Abstract
Genetic screening of yeast for sld (synthetic lethality with dpb11) mutations has identified replication proteins, including Sld2, -3, and -5, and clarified the molecular mechanisms underlying eukaryotic chromosomal DNA replication. Here, we report a new replication protein, Sld7, identified by rescreening of sld mutations. Throughout the cell cycle, Sld7 forms a complex with Sld3, which associates with replication origins in a complex with Cdc45, binds to Dpb11 when phosphorylated by cyclin-dependent kinase, and dissociates from origins once DNA replication starts. However, Sld7 does not move with the replication fork. Sld7 binds to the nonessential N-terminal portion of Sld3 and reduces its affinity for Cdc45, a component of the replication fork. Although Sld7 is not essential for cell growth, its absence reduces the level of cellular Sld3, delays the dissociation from origins of GINS, a component of the replication fork, and slows S-phase progression. These results suggest that Sld7 is required for the proper function of Sld3 at the initiation of DNA replication.
Keywords: Cdc45, cell cycle, DNA replication, Sld3, yeast
Introduction
Eukaryotic chromosomes must be duplicated once and only once per cell cycle to maintain the integrity of their genetic information. To ensure the accurate timing of chromosomal duplication and to prevent rereplication during the cell cycle, the initiation of DNA replication is strictly regulated. DNA replication is initiated from specific regions called ‘origins’, which are scattered in large numbers along chromosomes. The six-subunit origin recognition complex (Orc) associates with replication origins throughout the cell cycle in budding yeast. Mcm, a DNA helicase comprising Mcm2–7, is loaded onto the Orc-bound origins to form the prereplicative complex (pre-RC). This loading requires Cdc6 and Cdt1, and occurs from late M to G1 phase. The pre-RC-formed origins are activated by two different protein kinases, cyclin-dependent kinase (CDK) and the Dbf4-dependent Cdc7 kinase (Labib, 2010). Once an origin has been activated, the pre-RC disassembles, and its reassembly is prevented until the next cell cycle (Bell and Dutta, 2002; Ghaemmaghami et al, 2003; Schwob and Labib, 2006; Sivaprasad et al, 2006; Walter and Araki, 2006; Masai et al, 2010). The process by which replication origins are activated was unclear until the replication proteins involved in this step were identified and characterized.
Screening for the yeast sld (synthetic lethality with dpb11-1) mutations (Kamimura et al, 1998) paved the way for the clarification of the initiation step in chromosomal DNA replication after the formation of the pre-RC. This screening isolated the sld1, -2, -3, -4, -5, and -6 mutations, which cannot be combined with the dpb11-1 mutation in the same cell. The sld1 mutation occurs in the third largest subunit, Dpb3, of DNA polymerase ε (Pol ε). This polymerase synthesizes the leading strand and functions in the checkpoint at the replication fork, and in the initiation of DNA replication (Masumoto et al, 2000; Dohrmann and Sclafani, 2006; Lou et al, 2008; Pursell and Kunkel, 2008). The DPB11 gene was originally isolated as a multicopy suppressor of mutations in Pol ε (Araki et al, 1995). The sld2 and sld3 mutations were identified in previously unidentified replication proteins, Sld2 and Sld3, both of which interact with Dpb11 when they are phosphorylated by CDK (Wang and Elledge, 1999; Kamimura et al, 1998, 2001; Masumoto et al, 2002; Tanaka et al, 2007; Zegerman and Diffley, 2007). These phosphorylation-dependent interactions are essential and represent the minimal requirement for the CDK-dependent activation of DNA replication (Tanaka et al, 2007; Zegerman and Diffley, 2007; Tanaka and Araki, 2010). sld4 is a mutation in Cdc45, a protein required for the initiation and elongation steps of DNA replication. Sld3 and Sld4 (Cdc45) form a complex throughout the cell cycle, which associates in a mutually dependent manner with the pre-RC-formed origins. This association occurs with early-firing origins in G1 and with late-firing origins in S phase (Kamimura et al, 2001; Kanemaki and Labib, 2006). The sld5 mutation occurs in a subunit of GINS, which consists of Sld5, Psf1, Psf2, and Psf3. GINS functions in the initiation and elongation stages of chromosomal DNA replication (Kanemaki et al, 2003; Kubota et al, 2003; Takayama et al, 2003; Labib and Gambus, 2007). Cdc45 and GINS associate with Mcm to form an active replicative helicase, the Cdc45–Mcm–GINS (CMG) complex (Gambus et al, 2006; Moyer et al, 2006; Pacek et al, 2006; Ilves et al, 2010). The sld6 mutation occurs in Rad53 kinase, which functions at checkpoints and in the regulation of the initiation of DNA replication (Dohrmann and Sclafani, 2006).
