Abstract
To clarify the role of a number of mRNA processing factors in transcription elongation, we developed an in vivo assay for direct analysis of elongation on chromatin. The assay relies on two substrates containing two G-less cassettes separated by either a long and GC-rich or a short and GC-poor DNA sequence (G-less-based run-on (GLRO) assay). We demonstrate that PAF, THSC/TREX-2, SAGA, the exosome component Rrp6 and two subunits of cleavage factor IA (Rna14 and Rna15) are required for efficient transcription elongation, in contrast to some results obtained using other assays. Next, we undertook a mutant screen and found out that the Nup84 nucleoporin complex is also required for transcription elongation, as confirmed by the GLRO assay and RNA polymerase II chromatin immunoprecipitations. Therefore, in addition to showing that the GLRO assay is a sensitive and reliable method for the analysis of elongation in vivo, this study provides evidence for a new role of the Nup84 complex and a number of mRNA processing factors in transcription elongation that supports a connection of pre-mRNA processing and nuclear export with transcription elongation.
Keywords: G-less run-on, mRNA factors, Nup84, RNAPII, transcription elongation
Introduction
Eukaryotic RNA polymerase II (RNAPII)-mediated transcription elongation is a dynamic and highly regulated stage of the transcription cycle that is functionally linked to mRNA processing, including capping, splicing, 3′-end processing, surveillance and export. The carboxy-terminal domain (CTD) of the RNAPII has a key role in this coupling, which is necessary for the efficient formation of an export competent ribonucleoparticle (mRNP) (Bentley, 2005; Luna et al, 2008; Selth et al, 2010). During elongation, specific factors associate with RNAPII modulating its catalytic activity to help it overcome situations derived from transient pausing, arrest and termination. In addition, elongation through chromatin demands nucleosome remodelling and/or histone modification via functionally specialized factors (Li et al, 2007). In the last few years, a number of factors acting at different levels in mRNP biogenesis and export have been reported to impact on transcription elongation. However, whether or not many of these factors have an active role in elongation in vivo is yet elusive.
Genetic and biochemical approaches, both in vivo and in vitro, have been used to study transcription elongation. In vitro there are at least two types of methods for the analysis of transcription elongation. The first is based on purified RNA polymerase engaging elongation directly on an oligonucleotide with a dC-tail (Kadesch and Chamberlin, 1982). The second one was set for the analysis of elongation in naked DNA using yeast whole cell extracts (WCEs) and a plasmid with two G-less cassettes (Rondon et al, 2003). Different in vivo methods have been used to study transcription elongation. In transcriptional run-on assays, the nascent pre-mRNA is labelled with a pulse of radioactive UTP in permeabilized cells. RNA is then analysed by hybridization to immobilized strand-specific probes (Warner, 1991). Another method extensively employed is chromatin immunoprecipitation (ChIP) analyses of RNAPII distribution across a gene (Mason and Struhl, 2005). None of these assays consider that elongation might be differently affected depending on the sequence, the GC:AT content or the length of the DNA template, even though these features have been shown to influence the efficiency of transcription elongation (Chavez et al, 2000; Gallardo and Aguilera, 2001; Rondon et al, 2003, 2004). Conversely, although constructs based on open reading frames (ORFs) with different length and GC content placed under a GAL1 promoter have been used to infer elongation efficiency by northern analysis (Chavez et al, 2001; Luna et al, 2005), they only provide a first but not definitive answer as the results do not exclude a putative impact of RNA stability.
Despite the availability of different in vivo assays, their lack of specificity on transcription elongation yield results that in some cases are unclear or different depending on the assay employed. That is the case of the PAF and THSC complexes. PAF is a five-subunit complex containing Paf1, Cdc73, Ctr9, Rtf1 and Leo1, which seems to orchestrate different mRNP biogenesis processes. It coordinates chromatin modification during transcription elongation via interaction with histone methylases and ubiquitinylases (Krogan et al, 2003; Wood et al, 2003), and it is involved in the 3′-end formation of polyadenylated and non-polyadenylated RNAPII transcripts (Penheiter et al, 2005; Sheldon et al, 2005). The role of PAF in transcription elongation was proposed on the basis of its physical interaction with RNAPII, genetic and physical interactions with factors such as Spt4–Spt5 and Spt16–Pob3, and its recruitment to the ORF of transcribed genes (Costa and Arndt, 2000; Krogan et al, 2002; Pokholok et al, 2002; Squazzo et al, 2002). Mutations in the Paf1 and Cdc73 subunits, but not in Rtf1 and Leo1, reduce the transcription-elongation efficiency in vitro (Rondon et al, 2004). However, loss of PAF components does not result in an altered distribution of elongating RNAPII (Mueller et al, 2004; Mason and Struhl, 2005).
THSC, also termed TREX-2, is a conserved multifunctional complex formed by Thp1, Sac3, Sus1 and Cdc31, which works at the transcription–mRNA export interface as defined in the yeast Saccharomyces cerevisiae. THSC is located at the nuclear pore complex (NPC) and is required for mRNA export and gene tethering (Fischer et al, 2002, 2004; Gallardo et al, 2003; Rodriguez-Navarro et al, 2004; Cabal et al, 2006). Northern and ChIP analyses of long and GC-rich sequences in THSC mutants point to a role in transcription elongation. However, this has not been corroborated by the G-less in vitro assay (Gonzalez-Aguilera et al, 2008). As THSC is located at the nuclear periphery, it is an open question whether the effect of this complex on transcription is only relevant when coupled to mRNA export, but not in cell extracts in which the nuclear membrane is disrupted. Other factors with known functions in transcription initiation or mRNA metabolism recently shown to be involved in elongation are several subunits of the SAGA complex, the Rrp6 component of the nuclear exosome, and subunits of the mRNA 3′-end processing complex cleavage factor IA (CFIA) (Luna et al, 2005; Govind et al, 2007).
In order to clarify and ascertain the role of these factors in transcription elongation, we developed an in vivo assay for a direct and sensitive analysis of transcription elongation on chromatin and in an intact nuclear structure. Importantly, this assay takes in consideration different features of the template that influence elongation as the length and GC content. After validating the assay with known bona fide transcription-elongation mutants such as spt4 and rpb9, we analysed a number of mutants of the PAF, THSC and SAGA complexes, the nuclear exosome, and the 3′-end processing complex CFIA. Taking advantage of this assay, we undertook a genetic screen for putative transcription-elongation mutants and showed that the Nup84 complex of the NPC is required for transcription elongation. Our results, in addition to showing that the G-less-based run-on assay (GLRO) assay is a highly reliable method for the in vivo analysis of transcription elongation, provide novel and unambiguous conclusions about the involvement of the analysed factors in transcription elongation and serves to define a new role for the Nup84 complex in RNAPII elongation. This strengthens the idea of a functional relationship between nuclear export and transcription elongation, demonstrating an impact of the NPC in transcription elongation.
