labelled D. rerio; wild-type, wt, versus mutant, mut). The identities of the splicing products are depicted on the right of the gels. M, DNA size markers (sizes in bp). (A) Western blot of U1C knockdown in HeLa cells. At 72 h after siRNA transfection, whole-cell lysates were prepared and analysed by SDS–PAGE and western blot, detecting U1C and γ-tubulin. HeLa cells after U1C knockdown (ΔU1C), and as controls, luciferase-siRNA-treated (ΔLuc) and untransfected HeLa cells (−) were compared. (B) In vivo alternative splicing of two exon-skipping targets, zgc:112089 (lanes 1–4), and c2orf24 (lanes 5–10). Exon-skipping ratios are given in percentage below each lane. For c2orf24, the wild-type (wt) and a mutant construct (mut) were analysed (lanes 7–10); as shown in the schematic on the right, a U-rich element located 86 nt downstream of the 5′ splice site of exon 6 was mutated to a C-rich element; the asterisks point to unspecific PCR products. (C) In vivo splicing of two examples for alternative 5′ splice-site choice, zgc:162329 (lanes 1–4), and ilf3 (lanes 5–10). The use of the distal 5′ splice site is quantitated in percentage below each lane. For ilf3, the wild-type (wt) and a mutant construct (mut) were analysed (lanes 7–10); in the mutant construct, the U-stretch 19 nt downstream of the proximal 5′ splice site was substituted by a C-stretch (schematically shown on the right).
In order to identify sequence motifs that are common to the U1C-dependent alternative exons, we performed a computational analysis of intronic sequences flanking U1C-dependent versus U1C-independent cassette exons. We focussed on the 230 predicted single-exon skipping targets, from which 176 target exons were selected for further analysis that have GU 5′ splice sites and a downstream intron with a minimal length of 135 bp. This length requirement ruled out the possibility that the introns selected may be biased towards short introns. The alternative exons were compared with two control sets of exons: first, to their corresponding upstream exons, and second, to all zebrafish refGene exons, except for the first and last exons in each gene. We compared the following features: 5′ splice-site strength; sequence motifs enriched in exons; sequence motifs enriched in the first 100 nt of introns (see Supplementary data). As a result, no significant correlation was found between the 5′ splice-site strength and the distribution of regulated versus the other exons, nor could we detect any significantly enriched motif within the alternative exons (data not shown). However, this analysis revealed a significant enrichment of uridine stretches with a minimal length of four nucleotides within the first 100 nt downstream of position +6. To test whether this U-rich elements are involved in U1C-dependent 5′ splice-site recognition and selection, mutated versions of the c2orf24, ilf3 and zgc:112089 (see Supplementary Figure S3A) minigene constructs were generated for in vivo splicing analysis after U1C knockdown.
First, a U-rich element of 17 nt length, located 86 nt downstream of the 5′ splice site of c2orf24 exon 6, was mutated to a C-rich element, substituting each uridine by a cytidine (Figure 6B, lower schematic on the right). Figure 6B demonstrates that this substitution reduced the effect of the U1C knockdown on exon skipping in comparison to the wild-type sequence (6.6 and 25.2% for the wild-type versus 8.5 and 19.8% for the mutant; compare lanes 8 and 9 with 10 and 11, respectively), suggesting that the U-element is functionally relevant for the activator role of U1C in exon inclusion. The effect in this case became apparent only in the absence of U1C, when both skipping and inclusion isoforms appear, but not in the control knockdown, when inclusion was close to 100% (lanes 7 and 9).
Second, we mutated a short U-stretch, located 19 nt downstream of the proximal 5′ splice site of ilf3 exon 15, to a C-stretch of the same length (Figure 6C, lower schematic on the right). Clearly, the mutation of the U-stretch resulted in an enhanced usage of the distal 5′ splice site in both the U1C knockdown and the control cells (compare lanes 7/8 with 9/10). Consistent with the mutational analysis of exon 6 skipping in c2orf24, the effect of U1C knockdown was clearly detectable for the wild-type construct (13.8 versus 23.7%; lanes 7 and 8), but not significant for the mutant derivative (35.3 versus 38.8%, lanes 9 and 10). We conclude that in both cases analysed, the effect of the U-stretch mutant depended on U1C, arguing for a functional collaboration of U1C and the U-rich element.