Dpb11 has two pairs of tandem BRCT domains, known as the phosphopeptide-binding domains (Glover et al, 2004). The N-terminal pair of BRCT domains binds to phosphorylated Sld3, and the C-terminal pair binds to phosphorylated Sld2 (Masumoto et al, 2002; Tanaka et al, 2007; Zegerman and Diffley, 2007). Their interactions are essential for the activation of origins, but the mechanism underlying the activation of origins by these interactions is unclear. One clue comes from the identification of the preloading complex (pre-LC). This complex contains Pol ε, GINS, Dpb11, and Sld2, and its formation depends on the interaction between Dpb11 and Sld2, but not on the pre-RC. Thus, the phosphorylation-dependent interaction between Dpb11 and Sld2 has a role in the formation of the pre-LC. Although the role of the interaction between Dpb11 and Sld3 is unknown, it has been proposed that GINS in the pre-LC is recruited to replication origin to form the CMG complex via the interaction between Dpb11 in the pre-LC and phosphorylated Sld3 on origins (Muramatsu et al, 2010; Tanaka and Araki, 2010). At the initiation of DNA replication, CMG, Dpb11, Sld2, and Sld3 dissociate from origins and the CMG complex moves with the replication fork.
In this study, we screened for novel sld mutations using the dpb11-24 allele. This dpb11 allele has a mutation in the N-terminal pair of tandem BRCT domains of Dpb11, whereas the dpb11-1 mutation used in the previous screening is located in the C-terminal BRCT domains. This new round of screening led to the isolation of the sld7 mutation, which occurs in a previously uncharacterized gene, and other mutations in known replication genes. Although it is not essential, the SLD7 gene has an important role in chromosomal DNA replication, as do the other SLD genes. The Sld7 protein forms a tight complex with Sld3 throughout the cell cycle, stabilizes Sld3, and reduces the affinity of Sld3 for Cdc45. The absence of Sld7 delays the dissociation of GINS from the origin and the progression of S phase. Based on these data, we discuss a possible function for Sld7.
Results
SLD7 gene is not essential but is important for cell growth and chromosomal DNA replication
To gain insight into the molecular mechanisms underlying chromosomal DNA replication, we rescreened for further sld mutations, which cannot be combined with the dpb11-24 mutation in a cell, as described previously (Kamimura et al, 1998). We isolated six mutations from 32 000 mutagenized colonies. Five of these occurred in known genes (two in SLD3, and one each in DPB2, MCM3, and MCM10), and the remaining mutation occurred in an unknown gene (YOR060c, Saccharomyces Genome Database) (Valens et al, 1997). We named this gene SLD7 (synthetic lethality with Dpb11-24 7) and characterized it further. This gene encodes a 29-kDa protein comprising 257 amino acids. The Sld7 protein has no sequence motifs that indicate its function. Using the BLAST and FASTA homology search programs, we found possible homologues of the Sld7 protein in Saccharomyces yeasts and in closely related genera, such as Zygosaccharomyces, Kluyveromyces, Eremothecium, and Pichia (Supplementary Figure S1). However, a homologue of the Sld7 protein could not be found in fission yeasts or in organisms other than yeasts. The isolated mutation (sld7-1) involves the substitution of arginine for glycine at position 42 of the Sld7 protein. The glycine 42 residue is conserved in the Sld7 homologues found in Zygosaccharomyces and Eremothecium species (Supplementary Figure S1).