Results
A new G-less cassette-based run-on assay, GLRO, for the direct analysis of transcription elongation in vivo
We have developed a run-on assay for transcription elongation that allows quantification of nascent mRNA directly, without any need of hybridization or PCR amplification. The assay is based on a set of two constructs that differ in the length and GC content of the DNA sequence to be transcribed. The comparative analysis of the transcription-elongation efficiency through a long versus a short DNA fragment is essential as long transcripts are more sensitive to defects in elongation than short ones. We will refer to this assay as GLRO (G-less cassette-based run-on assay). The GLRO constructs are based on the CYCds plasmid (Steinmetz and Brow, 2003) bearing two G-less cassettes of 262 nt and 132 nt separated by a 243-nt CYC1 fragment as spacer sequence. This construct is transcribed from the strong constitutive TDH3 promoter. In this plasmid, we cloned a 2-kb fragment of the LacZ gene between the two G-less cassettes to generate the CYC-LacZ construct. The length and high GC content of lacZ makes transcription through this sequence poorly efficient in mutants impairing elongation (Chavez et al, 2000; Rondon et al, 2003). The GLRO assay was first performed in wild-type cells transformed with the plasmid-borne CYCds and CYC-lacZ constructs, from now on termed GLRO-short and GLRO-long, respectively (Figure 1A). Briefly, after in vivo labelling of the nascent mRNA in the run-on reaction, the resulting transcripts were purified and treated with RNase T1 to degrade all G-containing sequences, rendering the two G-less cassettes as two intact fragments that were resolved by polyacrylamide gel electrophoresis (PAGE). Transcription-elongation efficiency was measured as the ratio of 32P incorporated into the 132-nt-long versus the 262-nt-long G-less cassette for each construct. We confirmed by northern that the lacZ inserted between the two G-less cassettes in GLRO-long is correctly transcribed in wild-type cells (data not shown). GLRO values in the different mutants are normalized to the corresponding wild-type levels for both the GLRO-short and GLRO-long constructs.
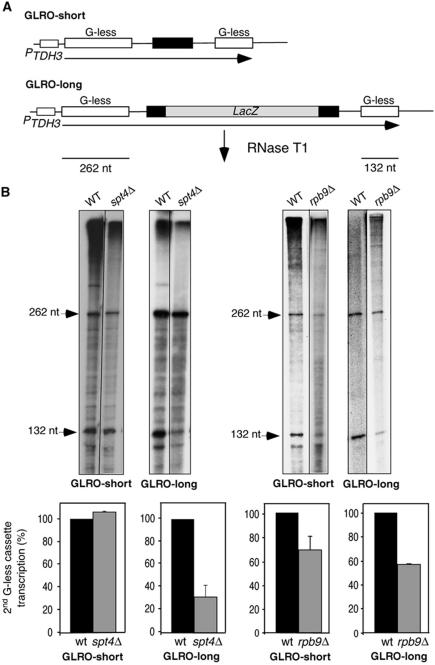
Figure 1.
Characterization of the G-less cassette-based run-on transcription-elongation assay (GLRO) in wild-type and spt4Δ and rpb9Δ mutants. (A) Scheme of the tandem G-less cassette constructs used for GLRO analysis. Black rectangles represent sequences derived from CYC 3′-flanking region, and the white and grey rectangles correspond to the two G-less cassettes and the 2-kb lacZ fragment, respectively. (B) GLRO analysis of wild-type and spt4Δ and rpbΔ9 mutants transformed with the GLRO-short and GLRO-long systems. Transformants were grown in SC-leu medium to exponential phase and run-on transcription assays were performed as described in Materials and methods. The transcription run-on products were digested with RNase T1 and resolved in a 6% PAGE. A representative acrylamide gel is shown. For each sample, the ratio of total counts incorporated into the distal versus the proximal G-less cassette was normalized against the ratio for the same construct in the wild-type strain. The mean value and s.d. of three independent experiments are shown.
To validate the GLRO assay, we measured transcription elongation in null mutants of genes with a known role in transcription elongation like SPT4 and RPB9. Spt4 (Spt4–Spt5/DSIF complex) is involved in RNAPII promoter-proximal pausing and it also has a positive role in transcription elongation as shown by 6-azauracil (6-AU) sensitivity, northern analysis, RNAPII ChIP and in vitro transcription-elongation assays (see Supplementary Table I; Hartzog et al, 1998; Rondon et al, 2003). Rpb9 is a non-essential subunit of RNAPII that functions in transcription initiation and elongation and is important for transcription fidelity (Hull et al, 1995; Hemming et al, 2000; Nesser et al, 2006). Rpb9 in vivo relevance in elongation was determined by 6-AU sensitivity and by its ability to interact with the transcription-elongation factor TFIIS (Awrey et al, 1997; Hemming et al, 2000). Transcription-elongation efficiency (transcription of the second G-less cassette versus the first one) of spt4Δ was 30% of the wild type for the GLRO-long and similar to the wild type for the GLRO-short construct (Figure 1B). This result is consistent with the decrease in efficiency (<25% with respect to the wild type) previously determined in vitro with an assay based on other two G-less cassette constructs (Rondon et al, 2003).
Next, we assayed the transcription-elongation efficiency of rpb9Δ, using both the in vivo run-on systems (Figure 1B) and the in vitro G-less assay (Supplementary Figure S1). In this case, the rpb9Δ mutant reduces transcription elongation to 60–70% of the wild-type values in both the GLRO-long and GLRO-short systems. In addition, a reduction to 50% of the wild-type elongation rate was also observed in vitro using rpb9Δ WCEs (Supplementary Figure S1). Together, these results indicate that the GLRO assay is sensitive enough to detect a decrease in elongation rate.