Discussion
The precise recognition of the 5′ splice site by the U1 snRNP is a prerequisite for correct spliceosome assembly, pre-mRNA splicing, and alternative splicing decisions. This early event in 5′ splice-site recognition precedes the interactions of both the U6 and U5 snRNAs with neighbouring intronic and exonic positions around the 5′ splice site. Such multiple, sequential checkpoints in spliceosome assembly guarantee the fidelity of splice-site choice; at the same time, they introduce new potential regulatory steps in alternative splicing. As discussed in the yeast system, there may be yet an additional, earlier step before the classical base pairing between the U1 snRNA and the 5′ splice-site region of the pre-mRNA: an interaction of the U1 snRNP-specific protein U1C with the 5′ splice site, which is independent of RNA–RNA base pairing (Zhang and Rosbash, 1999; Du and Rosbash, 2001, 2002). However, whether this holds for higher eukaryotes, is controversial (Muto et al, 2004). Alternatively, this U1C-5′ splice-site contact may occur within the U1 snRNP. Therefore, we asked whether this very early event at the 5′ splice site may represent an additional regulatory step in 5′ splice-site selection, searching for U1C-dependent alternative splicing events.
Here, we have described a U1C mutant zebrafish, hi1371, in which we detected a stable, but U1C-deficient U1 snRNP (Figures 1 and 2). U1C deficiency in the zebrafish mutant resulted in early developmental defects (Figure 5A); however, this does not reflect a general splicing block, which causes a developmental arrest as early as 4.5 h post-fertilization (hpf) (König et al, 2007); even a 50% reduction of the major spliceosomal snRNPs is lethal within 24 hpf (Strzelecka et al, 2010). Surprisingly, U1C mutant embryos appear to develop quite normally until day 2 and are viable for about 5 dpf. Therefore, we hypothesized that sufficient residual splicing activity remained: rather than a general splicing failure, there may be a shift in 5′ splice-site selection, favouring 5′ splice sites that are U1C independent or at least less U1C dependent than others; as a result, accumulating alternative splicing changes and splicing defects would account for the mutant phenotype.
We tested this hypothesis by a genomewide RNA-Seq analysis, comparing wild-type and U1C mutant zebrafish embryos. This uncovered a large set of U1C-dependent target genes that exhibit specific alterations in alternative splicing (Figures 3 and 4; Supplementary Figure S2). There are about 3.5-fold more cases of U1C-dependent alternative 5′ splice sites compared with 3′ splice sites, strongly supporting our hypothesis, that U1C has a direct influence on 5′ splice-site selection. In ∼95% of the single- and multiple-exon skipping cases, U1C-deficiency results in an increase in skipping, arguing for an activator role of U1C in 5′ splice-site selection. Additionally, we see a preference for distal over proximal sites among the U1C-dependent 5′ splice sites, so that in the absence of U1C, proximal 5′ splice sites are favoured. Although competing 5′ splice sites can be bound simultaneously by separate U1 snRNPs, usually the downstream one is preferentially used for splicing (Eperon et al, 1993). The concentration of the antagonistic factors SF2/ASF and hnRNPA1, and intrinsic sequence features that influence U1 snRNA complementarity and U1 snRNP occupancy, can enhance the use of the upstream 5′ splice site (Mayeda and Krainer, 1992; Eperon et al, 2000; Roca et al, 2005). Therefore, U1C might be important to enhance U1 snRNP binding to the distal 5′ splice site, switching splicing to this site in case of low U1 snRNP occupancy, which may provide a physiologically significant mechanism to react to various levels of endogenous U1 snRNP.