To examine whether the SLD7 gene is essential for cell growth, one of the SLD7 gene copies in a leu2/leu2 diploid cell was replaced with sld7Δ∷LEU2. Sporulation and tetrad dissection of the resultant SLD7/sld7Δ∷LEU2 cells yielded two large and two small colonies (Figure 1A). All the large colonies were Leu–, whereas all the small colonies were Leu+, indicating that the small colonies were derived from an sld7Δ∷LEU2 allele. Two of 36 Leu– spores (wild type) and 15 of 36 Leu+ spores (sld7Δ∷LEU2) did not form colonies. The viability of the Leu+ spores varied across experiments (50–90%). Microscopic observations revealed that some of these lethal spores carrying the sld7Δ∷LEU2 allele died without germinating, whereas others died after several divisions. The dissection of tetrads derived from SLD7/sld7Δ∷LEU2 cells carrying the YEp195SLD7[URA3] plasmid revealed that 90% of the spores formed colonies. All the clones grew on 5-fluoroorotic acid plates, which only allow the growth of cells that have lost YEp195SLD7[URA3], although the sld7Δ (Leu+Ura–) cells grew slightly more slowly on the plates than did the wild-type cells. Therefore, the SLD7 gene is important but not essential for cell growth. Although the null mutant of SLD7, constructed in a systematic high-throughput deletion study, was reported to be inviable (Winzeler et al, 1999), the same construct obtained from EUROSCARF was viable in our hands. The sld7Δ cells grew more slowly than the wild-type cells in liquid YPD medium (doubling time for the wild type was 77.5 min and for sld7Δ was 86.2 min at 30°C), as observed on the plates.
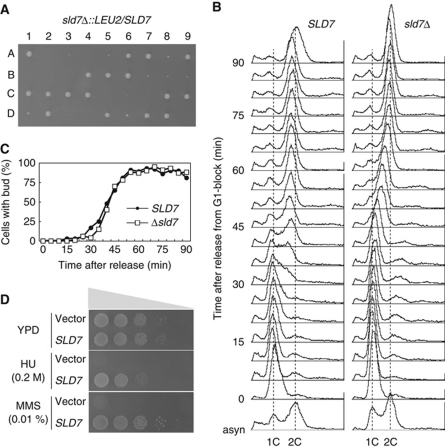
Figure 1.
Important functions of Sld7 in chromosomal DNA replication. (A) sld7Δ∷LEU2/SLD7 diploid cells were sporulated and dissected. The spores were incubated on YPD plates at 30°C for 3 days. (B) DNA replication in sld7Δ cells. The YTT1 (sld7Δ) and wild-type (WT: YYK9) cells were synchronized in G1 phase with α-factor and released from G1 arrest at 25°C. Aliquots of the cells were sampled at 5 min intervals, and their DNA content was measured by flow cytometry. (C) The population of cells with buds after release from the G1 block in the same culture, as in (B). (D) Sensitivity of sld7Δ cells to HU and MMS. Plasmids with or without the SLD7 gene were introduced into sld7Δ cells. Tenfold serial dilutions of the cells were spotted onto YPD plates containing 0.2 M HU or 0.01% MMS. The cells were incubated at 25°C for 4 days.
The sld7Δ mutation was synthetically lethal with mutations in DPB11 (dpb11-1 and dpb11-24), SLD genes (drc1-1 [sld2], sld5-12, sld3-5, and cdc45-27 [sld4]), Pol ε (pol2-11), and GINS (psf1-1), but not with mutations in components of the pre-RC (mcm2-1, mcm3-1, orc2-1, and orc5-1) or in the checkpoint gene (ddc1Δ). These genetic interactions suggest that Sld7, like other Sld proteins, functions in DNA replication. Therefore, we examined the chromosomal DNA replication in sld7Δ cells with flow cytometry. The sld7Δ cells arrested in G1 phase with α-factor were released at 25°C, and their DNA content was measured. The DNA content of the wild-type cells increased after 30 min and reached 2C 45 min after release (Figure 1B), whereas that of the sld7Δ cells increased after 40 min and reached 2C 70 min after release, taking about twice as long as the wild-type cells to complete their DNA replication. The buds appeared at the same time in both cells, suggesting that the cell-cycle events at the G1/S boundary are normal in sld7Δ cells, except for DNA replication (Figure 1C). Although the sld7Δ cells transiently exposed to hydroxyurea (HU) or methyl methanesulfonate (MMS) had slightly reduced viability compared with that of the wild-type cells, like many mutants defective in DNA replication, their growth on plates was inhibited by HU or MMS more severely than was that of the wild-type cells (Figure 1D). Therefore, Sld7 seems to function in cooperation with the Dpb11 and Sld proteins to ensure efficient chromosomal DNA replication.