Transcription elongation is defective in PAF mutants
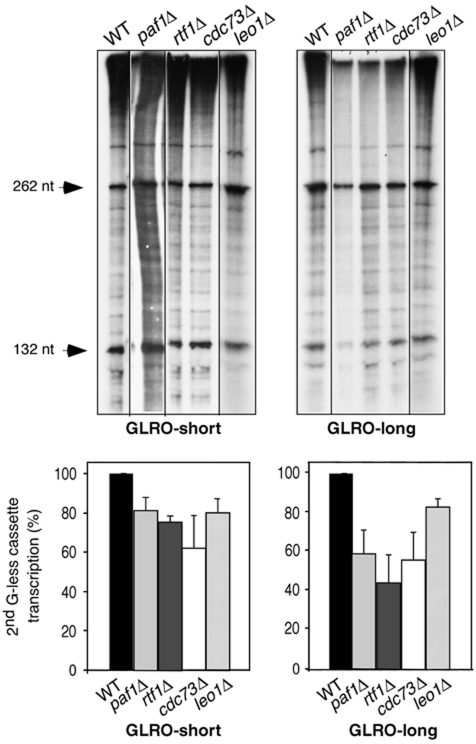
The PAF complex has several roles in mRNP biogenesis from transcription elongation to transcription termination and 3′-end mRNA processing (Mueller et al, 2004; Penheiter et al, 2005; Sheldon et al, 2005). Evidence supports a hierarchical and specialized role for the different subunits. It has previously been shown that ablation of any of the PAF-subunit genes severely decreased the mRNA levels of the long and GC-rich lacZ ORF driven from a GAL1 promoter, whereas it did not affect transcription of short mRNAs driven from that same promoter (Rondon et al, 2004). These data suggest a defect in transcription elongation rather than in initiation, otherwise the defect should have been the same for both constructs. However, analysis of in vitro transcription elongation indicated that only two subunits, Paf1 and Cdc73, had a detectable function in transcription elongation (Rondon et al, 2004). In contrast, when transcription elongation was assessed in vivo by RNAPII distribution across a long gene, no defect was observed in any of the PAF mutants (Mason and Struhl, 2005). Therefore, we decided to test the effect of different PAF mutants in the GLRO assays. The paf1Δ, cdc73Δ and rtf1Δ strains showed a slight reduction in the GLRO-short system and a stronger reduction in the GLRO-long with levels of 50–60% of the wild type (Figure 2). Transcription levels were also slightly reduced the GLRO-long system in leo1Δ, although not as strongly as in the other mutants. Overall, our results confirm that, indeed, PAF has a clear role in transcription elongation in vivo.
Figure 2.
GLRO analysis of PAF mutants. Wild-type (BY4741), paf1Δ, rtf1Δ, cdc73Δ and leo1Δ isogenic strains were transformed with the GLRO-short and GLRO-long constructs and transcription run-on assays were performed. A representative assay is shown. The mean value and s.d. of three independent experiments are shown. Other details are as in Figure 1.
Differential effect of THSC mutations in transcription elongation
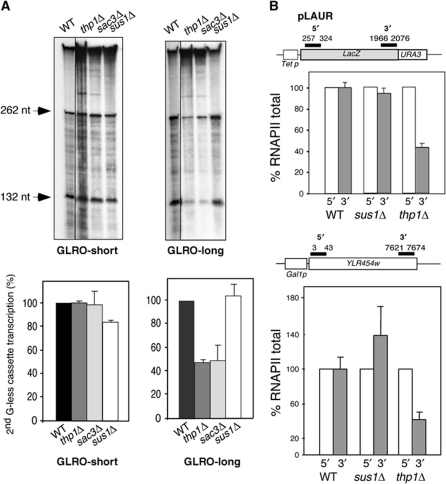
THSC mutants are impaired in transcription elongation in vivo, as determined by northern analysis of long versus short genes and RNAPII ChIPs, but they fail to show a significant elongation defect in vitro in contrast to other mutants with similar in vivo effects (Gallardo et al, 2003; Gonzalez-Aguilera et al, 2008). Therefore, we sought to analyse transcription in THSC mutants with the GLRO assay. Interestingly, the sac3Δ and thp1Δ strains diminished transcription-elongation efficiency to 50–60% of the wild-type levels only in the GLRO-long construct, whereas sus1Δ showed no decrease (Figure 3A). These results confirm that THSC has a role in transcription elongation that is only appreciated in vivo when transcription and mRNA export are coupled.
Figure 3.
Analysis of transcription elongation in THSC. (A) GLRO analysis of THSC mutants. Transcription run-on assays of thp1Δ, sac3Δ, sus1Δ, mutants carrying the plasmids GLRO-short and GLRO-long. (B) RNAPII occupancy in sus1Δ mutant. ChIP analyses in wild-type (BY4741), thp1Δ and sus1Δ strains transformed with the pLAUR expression system. ChIP analyses in wild-type, thp1Δ and sus1Δ strains carrying the GAL1p::YLR454w fusion construct located at the endogenous YLR454w chromosomal locus. The scheme of the gene and the PCR-amplified fragments are shown. Numbers indicate the primer position respect to the first ATG of the gene. The sequence is provided in Supplementary Table II. The DNA ratios between the 5′ and 3′ regions were calculated from their signal relative to the signal of the intergenic region. The recruitment data shown refer to the value of the 5′ region normalized to 100%. ChIPs were performed from three independent cultures, and quantitative PCRs were repeated three times for each culture.
Sus1 is a factor present in two protein complexes: THSC and SAGA, a complex involved in transcription initiation of a subset of genes (Rodriguez-Navarro et al, 2004). As previous studies suggested that sus1Δ has a defect in transcription elongation in vivo (Pascual-Garcia et al, 2008), we wanted to confirm by other means the lack of effect shown by sus1Δ in the GLRO assay. Thus, we measured RNAPII occupancy across the pLAUR expression system that contains a 4.15-kb lacZ-URA3 translational fusion under the tet promoter (Figure 3B) (Jimeno et al, 2002). THSC mutants transcribe poorly the pLAUR construct, as shown by northern (Gallardo et al, 2003; Gonzalez-Aguilera et al, 2008). Indeed, RNAPII occupancy at the 3′-end of lacZ gene was reduced 50% with respect to the 5′-end in thp1Δ, consistent with previous in vivo data. Importantly, no reduction was observed in a sus1Δ mutant. Similarly, ChIP analyses in the YLR454w gene fused to the GAL1 promoter showed a decrease in the RNAPII signal at the 3′-end in thp1Δ cells but not in sus1Δ cells (Figure 3B). These results differ from the small decrease in RNAPII level reported for the YLR454w ORF, probably due to experimental conditions (Pascual-Garcia et al, 2008). We conclude that Sus1, even though it is recruited along the ORF of genes in a transcription-dependent manner (Pascual-Garcia et al, 2008), does not significantly affect transcription elongation.