Are the diverse effects we observe all caused directly by U1C deficiency? Two lines of evidence strongly argue for this: first, injection of in vitro transcribed U1C cRNA rescued early developmental defects (Figure 5A and B), confirming the U1C specificity of the phenotypic effects. More importantly in terms of U1C function, wild-type splicing patterns for validated target genes were restored (Figure 5C), which clearly demonstrates that the alternative splicing changes and splicing defects observed depend directly on U1C protein. Second, we were able to confirm these U1C-specific effects in a heterologous system: using four zebrafish minigene constructs (exon skipping: zgc:112089 and c2orf14; alternative 5′ splice sites: zgc:162329 and ilf3), the splicing alterations after siRNA-mediated U1C knockdown mimicked the effects observed in the zebrafish mutants. In addition, a functional knockdown of the U1 snRNP with an antisense morpholino oligonucleotide blocking the 5′ end of the U1 snRNA did not show any significant changes in the alternative splicing patterns of the minigene constructs (Supplementary Figure S5). Taken together, these results clearly validate the splicing defects as primary, U1C-linked and -specific events.
Nevertheless, U1C—like other intrinsic spliceosomal components—presumably acts in every splicing event, alternative or constitutive. As this study has shown, however, U1C regulates alternative splicing of a distinct group of genes. In addition, previous studies based on a large-scale RNAi screen in Drosophila as well as on systematic screening of mutant yeast strains demonstrated that loss or mutation of certain core spliceosomal components can result in target-specific splicing alteration (Clark et al, 2002; Park et al, 2004; Pleiss et al, 2007). Thus the question arises: How can the loss of a general splicing factor affect a specific set of target genes? Tissue-specific pathological phenotypes in human diseases, such as Retinitis Pigmentosa (McWhorter et al, 2003; Winkler et al, 2005; reviewed by Mordes et al, 2006; Boon et al, 2009) or spinal muscular atrophy (Zhang et al, 2008; Linder et al, 2011), illustrate that reduced levels or the loss of a general splicing factor can cause cell-type-specific defects. The mechanistic basis for this is still completely unclear to date. We suggest that the U1C mutant zebrafish can be considered as another, new model system for investigating this general question of human disease mechanisms.
What is the molecular basis for splicing alterations in this specific set of about 350 U1C-dependent target genes? We consider two possibilities: first, there may be an intrinsic characteristic of these 5′ splice sites that makes them particularly U1C dependent. Second, the splice-site-specific effect may be mediated through interaction with other, trans-acting factors. Regarding the first explanation, we have carefully compared the sequences and strengths in this set of 5′ splice sites, but were unable to identify a common feature. Therefore, the second possibility, a mediator function to U1C, becomes more likely. Focussing on the downstream intronic region, we searched for common sequence elements associated with the U1C-regulated 5′ splice sites in zebrafish, and indeed found there an enrichment of U-rich elements. We were able to validate a functional role of the U-stretch, using mutational analysis of three exemplary zebrafish minigenes, and HeLa cell transfection after U1C siRNA-mediated knockdown. We have shown that the U1C dependency of alternative splicing of the zebrafish target genes can be reproduced in human cells, indicating that the functional role of U1C in alternative splicing regulation is conserved. However, the conservation does not extend to target gene specificity. We tested several human orthologues of our zebrafish-specific exon-skipping targets, for their splicing pattern after U1C knockdown in HeLa cells; no U1C-dependent effects on alternative splicing were observed (Supplementary Figure S6).
Our findings of U-rich elements that are functionally linked to U1C-dependent 5′ splice-site choice are reminescent of earlier work by Aznarez et al (2008). They detected a significant enrichment of U-rich motifs downstream of weak cassette exons, postulating a widespread role of these elements in promoting exon inclusion in the human system. From our experiments in the heterologous HeLa system, we conclude that U1C acts as an activator for certain 5′ splice sites, mediating its effect through associated U-stretches. According to SELEX and crosslinking data, the cytotoxic granule-associated RNA-binding protein TIA-1/TIAl-1 (Förch et al, 2000) binds specifically to such U-rich motifs, facilitating the recognition of the corresponding 5′ splice sites through direct interactions with U1C and through U1 snRNP recruitment (Förch et al, 2002; Izquierdo et al, 2005; Aznarez et al, 2008). Our results support the notion that at least some cases of U1C-dependent splicing regulation may operate through interactions of U1C with TIA-1, the latter factor binding to intronic U-rich elements. In comparison to previous studies, we extend the function of those U-rich elements to the regulation of alternative 5′ splice sites.