Sld7 protein forms a complex with Sld3 protein
To clarify the function of Sld7, we used two-step immunoprecipitation (IP) of cells expressing Sld7 protein tandemly tagged with 3Flag–HA (Sld7–3Flag–HA) to identify proteins that bind to Sld7. The Sld7–3Flag–HA complex was immunoprecipitated from the extracts with anti-Flag M2 affinity agarose and then with an anti-HA matrix, after the proteins had been released from the M2 affinity beads with Flag peptides. A comparison with immunoprecipitates from extracts of wild-type cells showed that an 80-kDa protein coprecipitated specifically with Sld7–3Flag–HA (Figure 2A). The band containing this protein was excised from the gel after electrophoresis, digested with trypsin, and subjected to analysis by mass spectrometry. According to the spectra assigned to the peptides in this analysis, the protein was identified as Sld3. Western blots probed with anti-Sld3 antibodies confirmed that the Sld3 protein coprecipitated with the Sld7–3Flag–HA complex (Figure 2A).
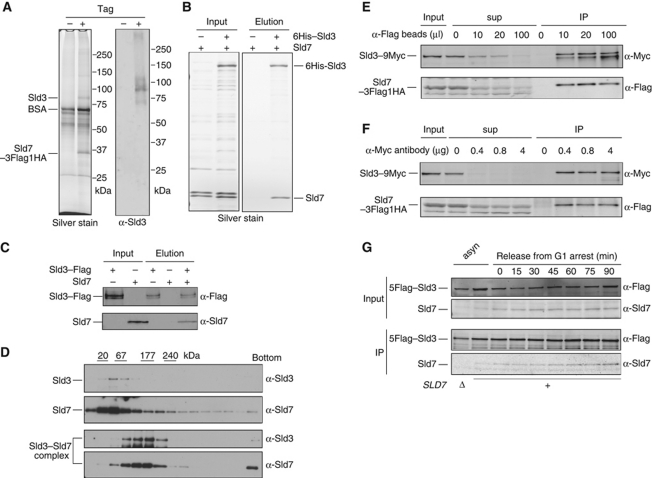
Figure 2.
Complex formation between Sld3 and Sld7. (A) YTT4 cells expressing Sld7–3Flag–HA were disrupted, and the Sld7–3Flag–HA protein was immunoprecipitated on anti-Flag M2 affinity agarose (Sigma) and released from the agarose with 3 × Flag peptide. The released Sld7–3Flag–HA complex was precipitated with anti-HA matrix (Roche) and released from the matrix with 3 × HA peptide. The 80-kDa protein that coprecipitated specifically with Sld7–3Flag–HA was subjected to mass spectrometric analysis. Western blots of the same sample were probed with anti-Sld3 antibodies. (B) Direct interaction between the purified 6His–Sld3 and Sld7 proteins. 6His–Sld3 and Sld7 proteins coexpressed in E. coli cells were adsorbed to Ni–NTA agarose (Qiagen) and eluted with imidazole. (C) Purified Sld3–Flag and Sld7 proteins were mixed. The proteins were bound to M2 anti-Flag agarose and eluted with 3 × Flag peptide. In all, 4% of the input (input) and 10% of the eluted sample (elution) were subjected to SDS–polyacrylamide gel electrophoresis, followed by western blotting with the indicated antibodies. (D) Sld3–Flag, Sld7, and 5Flag–Sld3–Sld7 were sedimented in a 15–30% glycerol gradient. The markers are chymotrypsinogen A (20 kDa), BSA (67 kDa), aldolase (177 kDa), and catalase (240 kDa). (E) Immunodepletion of the Sld7–3Flag–HA protein with various amounts of anti-Flag M2 beads. The crude yeast extracts were prepared from YTT5 cells (SLD7–3FLAG–HA SLD3–9MYC). After precipitation, the supernatants (sup) and precipitates (IP) were subjected to western blotting and probed with anti-Flag and anti-Myc antibodies. The input and samples corresponding to the same number of cells were applied to the gel. (F) Immunodepletion of the Sld3–9Myc proteins with various amounts of anti-Myc antibody (c-myc Ab-1 9E11; NeoMarkers) on Protein A beads. The same extracts prepared in (E) were used. After precipitation, the supernatants (sup) and precipitates (IP) were analysed as in (E). (G) Immunoprecipitation of Sld7–3Flag–HA from the extracts of synchronized cells using anti-Flag antibody. YTT4 cells expressing SLD7–3FLAG–HA were arrested in G1 phase with α-factor and then released from the G1 block. The immunoprecipitation in (E), (G), and (F) was specific for the interaction between Sld3 and Sld7 (Supplementary Figure S3).