SAGA mutations that impair transcription elongation
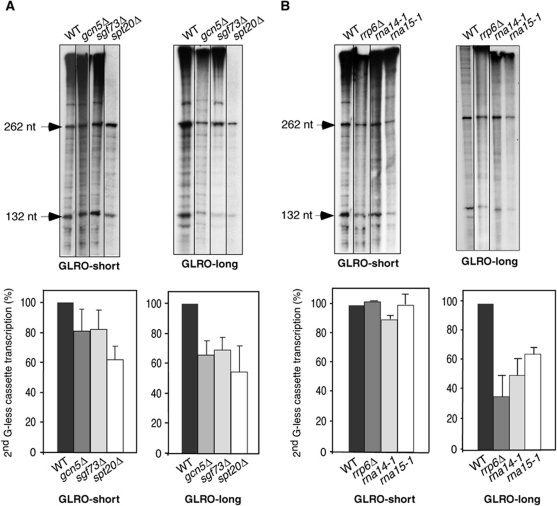
SAGA is a well-known transcription initiation factor with a possible role in elongation as several of its components are recruited along the genes (Govind et al, 2005, 2007). In order to explore this possibility, we decided to extend our analyses to some SAGA mutants, such as gcn5Δ, spt20Δ and sgf73Δ (Figure 4A). Gcn5 is an acetyltransferase, Spt20 has a role in the structural integrity of SAGA and Sgf73 is a subunit of the histone-deubiquitinating module that interacts with THSC (Candau et al, 1997; Grant et al, 1997; Kohler et al, 2008). Mutations in these factors did not substantially affect GLRO-short transcription with the exception of spt20Δ. However, in the GLRO-long construct we appreciated a decrease of the elongation rate in the three mutants (Figure 4A). These data are in agreement with the reduced RNAPII occupancy at the 3′-end of specific genes in gcn5 and spt20 mutants (Govind et al, 2005; Gaillard et al, 2009), and with the low efficiency of spt20Δ to transcribe in vitro the two G-less cassette system (<60% with respect to the wild type) (Gaillard et al, 2009). In summary, these results support the view of SAGA as a histone-modification complex implicated in transcription initiation and also in transcription elongation. It remains to be seen whether these subunits influence elongation as part of SAGA or independently.
Figure 4.
GLRO analysis of mutants of the SAGA, nuclear exosome and 3′-end processing factors. (A) GLRO analysis of SAGA mutants. Transcription run-on assays of wild-type, sgf73Δ, gcn5Δ and spt20Δ strains carrying GLRO-short and GLRO-long plasmids are shown. (B) GLRO analysis of nuclear exosome and 3′-end processing mutants. rrp6Δ, rna14-1, rna15-1, and a isogenic wild-type strain were transformed with the GLRO-short and GLRO-long plasmids and transcription run-on assays were performed. A representative assay is shown. The mean value and s.d. of three independent experiments are shown. Other details are as in Figure 1.
Transcription elongation diminishes in mutants of nuclear exosome components or 3′-end processing factors
Finally, we determined with the GLRO assay the transcription-elongation efficiency of mutants with previously shown in vivo and in vitro elongation defects, which were particularly intriguing given their known role in different mRNA metabolism steps. In particular, we analysed the implication of 3′-end mRNA processing and termination factors Rna14 and Rna15 and the nuclear exosome component Rrp6. In these three mutants, the transcription-elongation rate was notably diminished to 40–50% of the wild-type level in the GLRO-long assay, whereas no difference was observed in the GLRO-short assay (Figure 4B). These results are consistent with the decrease in the RNAPII occupancy observed by ChIP analysis and with the defect on transcription elongation reported in vitro with the two G-less assay (Luna et al, 2005). Therefore, the GLRO assays unambiguously identify CFIA and the nuclear exosome component Rrp6 as two factors that confer high efficiency of transcription elongation in vivo. The fact that the effect is only observed in the GLRO-long system suggests that the activity of these factors is only relevant in a late elongation stage as it seems to be the case also for Spt4 and THSC.
A novel role for the Nup84 complex in transcription elongation
As mentioned above, transcription through the bacterial lacZ gene in yeast is especially sensitive to mutations in elongation factors. Thus, mutations in several of the complexes required for elongation, as stated by the GLRO assay, also diminished lacZ mRNA accumulation in northern assays (Luna et al, 2005). Having developed the new GLRO assay as a sensitive method to assess the in vivo transcription-elongation efficiency, we sought to perform a genetic screen to identify new factors required for transcription elongation. By employing a Tn3 deletion library (Burns et al, 1994), we mutated a strain with a GAL1p::LacZ fusion integrated in Chromosome V and searched for a decrease in β-galactosidase (β-gal) activity. In all, 6160 colonies were screened by β-gal colony colour assay. After several selection steps that included quantification of β-gal activity, only three mutants unambiguously reduced the β-gal activity to 40% the wild-type level. However, in only one of the mutants, the reduction of β-gal co-segregated with the Tn3 marker and, therefore, was genetically linked to the Tn3 insertion. This low yield of candidates may be due to both a low genomic representation in the library used and, above all, the poor sensitivity of the β-gal colony colour assay, in contrast to northern analysis, to detect a decrease in lacZ transcription. Cloning and sequencing of the candidate revealed that the Tn3 transposon was inserted at the NUP84 promoter, disrupting its activity. Therefore, we continued the analysis with a nup84Δ strain.
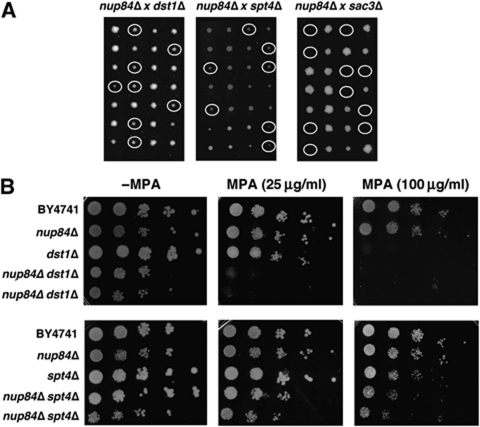
For further genetic evidence of the Nup84 involvement in RNAPII elongation in vivo we searched for possible genetic interactions of Nup84 with other factors implicated in elongation. As can be seen in Figure 5A, the double mutants nup84Δ dst1Δ and nup84Δ spt4Δ are viable in contrast to the double mutants nup84Δ sac3Δ consistent with non-overlapping roles of the Nup84 and THSC complexes in mRNA export. However, nup84Δ dst1Δ and nup84Δ spt4Δ increased the sensitivity to mycophenolic acid (MPA), a drug that compromises transcription elongation, with respect to the single mutants (Figure 5B). These results are, therefore, consistent with a role of Nup84 in RNAPII transcription elongation in vivo.
Figure 5.