In sum, we demonstrate that loss of the general splicing factor U1C does not result in a general splicing failure, but alters the alternative splicing patterns of a distinct group of target genes. Therefore, our results indicate a genomewide and target-specific role of U1C in 5′ splice-site recognition and selection, adding this intrinsic U1 snRNP protein to the growing list of splicing regulators and integrating it into larger splicing-regulatory networks. Our results also suggest that changing the abundance and/or snRNP assembly of U1C protein may provide a novel potential control mechanism to modulate alternative splicing. Finally, we have introduced here the zebrafish as a valuable new model organism for genomewide studies on alternative splicing regulation.
Materials and methods
Zebrafish culture
Zebrafish were maintained as described elsewhere (Mullins et al, 1994). Hi1371 mutants were obtained from ZIRC (Zebrafish International Resource Center) (Golling et al, 2002; Amsterdam et al, 2004). Fully viable heterozygous hi1371+/− individuals were interbred to obtain homozygous U1C mutant embryos.
Zebrafish U1C expression, antibody production, and western blotting
The full-length coding sequence of D. rerio U1C was PCR amplified and cloned into pETM11, using NcoI and KpnI restriction sites. Recombinant purified protein was used for rabbit immunization (Biogenes, Berlin, Germany). Antiserum obtained from the final bleeding was used for western blotting (1:1000).
For estimating endogenous U1C protein levels, embryos were homogenized in SDS-loading buffer, separated by 15% SDS–PAGE and analysed by western blotting; to control that equal amounts of total protein were loaded onto the gel, the membrane was Ponceau S stained after chemiluminescence detection.
Embryo lysates, glycerol gradient centrifugation, and northern blotting
D. rerio embryo lysates were prepared as described previously (Trede et al, 2007). The glycerol gradient ultracentrifugation procedure was adapted from Bell et al (2002), using 2 ml gradients (10–30%); centrifugation was carried out at 44 000 r.p.m. for 5 h (4°C), and 200 μl fractions were taken from the top to the bottom of the gradient. RNA from each fraction was isolated and analysed by 10% denaturing PAGE and northern blotting. To assay steady-state levels of the snRNAs, total RNA was prepared from 3 dpf embryos, using TRIzol (Invitrogen) and RNeasy kit (Qiagen), and analysed by 10% denaturing PAGE and northern blotting; bands were quantitated by TINA software, version 2.07d.
Quantitative real-time RT–PCR, RNA-Seq sample preparation, and validation RT–PCR
Total RNA from 3 dpf wild-type and mutant embryos was prepared by TRIzol reagent (Invitrogen) and RNeasy kit (Qiagen). Equal amounts of total RNA were subjected to reverse transcription, using the qScript cDNA synthesis kit (Quanta Biosciences). Control reactions were performed in the absence of reverse transcriptase.
For estimating endogenous U1C mRNA levels, aliquots were analysed by quantitative real-time PCR (Bio-Rad ICycler, Hercules, CA), using CYBR Green JumpStart Taq ReadyMix (Sigma, St Louis, MO) and primer sets specific for ZfU1C and β-actin mRNAs.
For Solexa high-throughput sequencing, the total RNA was processed by Illumina standard protocols to prepare the mRNA-Seq library, followed by sequencing in a single-read, 76-base mode on a GAIIx sequencer. Solexa RNA-Seq sequence-read data will be uploaded to the Sequence Read Archive at NCBI.