To examine whether the interaction between Sld7 and Sld3 is direct, 6His–Sld3 and Sld7 proteins were coexpressed in Escherichia coli cells, and the 6His–Sld3 was precipitated with Ni–NTA agarose (Figure 2B). In this assay, Sld7 protein was precipitated and eluted with the 6His–Sld3 protein. Sld3–Flag and Sld7, expressed separately in and purified from E. coli cells, also coprecipitated and coeluted when they were mixed (Figure 2C). Therefore, Sld7 protein binds directly to Sld3 protein. We also analysed the purified proteins by glycerol gradient sedimentation. When Sld3 and Sld7 were applied separately to a glycerol gradient and centrifuged, they sedimented in the fractions corresponding to 65 kDa (Figure 2D). When the Sld3–Sld7 complex purified from yeast was applied, Sld3 and Sld7 cosedimented in the fraction corresponding to 180 kDa, indicating that they had formed a complex.
Using coprecipitation, we also examined the proportions of cellular Sld3 and Sld7 that exist as the Sld3–Sld7 complex. As shown in Figure 2E and F, most of the Sld3–9Myc and Sld7–3Flag–HA proteins coprecipitated when the tagged proteins were precipitated with anti-Flag antibody or anti-Myc antibody. Moreover, the Sld3 and Sld7 proteins coprecipitated throughout the cell cycle (Figure 2G). Therefore, most of the Sld3 and Sld7 proteins in the cells form a complex throughout the cell cycle.
Sld7 binds to the N-terminal portion of Sld3
The Sld7-binding region of the Sld3 protein was determined using a two-hybrid assay. Interactions between Sld7 and Sld3 (residues 91–668) and Sld3 (residues 1–120) were observed, but not between Sld7 and Sld3 (residues 121–668) or Sld3 (residues 1–60) (Figure 3A). The interaction between Sld3 (residues 1–90) and Sld7 produced a very weak signal. These results suggest that the Sld7 protein binds to at least two regions of the Sld3 protein, upstream and downstream from amino acid 90, including the N-terminal portion of the Sld3 protein, within amino acids 60–120, which is not essential for cell growth (Figure 3A). The two-hybrid assay also revealed that the central portion of Sld3 (residues 150–430) interacts with Cdc45 (Figure 3A).