Genetic interactions of Nup84 with transcription-elongation and mRNA export factors. (A) Tetrad analyses of different crosses between nup84Δ and dst1Δ, spt4Δ and sac3Δ strains. Circles indicate the positions of the double mutants. Only the double nup84Δ sac3Δ are inviable. (B) Micophenolic acid (MPA) sensitivity of nup84Δ, dst1Δ and spt4Δ single and double mutants as determined by 10-fold serial dilutions on SC plates containing the indicated concentrations of MPA.
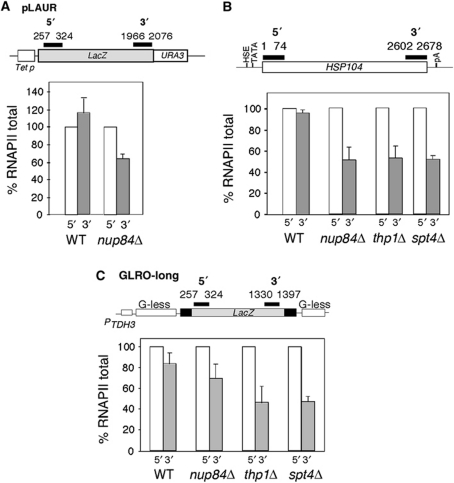
To obtain molecular evidence of the Nup84 effect in transcription elongation, we first confirmed by northern analysis that lacZ transcription was reduced in a nup84Δ mutant (data not shown). Then, we determined by ChIP the RNAPII levels at the pLAUR system. As expected, the observed decrease in lacZ mRNA correlates with a reduction in RNAPII occupancy at the 3′-end of the pLAUR system versus the 5′-end (Figure 6A), indicating that Nup84 contributes to RNAPII transcription elongation through the bacterial lacZ gene. Similar results were obtained in the endogenous HSP104 gene and the GLRO-long system, indicating that the effect in elongation is not sequence specific (Figure 6B and C). The reduction of RNAPII occupancy observed at the 3′-end region of HSP104 in the nup84Δ mutant was as severe as in the spt4Δ or thp1Δ (50%) while in the GLRO-long system it was less pronounced (70%) (Figure 6B and C). Altogether the RNAPII distribution analysis is consistent with a general defect in transcription elongation.
Figure 6.
RNAPII distribution along different genes in nucleoporin mutants. (A) RNAPII ChIP analysis in the pLAUR system in the nup84Δ and an isogenic wild-type strains. (B) RNAPII ChIP analysis in the HSP104 endogenous gene in the nup84Δ, thp1Δ and spt4Δ mutants and their isogenic wild-type strain (C) RNAPII ChIP analysis in the GLRO-long system in the nup84Δ, thp1Δ and spt4Δ mutants and their isogenic wild-type strain. Other details are as in Figure 3.
Next, we took advantage of the newly developed GLRO assay to directly determine the impact of nup84Δ in transcription elongation. As can be seen in Figure 7A, nup84Δ deletion affects elongation through the GLRO-long construct although to a lesser extent than mutations in PAF, THSC or Spt4 (70% the wild-type level in nup84Δ and 30–50% in PAF, THSC or Spt4 mutants). These results are consistent with RNAPII ChIP analysis in the GLRO-long construct, confirming once more the reliability and high sensitivity of this assay.
Figure 7.
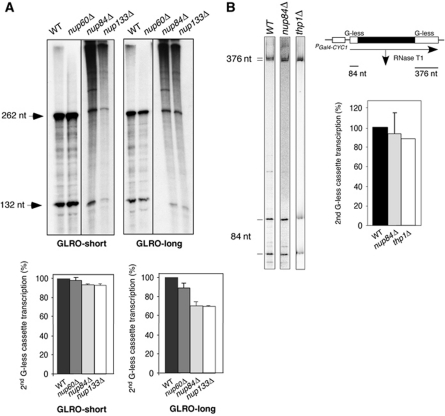
GLRO analysis of nucleoporin mutants. (A) GLRO analysis was performed in nup84Δ, nup133Δ, nup60Δ and an their isogenic wild-type strain on both the GLRO-short and GLRO-long systems. The mean value and s.d. of three independent experiments are shown. Other details are as in Figure 1. (B) In vitro transcription-elongation assays of wild-type and nup84Δ WCEs. Scheme of the two G-less cassette systems pGCYC1-402 used for the in vitro transcription-elongation assay. RNase T1 treatment of the mRNA driven from the GAL4-CYC1 promoter renders two fragments corresponding to the two G-less cassettes. In vitro transcription assays of WCEs from BY4742, nup84Δ and thp1Δ isogenic strains are shown. Each reaction was stopped after 40 min, treated with RNaseT1, and run in a 6% PAGE. Efficiency of transcription elongation was determined as the percentage of total transcripts that covered the 376-nt G-less cassette with respect to the transcripts that covered the 84-nt cassette. The signal was normalized with respect to the U content of each G-less cassette (161 U residues in the long cassette and 24 in the short one). The mean value and s.d. of at least three independent experiments are shown for WT and nup84Δ.
Nup84 is a structural component of the NPC that together with Nup85, Nup120, Nup133, Nup145C, Sec13 and Seh1 forms the Nup84 complex (Strambio-De-Castillia et al, 2010). To assay whether RNAPII elongation requires the integrity of this complex or the whole NPC, we tested the effect on elongation caused by ablation of Nup133, a member of the Nup84 complex, and Nup60, which does not belong to this complex. Figure 7A shows that similarly to nup84Δ, the elongation rate in the nup133Δ mutant decreased to 70% of the wild-type value only in the GLRO-long system. In contrast, nup60Δ showed no significant effect on transcription elongation. We conclude, therefore, that disruption of the Nup84 complex specifically hinders transcription elongation.
Finally, we assayed whether the effect of nup84Δ on transcription is seen only in vivo, in conditions in which transcription is coupled to mRNA export in the presence of functional NPCs, or whether it might also be observed in WCEs in which the nuclear envelope has been disrupted. As can be seen in Figure 7B, Nup84 is not required for RNAPII transcription elongation in vitro. This result is, therefore, similar to that previously obtained with THSC mutants, which are also affected in mRNA export, with a clear effect in transcription elongation in vivo but not in vitro (Figure 3A) (Gonzalez-Aguilera et al, 2008). Overall, these data support a role of the Nup84 complex in transcription elongation that is linked to its role in mRNA export at the NPC.