For validation PCRs, various target-gene-specific primer sets spanning the region of interest were designed, using the design programme primer3 version 0.4.0 (http://frodo.wi.mit.edu/primer3/). All oligonucleotide sequences are listed in Supplementary Table S8.
cRNA injection and rescue verification
The coding sequence (including the first 215 nt of the 3′ untranslated region; 3′-UTR) of D. rerio U1C was amplified by RT–PCR on total RNA isolated from 3 dpf embryos and cloned into BSK-A (Gebauer et al, 1994), using SacI and XbaI restriction sites. The vector was linearized with HindIII, and m7GpppG-capped cRNA was generated by T3 RNA polymerase (Fermentas). Additionally, the cRNA was 3′ polyadenylated during in vitro synthesis, because a 73-nt long adenosine stretch had been introduced into the vector downstream of the polylinker. In a similar way, the coding sequence of the firefly luciferase was cloned and the control cRNA was generated. cRNAs were injected at a concentration of 1 μg/μl into hi1371 embryos of the one-cell stage. At 2.5 days after injection, the rescue was monitored such that phenotypically wild-type embryos were sorted out of those with mutant-specific defects and selected for simultaneous isolation of DNA and RNA (TRIzol; Invitrogen). Genomic DNA was subjected to genotyping PCR, independently detecting both the wild-type and mutant ZfU1C allele. Reverse-transcribed total RNA (qScript cDNA synthesis kit; Quanta Biosciences) was used for PCR either to detect simultaneously endogenous and injected U1C mRNA or to check splicing patterns of validated targets genes, using the respective validation RT–PCR primer sets (Supplementary Table S8); β-actin served as a control. Ethidium bromide-stained bands were quantitated, using the GeneTools software provided with the G:BOX gel documentation system from SynGene. Since the cRNA comprises only the first ∼200 nt of the natural 3′-UTR, the use of two different reverse primers within the common upstream and the endogenous mRNA-specific downstream half of the 3′-UTR in combination with the same forward primer allowed to distinguish between endogenous and injected U1C mRNA. A 311-bp product obtained by RT–PCR is derived specifically from the endogenous mRNA, whereas the 189-bp product could be amplified from either endogenous or injected mRNA. Note that the 189-bp signals we obtained for the injected embryos could be exclusively assigned to the exogenous cRNA, because uninjected U1C mutant individuals did not give a product of that size (see Figure 5C, third top panel; compare lanes mut and rescue 1–3).
siRNA knockdown and in vivo splicing analysis in HeLa cells; U1 snRNP affinity purification
One day before siRNA transfection, HeLa cells were seeded in 6-cm culture dishes (2.2 × 105 cells per dish). siRNA duplexes, specific for the human U1C 3′-UTR (5′-AGGCCUUAUUGUAUCGGUU[dT][dT]), and firefly luciferase mRNA (5′-CGUACGCGGAAUACUUCGA[dT][dT]) were transfected (at a final concentration in culture medium of 40 nM) with Lipofectamine 2000 (Invitrogen). Three days after siRNA transfection, zebrafish-derived minigene constructs (5 μg per dish) were transfected, using FuGeneHD (Roche). After another 24 h, total RNA was isolated, using Trizol (Invitrogen) and treated with RQ1-DNase (Promega). After reverse transcription (qScript cDNA synthesis kit; Quanta Biosciences), PCR was performed, using gene-specific primers to examine the splicing patterns. To be semiquantitative, all PCR reactions were performed in the linear amplification range. Knockdown efficiencies were assessed by western blot with monoclonal antibodies against U1C (4H12; Santa Cruz), and as a control, γ-tubulin (GTU-88; Sigma).
All minigene constructs were amplified on genomic DNA from 3 dpf D. rerio wild-type embryos and cloned into pcDNA3 vector, using KpnI and XhoI restriction sites for zgc:112089, c2orf24, and ilf3, or EcoRI and XbaI restriction sites for zgc: 162329.
Exon-skipping constructs: Zgc:112089 exons 1–3 were PCR amplified such that exon 1 was shortened by the first 150 bp, and c2orf24 exons 5–7 were amplified by two-step PCR such that full-length intron 5 was retained and intron 6 was shortened by 342 bp (retaining ∼150 bp downstream and upstream of the 5′ and 3′ splice sites, respectively; see schematic in Figure 6B). For mutational analyses, T-rich elements located downstream of the 5′ splice sites of the middle exons of minigenes zgc:110289 (28 bp downstream of exon 2) and of c2orf24 (86 bp downstream of exon 6) were substituted by C-rich elements of the same length.