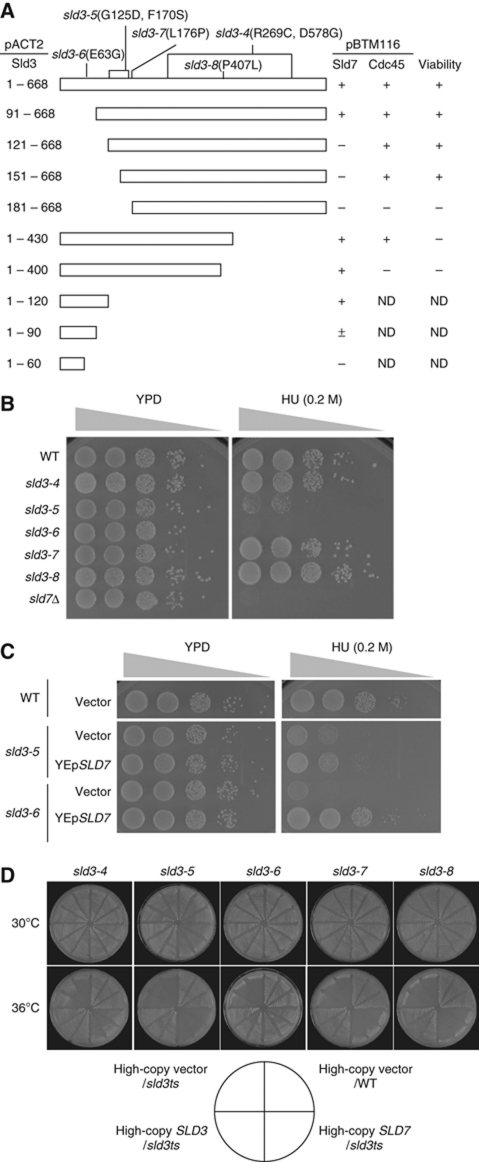
Figure 3.
Interaction between Sld3 and Sld7. (A) The regions of Sld3 that interact with Sld7 and Cdc45 were determined. Various fragments derived from Sld3 were cloned into pACT2. The resultant plasmids and either pBTM116–Sld7 or pBTM116–Cdc45 were introduced into the yeast strain L40, and the interaction was detected by lacZ expression with colour. To determine whether the fragment is sufficient for cell growth, YCplac22 or pRS313 bearing various fragments of SLD3 with its own promoter was introduced into YYK13 cells (sld3Δ∷LEU2[YEp195SLD3]) (Kamimura et al, 2001), and the resultant transformants were streaked onto FOA plates, which allow the growth of transformants expressing active Sld3 from YCplac22 or pRS313. (B) The HU sensitivity of the five temperature-sensitive mutants of SLD3: sld3-4, -5, -6, -7, and -8. Tenfold serial dilutions of the mutant cells were spotted onto YPD plates containing 0.2 M HU. The cells were incubated at 25°C for 4 days. (C) High-copy plasmid (YEp) with or without the SLD7 gene was introduced into the sld3-5 and sld3-6 cells. Then, 10-fold serial dilutions of the cells were spotted onto YPD plates containing 0.2 M HU and incubated at 25°C for 4 days. (D) High-copy SLD7 suppressed the temperature-sensitive growth of the sld3-6 mutant, but not that of the sld3-4, -5, -7, or -8 mutant. The cells carrying YEplac195 (vector) or YEplac195SLD7 (YEpSLD7) were incubated at 30 or 36°C for 3 days.
We have previously reported five temperature-sensitive mutations of SLD3: sld3-4, -5, -6, -7, and -8 (Figure 3A; Kamimura et al, 2001). Among these mutations, sld3-5 and sld3-6 occur in the N-terminal portion of the protein and conferred sensitivity to HU, as did sld7Δ (Figure 3B). High-copy SLD7 suppressed the HU sensitivity and temperature-sensitive growth of sld3-6 cells (Figure 3C and D). In contrast, a previous study showed that high-copy CDC45 suppressed the temperature-sensitive growth conferred by the sld3-4, -5, -7, and -8 mutations, which occur in the central portion of the protein, but not that conferred by the sld3-6 mutation (Kamimura et al, 2001). Furthermore, Sld3 lacking the region that interacts with Sld7 also conferred sensitivity to HU and this sensitivity was not suppressed by high-copy SLD7 (Supplementary Figure S4). These results are consistent with the observation that Sld7 and Cdc45 interact with the N-terminal and central portions of Sld3, respectively.