Discussion
In this study, we report a new role of the Nup84 complex in transcription elongation and revisit the implications of PAF, THSC/TREX-2, SAGA, Rrp6 and CFIA in this process. First, we developed a novel in vivo GLRO assay that directly measures RNAPII-elongation capacity. With this system, we tested the effect of different mutations in complexes previously connected to transcription elongation. We determined that PAF, THSC/TREX-2, SAGA, the exosome component Rrp6 and two subunits of CFIA (Rna14 and Rna15) are required for efficient elongation. Second, we demonstrated that the structural component of the nuclear pore, the Nup84 complex, promotes RNAPII elongation.
It is intuitive that the longer a sequence is the highest the chances are of RNAPII encountering difficulties to proceed. Indeed, we have previously noticed that RNAPII elongation through a long gene is more demanding than through a short one (Chavez et al, 2001). Moreover, the intrinsic characteristics of some sequences, like the bacterial lacZ gene with a high GC content and a diffuse chromatin positioning (Chavez et al, 2001), makes it more difficult for RNAPII to elongate through it. This feature made lacZ an excellent reporter to assess RNAPII ability to elongate (Rondon et al, 2003). In this study, we inserted the lacZ sequence between two G-less cassettes to generate a reporter highly sensitive to elongation defects (GLRO-long). Thus, in a mutant of a bona fide elongation factor like Spt4, transcription is not affected in the GLRO-short system, with a transcriptional unit of <1 kb, but it is affected in the 3-kb transcriptional unit of the GLRO-long system that includes the lacZ sequence. This is also the case for mutations in factors with a known role in mRNA processing, export or surveillance as THSC, CFIA, Rrp6 and the Nup84 complex. Conversely, ablation of the non-essential Rpb9 subunit of RNAPII or the Spt20 subunit of SAGA diminished elongation efficiency in both systems but to a lesser extent than spt4Δ. This indicates that, according to our assays, Rpb9 and Spt20 have an active and positive role in transcription elongation along the entire ORF length. On the contrary, Spt4, THSC, CFIA, Rrp6 and Nup84 exert their elongation role only at promoter distal regions, suggesting that at least two different functional stages exist during transcription elongation. Other factors analysed, the Gcn5 and Sgf73 components of SAGA and Paf1, Rtf1 and Cdc73 subunits of PAF complex, would belong to an intermediate class with a mild effect in the GLRO-short system that is exacerbated in the GLRO-long construct. The existence of different classes of transcription-elongation defects opens new questions to our understanding of transcription-elongation phases that would need further investigation.
There are two main causes for which transcription elongation through the GLRO-long system could be more sensitive than through the GLRO-short system. First, the length of the DNA to be transcribed, because the torsional stress generated by the advance of the RNAPII is higher in long versus short genes. This increases the levels of superhelicity ahead of the RNAPII inhibiting its progression (Wang, 2002). Second, the length of the nascent RNA molecule, which makes elongation more dependent of particular RNA binding and processing factors (Luna et al, 2008). A long pre-mRNA molecule may demand a large amount of such factors, whereas short pre-mRNAs required less so that their impact in elongation may be either lower or non-detectable. Other elements that may increase sensitivity to transcriptional defects in the GLRO-long system are the GC content and the chromatin structure. Thus, the high GC content of lacZ may harden progression of the RNAPII due to a larger energy requirement to open the two strands at the RNAPII active site. In addition, the different chromatin structure of lacZ could also influence elongation enhancing the defects caused by the mutations assayed. Finally, the possibility that the GLRO-long construct may have sequence or nucleosome-dependent transcriptional pausing sites more difficult to be traversed by the RNAPII cannot be discarded. In any case, it is worth noticing that we obtained similar results with a GLRO-long construct containing the yeast YAT1 gene instead of lacZ (data not shown), implying that our results are not DNA sequence specific. Therefore, it is clear that the GLRO-long construct provides a highly sensitive assay for the analysis of transcription elongation. We postulate that the role of the factors showing a defect in this assay is to add ‘processivity’ to the transcription machinery.
Transcription elongation has been extensively studied during the last decade through genetic, biochemical and molecular integrated approaches (see Supplementary Table I; Selth et al, 2010). Hypersensitivity to drugs that diminish the nucleotide pools such as 6-AU and MPA has been used as a first indication that a protein could participate in transcription elongation (Hampsey, 1997; Riles et al, 2004). In the yeast S. cerevisiae, this relationship can be strengthened by determining a synergistic effect with mutations in known transcription-elongation factors, such as the TFIIS mutation dst1Δ (Lennon et al, 1998). At the biochemical level, isolation of proteins interacting with RNAPII helped identify new elongation factors (Otero et al, 1999; Krogan et al, 2002). At the molecular level, physical interaction with chromatin together with changes in RNAPII profile constitutes a reliable body of results pointing to a role in transcription elongation in vivo (Ahn et al, 2004; Kim et al, 2004). However, the implication of some protein complexes such as PAF or THSC remained controversial as different assays yielded contradictory results (see Supplementary Table I). Thus, PAF complex co-localization with RNAPII across the genome together with its physical and genetic interactions with known elongation factors such as Spt4–Spt5 and Spt16–Pob3 suggested a role in elongation (Krogan et al, 2002; Squazzo et al, 2002; Kim et al, 2004; Mueller et al, 2004). Indeed, PAF function in the assembly of histone-modification enzymes (Krogan et al, 2003; Wood et al, 2003) implies that it may facilitate elongation by acting directly on chromatin. However, at least two PAF subunits seem to promote elongation independently of whether the DNA substrate is naked or in a chromatin state, as indicated by in vitro experiments on naked DNA (Rondon et al, 2004). In spite of what is in principle expected for mutations in an elongation factor, RNAPII distribution across a long gene in PAF mutants does not differ from the wild type (Mason and Struhl, 2005). Similarly, mutations in the THSC complex decrease lacZ mRNA accumulation pointing to a role in elongation that is not evident in an in vitro assay with WCE (Gallardo et al, 2003; Gonzalez-Aguilera et al, 2008). In this study, we indeed ascertain that PAF and THSC integrity is needed for processive elongation. This result is consistent with the recent observation that human PAF complex is required for transcription elongation in vitro (Kim et al, 2010). Therefore, we can conclude that RNAPII ChIPs and in vitro WCE-based assays are limited in their capacity to detect elongation defects occurring in a physiological state. ChIP assays measures the distribution of proteins through a gene at given times but does not provide information about the relative activity of these proteins. On the other hand, in vitro assays uncouple transcription and mRNA export, limiting the capacity of this method to identify any possible feedback effect of mRNA export on elongation. It is worth noting that Thp1 and Sac3, the THSC/TREX-2 subunits with the highest effect in transcription and hyperrecombination (Gonzalez-Aguilera et al, 2008), are the only subunits with a clear impact on transcription elongation in vivo as determined by ChIP and the GLRO assay. Instead, Sus1, a subunit of both the THSC and SAGA complexes (Rodriguez-Navarro et al, 2004), although involved in transcription initiation, does not have a significant effect in transcription elongation. As previously proposed, we consider that any possible effect of Sus1 in elongation may be mediated by disruption of the THSC complex (Luna et al, 2009).