Alternative 5′ splice-site constructs: Zgc:162329 exons 1–2 (retaining ∼150 bp downstream and upstream of the 5′ and 3′ splice sites, respectively), and ilf3 exons 15–16 (including full-length intron 15) were PCR amplified; for mutational analysis a short T-stretch downstream of the proximal 5′ splice site of exon 15 of ilf3 was substituted by a C-stretch.
All oligonucleotide sequences are listed in Supplementary Table S8. Ethidium bromide-stained bands were quantitated, using the GeneTools software provided with the G:BOX gel documentation system from SynGene.
Affinity purification of U1 snRNPs (Supplementary Figure S4C) from HeLa whole-cell lysates was done according to Palfi et al (2005); affinity-selected RNAs were identified by northern blotting, and purified proteins were analysed by western blot, using monoclonal antibodies against U1C (4H12, Santa Cruz) and U1-70K (H111, Synaptic Systems).
Antisense morpholino transfection, RNase H protection assay, and silver staining
1.5 × 106 HeLa cells were transfected with 100 μM antisense morpholino oligonucleotide (U1 5′-GGTATCTCCCCTGCCAGGTAAGTAT-3′ and Ctr 5′-CCTCTTACCTCAGTTACAATTTATA-3′; Kaida et al, 2010), using the Nucleofector Solution R (Lonza) and Nucleofector programme I-013 according to the manufacturer's instructions. After transfection, cells were transferred to 6 cm dishes, and after 8 h zebrafish-derived minigene constructs (5 μg per dish) were transfected, using FuGeneHD (Roche). The efficiency of morpholino-mediated U1 snRNA inhibition was assessed by an RNase H protection assay (Kaida et al, 2010). Total cell extracts were prepared by freezing the cell suspension in RNase H reaction buffer in liquid nitrogen. After thawing, the cell extract was incubated with 5 μM antisense DNA oligonucleotide (5′-CAGGTAAGTAT-3′) and 1.5 U RNase H (Promega) for 30 min at 37°C. RNA was isolated by phenol extraction and ethanol precipitation and analysed on a 10% denaturing polyacrylamide gel followed by silver staining (Supplementary Figure S5).
Supplementary Material
Acknowledgments
We thank Barry Paw and the members of our group for discussions and comments on the manuscript, Lisa Thalheim for contributing the algorithm for unbiased sequence alignment, and David Ibberson and Richard Carmouche from the EMBL GeneCore sequencing team for excellent support. The results in this work are part of a dissertation (TD Rösel) submitted at the Justus Liebig University in Giessen. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to AB), the Federal Ministry for Education and Research (BMBF NGNF-2 programme; to AB), and the European-Commission-funded Network of Excellence EURASNET (to AB).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ackermann GE, Paw BH (2003) Zebrafish: a genetic model for vertebrate organogenesis and human disorders. Front Biosci 8: d1227–d1253 [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Hopkins N (2006) Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet 22: 473–478 [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N (2004) Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA 101: 12792–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ (2008) A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res 18: 1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ (2010) Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A (2002) p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 21: 2724–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dov C, Hartmann B, Lundgren J, Valcárcel J (2008) Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem 283: 1229–1233 [DOI] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Boon KL, Xiao S, McWhorter ML, Donn T, Wolf-Saxon E, Bohnsack MT, Moens CB, Beattie CE (2009) Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects. Hum Mol Genet 18: 3615–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA (2002) Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle M, Baralle FE (2006) Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res 34: 3494–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Stands L, Staley JP, Jackups RR Jr, Latus LJ, Chang TH (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7: 227–232 [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M Jr (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296: 907–910 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G (2009) RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA (1994) Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869 [DOI] [PubMed] [Google Scholar]
- Du H, Rosbash M (2001) Yeast U1 snRNP-pre-mRNA complex formation without U1snRNA-pre-mRNA base pairing. RNA 7: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Rosbash M (2002) The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90 [DOI] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR (1993) Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J 12: 3607–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Cáceres JF, Hayward DG, Krainer AR (2000) Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol 20: 8303–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J (2000) The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Förch P, Puig O, Martínez C, Séraphin B, Valcárcel J (2002) The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J 21: 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Xu W, Cooper GM, Richter JD (1994) Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J 13: 5712–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N (2002) Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet 31: 135–140 [DOI] [PubMed] [Google Scholar]