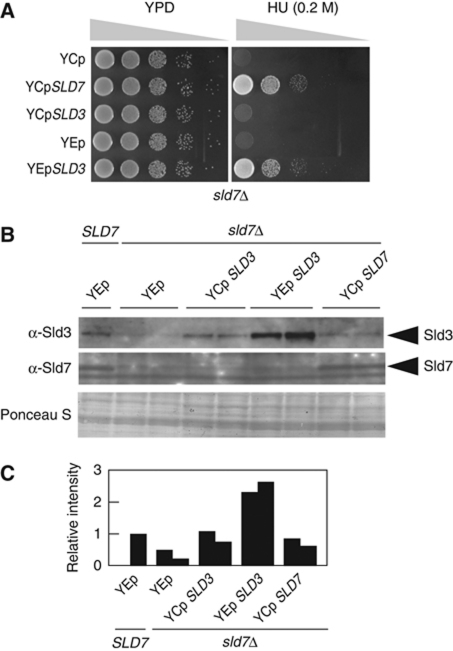
We also examined whether an increased dose of the SLD3 gene restores the HU sensitivity of the sld7Δ mutant cells. As shown in Figure 4A, the SLD3 gene on a high-copy plasmid restored the HU sensitivity of the sld7Δ mutant cells, whereas the SLD3 gene on a low-copy plasmid did not. The level of the Sld3 protein in sld7Δ cells was reduced and was restored by the introduction of SLD3 on a low-copy plasmid. The introduction of SLD3 on a high-copy plasmid more than doubled the Sld3 level (Figure 4B and C).
Figure 4.
Reduced Sld3 protein levels in sld7Δ cells. (A) The SLD3 gene on the high-copy plasmid YEplac195 (YEp) or on the low-copy plasmid YCplac33 (YCp) was introduced into YTT1 cells (sld7Δ). Tenfold dilutions of the cells were spotted onto YPD plates containing 0.2 M HU and then incubated at 25°C for 4 days. (B, C) Restoration of the Sld3 protein levels in sld7Δ cells by the introduction of SLD3 on a plasmid. The cells carrying the indicated plasmids were disrupted as described in ‘Materials and methods’ and subjected to SDS–polyacrylamide gel electrophoresis, followed by western blotting and probing anti-Sld3 and anti-Sld7 antibodies.
Sld7 associates with replication origins in an Sld3-dependent manner
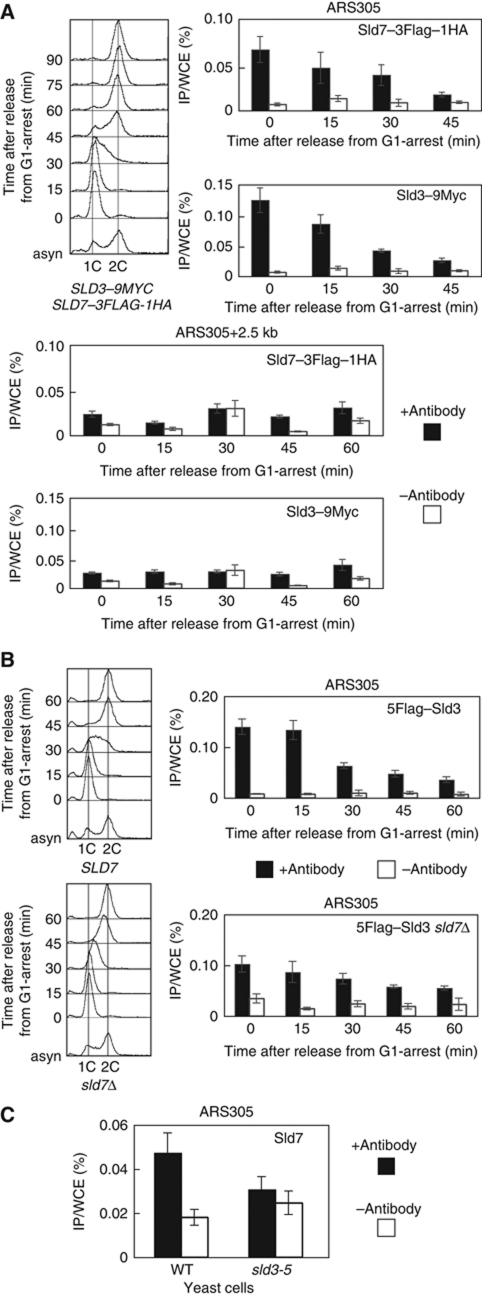
The Sld3 protein associates with early-firing origins in G1 phase and with late-firing origins in S phase, and dissociates from origins when DNA replication starts (Kamimura et al, 2001; Kanemaki and Labib, 2006). Because the Sld7 protein forms a complex with the Sld3 protein throughout the cell cycle, it was expected to associate with replication origins. Therefore, we confirmed the association between Sld7 and origins using a chromatin IP (ChIP) assay. Cells expressing Sld3–9Myc and Sld7–3Flag–HA were arrested in G1 phase with α-factor and released at 25°C. Aliquots of the cells were sampled, and cross-linked DNA–protein complexes were immunoprecipitated using anti-Flag and anti-Myc antibodies. As shown previously, the Sld3–9Myc protein associated with ARS305, an early-firing origin, 0–30 min after release from the G1 block (Figure 5A), which corresponds to the period from late G1 to very early S phase on flow cytometry (Figure 5A). The origin-association signals of the Sld3–9Myc proteins decreased gradually. The ChIP assay of the Sld7–3Flag–HA protein showed exactly the same pattern as that of the Sld3–9Myc protein (Figure 5A). In the region neighbouring ARS305, the signals were at the background level. These results indicate that the Sld7 protein, as a component of the Sld3–Sld7 complex, associates with early-firing origins in G1 phase, and then dissociates gradually from origins, and that this protein does not move with the replication fork.