Given that pre-mRNA processing occurs co-transcriptionally, defects in mRNP biogenesis could have a feedback in transcription. We believe that this is the case of THSC, CFIA and Rrp6 that work on mRNA export, 3′-end processing/termination and nuclear RNA surveillance, respectively (Birse et al, 1998; Libri et al, 2002; Gonzalez-Aguilera et al, 2008). The nature of these feedback mechanisms is still obscure. One possibility is that the pre-mRNA processing factors that interact with RNAPII modulate its elongating capacity. Alternatively, the defect in elongation observed in mutants of pre-mRNA processing factors could be a side effect of the coupling between transcription and mRNA processing. Thus, the aberrant mRNPs generated co-transcriptionally could impede RNAPII elongation via activation of quality control mechanism that slowdown RNAPII or by physically blocking RNAPII elongation. An example of the latter was shown in the absence of the THO complex. The co-transcriptional pre-mRNA produced in these mutants hybridize with the template DNA originating R-loops that impair RNAPII elongation (Huertas and Aguilera, 2003; Tous and Aguilera, 2007). Notably, however, not all the mutations that hinder pre-mRNA processing diminish elongation (Luna et al, 2005). Further analysis is necessary to know how the mRNP is modified when elongation is affected and why only specific mRNA processing and export factors have an impact on elongation.
Since the Nup84 complex, as part of the NPC, is also involved in mRNP export (Doye et al, 1994; Heath et al, 1995; Dockendorff et al, 1997) it may influence transcription elongation in the same manner as other pre-mRNPs processing factors. Indeed, the Nup84 complex directly interacts with the mRNA exporter Mex67–Mtr2 (Yao et al, 2008) that is also needed for efficient transcription elongation (Jimeno et al, 2002). Besides, gene migration to the NPC during transcription opens the possibility of nucleoporins being directly implicated in this process (Brickner and Walter, 2004; Casolari et al, 2004; Cabal et al, 2006). Interestingly, all Nup84 complex components but Seh1 activate transcription when fused to a DNA-binding domain (Menon et al, 2005). This seems to be a conserved feature as the fusion of human NUP98 to the PMX1 homeobox activates transcription of several genes in some leukaemia patients (Hirose et al, 2008). Moreover, Drosophila Nup98, Nup62, Nup50, Sec13 and some nucleoporins containing Phe-Gly repeats associate with long chromosomal regions in polytene chromosomes co-localizing with RNAPII and active chromatin marks. Knocking down these nucleoporins affect expression of the genes contained in these regions (Capelson et al, 2010; Kalverda et al, 2010). Importantly, the interaction between these nucleoporins and active chromatin take place in the nucleoplasm far from the NPC (Kalverda et al, 2010). However, the lack of a defect in transcription elongation observed in nup84Δ WCE clearly indicates that Nup84 does not function in RNAPII elongation in the absence of an intact nuclear envelope. Our data suggest that the Nup84 complex has a positive role in transcription elongation as part of a functional NPC, consistent with the coupling between transcription and mRNA export (Rodriguez-Navarro and Hurt, 2011).
In summary, this study validates a novel and highly efficient method for the analysis of transcription elongation in vivo, clarifying some of the controversial effects on elongation previously reported for particular protein factors, such as different subunits of THSC or PAF. Furthermore, this study indicates that RNAPII elongation depends not only on bona fide transcription-elongation factors that may diminish RNAPII pausing and facilitate its progression through chromatin but also on specific factors with a main role in processing and export of the nascent mRNP. Importantly, we found that the Nup84 complex has a transcriptional-elongation role linked to its function in RNA export. Further work is needed to determine the mechanisms by which these factors can modulate RNAPII elongation, but our work unambiguously uncovers a link between mRNA processing and export functions and transcription elongation.
Materials and methods
Strains and plasmids
Yeast strains used were wild-type W303-1A (R Rothstein, Columbia, NY) and its isogenic mutants rna14-1 and rna15-1 (F Lacroute); wild-type YPH499 and its isogenic mutant YVV9 (rpb9Δ::HIS3) (Van Mullem et al, 2002) and wild-type BY4741 and its isogenic mutants dst1Δ (Y04411), spt4Δ (Y06986), cdc73Δ (Y05326), leo1Δ (Y02379), paf1Δ (Y05727), rtf1Δ (Y04611), rrp6Δ (Y01777), gcn5Δ (Y07285), sgf73Δ (Y0443), spt20Δ (Y07390), thp1Δ (Y01764), sac3Δ (Y03517), sus1Δ (YBR111w-A), nup84Δ (Y03813 and Y13813), nup133Δ (Y05998) and nup60Δ (Y00407) (Euroscarf, Frankfurt, DE). dst1Δ nup84Δ and spt4Δ nup84Δ double mutants were generated by crossing nup84Δ (Y13813) with either dst1Δ (Y04411) or spt4Δ (Y06986) in the BY4741 genetic background.
Plasmids pCM184-LAUR (Jimeno et al, 2002) and pGCYC1-402 (Rondon et al, 2003) were described previously. Plasmid pG-Leu-CYCds for transcription run-on analysis was kindly provided by D Brow (Steinmetz and Brow, 2003). Plasmid pCYC-LacZ (GLRO-long) was constructed by amplifying 2 kb from the lacZ gene using Pfu polymerase and the primers lacz-Up and lacz-Down (sequence indicated in Supplementary Table II). This fragment was cloned into pG-Leu-CYCds digested with XhoI after filling with Klenow fragment.
In vivo GLRO assays of transcription elongation
Wild-type and mutants strains harbouring G-less cassette plasmids, pG-Leu-CYCds (GLRO-short) or pCYC-LacZ (GLRO-long), were grown to an OD600 of 0.5 in SC-Leu at 30°C. Transcription run-on assays were carried out as previously described (Steinmetz and Brow, 2003). Dried gels were analysed with a Phosphorimager (FujiFLA5100) using ImageQuant software (Molecular Dynamics). For each sample, the ratio of total counts in the 132-nt band divided by total counts in the 262-nt band was determined and corrected with the U content (40 U residues in the short cassette and 102 U in the long one).
In vitro transcription-elongation assays
WCEs were prepared from yeast cells grown in YEPD at 30°C to an OD600 of 1. In vitro transcription was performed on pGCYC1-402 as described in Rondon et al (2003) with the modifications introduced by Mariconti et al (2010).