- Hage R, Tung L, Du H, Stands L, Rosbash M, Chang TH (2009) A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/Pre-mRNA interaction. Mol Cell Biol 29: 3941–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Bach M, Winkelmann G, Lührmann R (1990) U1-specific protein C needed for efficient complex formation of U1 snRNP with a 5′ splice site. Science 247: 69–72 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, Bilbao D, Valcárcel J (2005) Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell 19: 475–484 [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G (2010) U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H, Matter N, Bader R, Thiele W, Müller F (2007) Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 131: 718–729 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M (2005) Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Linder B, Dill H, Hirmer A, Brocher J, Lee GP, Mathavan S, Bolz HJ, Winkler C, Laggerbauer B, Fischer U (2011) Systemic splicing factor deficiency causes tissue-specific defects: a zebrafish model for retinitis pigmentosa. Hum Mol Genet 20: 368–377 [DOI] [PubMed] [Google Scholar]
- Lund M, Kjems J (2002) Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA 8: 166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T (2005) Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 1: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365–375 [DOI] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE (2003) Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol 162: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P Jr, Wu JY (2006) Pre-mRNA splicing and retinitis pigmentosa. Mol Vis 12: 1259–1271 [PMC free article] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Haffter P, Nüsslein-Volhard C (1994) Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol 4: 189–202 [DOI] [PubMed] [Google Scholar]
- Muto Y, Pomeranz Krummel D, Oubridge C, Hernandez H, Robinson CV, Neuhaus D, Nagai K (2004) The structure and biochemical properties of the human spliceosomal protein U1C. J Mol Biol 341: 185–198 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW (2003) The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25: 1147–1149 [DOI] [PubMed] [Google Scholar]
- Palfi Z, Hui J, Bindereif A (2005) Biotin-based affinity purification of RNA-protein complexes. In Handbook of RNA Biochemistry, RK Hartmann, A Bindereif, A Schön and E Westhof (eds), pp 676–693. Wiley, WIVCH, Verlag GmbH & Co.: KGaA, Weinheim [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR (2004) Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA 101: 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C (2007) Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol 5: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K (2009) Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Sachidanandam R, Krainer AR (2005) Determinants of the inherent strength of human 5′ splice sites. RNA 11: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B, Kretzner L, Rosbash M (1988) A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J 7: 2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano PG, Guthrie C (1988) 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev 2: 1258–1267 [DOI] [PubMed] [Google Scholar]
- Stark H, Dube P, Lührmann R, Kastner B (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409: 539–542 [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM (2010) Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17: 403–409 [DOI] [PubMed] [Google Scholar]
- Tang J, Abovich N, Fleming ML, Séraphin B, Rosbash M (1997) Identification and characterization of a yeast homolog of U1 snRNP-specific protein C. EMBO J 16: 4082–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA (1994) SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev 8: 2704–2717 [DOI] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S (2009) Alternative splicing and disease. Biochim Biophys Acta 1792: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede NS, Medenbach J, Damianov A, Hung LH, Weber GJ, Paw BH, Zhou Y, Hersey C, Zapata A, Keefe M, Barut BA, Stuart AB, Katz T, Amemiya CT, Zon LI, Bindereif A (2007) Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci USA 104: 6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB (2008) Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U (2005) Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev 19: 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Rümpler S, Klein Gunnewiek J, van Venrooij WJ, Lührmann R (1996) In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res 24: 4614–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Rosbash M (1999) Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev 13: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM (1986) A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.