ChIP of RNAPII
For ChIP experiments with the GAL1p::YLR454w fusion (Mason and Struhl, 2005), strains were grown in synthetic complete medium lacking tryptophan (SC-trp) with 2% glycerol–2% lactate to an OD660 0.5 and transcription was activated adding 2% galactose. Samples were taken after 4 h in galactose and ChIP assays were performed as described (Gonzalez-Aguilera et al, 2008). For ChIP experiments with pLAUR and the GRLO-long system, yeast cells were grown in selective SC-trp or SC-leu at 30°C to an OD600 of 0.5. For ChIP in the HSP104 gene, the strains were grown in yeast extract-peptone-dextrose (YPD) at 30°C to an OD600 of 0.5 and heat shocked by the addition of the appropriate volume of warm media followed by incubation at 37°C for 30 min. Immunoprecipitations were performed with monoclonal anti-Rpb1-CTD antibody 8WG16 (Berkeley antibody company) and Dynabeads Protein A (Invitrogen). The relative abundance of each DNA fragment was calculated normalizing IP/input ratios as described previously (Gonzalez-Aguilera et al, 2008). In all cases, ChIPs were performed from three independent cultures, and quantitative PCRs were repeated three times for each culture. The primers used are shown in Supplemental Table II.
Transposon-mediated mutagenesis screening
The haploid strain W303NK (GAL1p::lacZ integrated in the URA3 locus in W303-1A) was transformed with the transposon-mutagenized yeast genome library containing the Tn3::LEU2::lacZ construct (Burns et al, 1994). lacZ transcription was initially assayed adding top agar with X-gal to the plates (0.5% low-melting agarose in 0.25 M phosphate buffer, 1% SDS with 1.2 mg/ml X-gal). β-Galactosidase activity was quantified in the white candidates as described (Chavez and Aguilera, 1997). Genomic DNA from the candidate was digested with EcoRV or ClaI, ligated and amplified by PCR using a primer place at 5′-end of the left arm of Tn3. PCR products were in sequence to determine Tn3 insertion point.
Supplementary Material
Acknowledgments
We thank P Thuriaux and D Brow for providing strains and plasmids, H Gaillard for critical reading of the manuscript and D Haun for style supervision. This work was funded by grants from the Spanish Ministry of Science and Innovation (BFU2006-05260 and BFU2010-16372) and the Junta de Andalucía (BIO102 and CVI2549).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM (1997) Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem 272: 14747–14754 [DOI] [PubMed] [Google Scholar]
- Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ (1998) Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301 [DOI] [PubMed] [Google Scholar]
- Brickner JH, Walter P (2004) Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M (1994) Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev 8: 1087–1105 [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U (2006) SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Candau R, Zhou JX, Allis CD, Berger SL (1997) Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J 16: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW (2010) Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Chavez S, Aguilera A (1997) The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 11: 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Garcia-Rubio M, Prado F, Aguilera A (2001) Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 21: 7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN (1997) C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol 17: 906–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC (1994) A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J 13: 6062–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E (2004) Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol 6: 840–848 [DOI] [PubMed] [Google Scholar]
- Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E (2002) The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 21: 5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Wellinger RE, Aguilera A (2009) Methods to study transcription-coupled repair in chromatin. Methods Mol Biol 523: 141–159 [DOI] [PubMed] [Google Scholar]
- Gallardo M, Aguilera A (2001) A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics 157: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A (2003) Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J Biol Chem 278: 24225–24232 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Tous C, Gomez-Gonzalez B, Huertas P, Luna R, Aguilera A (2008) The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 19: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG (2005) Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol 25: 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25: 31–42 [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Hampsey M (1997) A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133 [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN (1995) Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol 131: 1677–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming SA, Jansma DB, Macgregor PF, Goryachev A, Friesen JD, Edwards AM (2000) RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J Biol Chem 275: 35506–35511 [DOI] [PubMed] [Google Scholar]
- Hirose K, Abramovich C, Argiropoulos B, Humphries RK (2008) Leukemogenic properties of NUP98-PMX1 are linked to NUP98 and homeodomain sequence functions but not to binding properties of PMX1 to serum response factor. Oncogene 27: 6056–6067 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Hull MW, McKune K, Woychik NA (1995) RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev 9: 481–490 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch TR, Chamberlin MJ (1982) Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem 257: 5286–5295 [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M (2010) Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371 [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG (2010) The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140: 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E (2008) Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10: 707–715 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon JC III, Wind M, Saunders L, Hock MB, Reines D (1998) Mutations in RNA II polymerase and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol 18: 5771–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH (2002) Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A (2008) Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma 117: 319–331 [DOI] [PubMed] [Google Scholar]
- Luna R, Gonzalez-Aguilera C, Aguilera A (2009) Transcription at the proximity of the nuclear pore: a role for the THP1-SAC3-SUS1-CDC31 (THSC) complex. RNA Biol 6: 145–148 [DOI] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A (2005) Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell 18: 711–722 [DOI] [PubMed] [Google Scholar]
- Mariconti L, Loll B, Schlinkmann K, Wengi A, Meinhart A, Dichtl B (2010) Coupled RNA polymerase II transcription and 3′ end formation with yeast whole-cell extracts. RNA 16: 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM (2005) Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA (2004) The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14: 447–456 [DOI] [PubMed] [Google Scholar]
- Nesser NK, Peterson DO, Hawley DK (2006) RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci USA 103: 3268–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 3: 109–118 [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodriguez-Navarro S (2008) Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev 22: 2811–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA (2005) A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Riles L, Shaw RJ, Johnston M, Reines D (2004) Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Hurt E (2011) Linking gene regulation to mRNA production and export. Curr Opin Cell Biol, doi: 10.1016/j.ceb2010.12.002 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A (2004) Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep 5: 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J 22: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ (2010) Transcript elongation by RNA polymerase II. Annu Rev Biochem 79: 271–293 [DOI] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM (2005) A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell 20: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J 21: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA (2003) Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 23: 6339–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP (2010) The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11: 490–501 [DOI] [PubMed] [Google Scholar]
- Tous C, Aguilera A (2007) Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun 360: 428–432 [DOI] [PubMed] [Google Scholar]
- Van Mullem V, Wery M, Werner M, Vandenhaute J, Thuriaux P (2002) The Rpb9 subunit of RNA polymerase II∣ binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J Biol Chem 277: 10220–10225 [DOI] [PubMed] [Google Scholar]
- Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Warner JR (1991) Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol 194: 423–428 [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Yao W, Lutzmann M, Hurt E (2008) A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J 27: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.