Figure 5.
Sld3-dependent association of Sld7 with chromatin. (A) Associations of Sld7–3Flag–HA and Sld3–9Myc proteins with chromatin. YTT5 cells expressing SLD3–9MYC and SLD7–3FLAG–HA were arrested in G1 phase with α-factor and released at 25°C. Aliquots of the cells were sampled at 15 min intervals and subjected to a ChIP assay. (B) YTT6 (5FLAG–SLD3) and YTT7 (5FLAG–SLD3 sld7Δ) cells were synchronized and subjected to a ChIP assay, as described in (A). The cells expressing 5FLAG–SLD3 progressed to S phase faster than the parental cells. (C) YYK19 (sld3-5) cells were grown at 25°C and α-factor was added to arrest the cells at G1, after which the cells were incubated at 37°C for a further 2 h. The cells were collected and subjected to a ChIP assay. The levels of Sld7 protein in the sld3-5 cells were almost the same as those observed in the wild-type cells (Supplementary Figure S5).
Sld3 associated with ARS305 from G1 phase, even in sld7Δ cells (Figure 5B). The reduced signals for the association between Sld3 and the origins might be attributable to the reduced level of Sld3 protein (Figure 4B and C). However, Sld3 in the sld7Δ cells dissociated from the origins slightly later than was observed in the wild-type cells. Therefore, the Sld3 protein associates with replication origins in the absence of the Sld7 protein, whereas the Sld7 protein is presumably required for a function of Sld3 other than its association with origins.
We then examined whether the association of Sld7 with origins is dependent on Sld3. Cells expressing temperature-sensitive alleles of sld3 and Sld7–5Flag were inviable or very sick, probably because of the synthetic effect. Therefore, we used anti-Sld7 antibodies to perform a ChIP assay of the G1-arrested cells at the nonpermissive temperature. As shown in Figure 5C, the association of the Sld7 protein with ARS305 was reduced in sld3-5 cells at the nonpermissive temperature. We have previously shown that the Sld3-5 protein does not associate with ARS305 efficiently at the nonpermissive temperature (Kamimura et al, 2001). Therefore, this result indicates that Sld7 alone does not associate with the origins, but seems to associate with them only as a complex with Sld3.
Sld7 affects the association of GINS with replication origins
When CDK is activated, the pre-LC, containing Dpb11, Sld2, Pol ε, and GINS, associates with replication origins through the interaction between CDK-phosphorylated Sld3 on origins and Dpb11, and Pol ε and GINS move with the replication fork when replication starts (Masumoto et al, 2000; Kanemaki and Labib, 2006; Muramatsu et al, 2010).
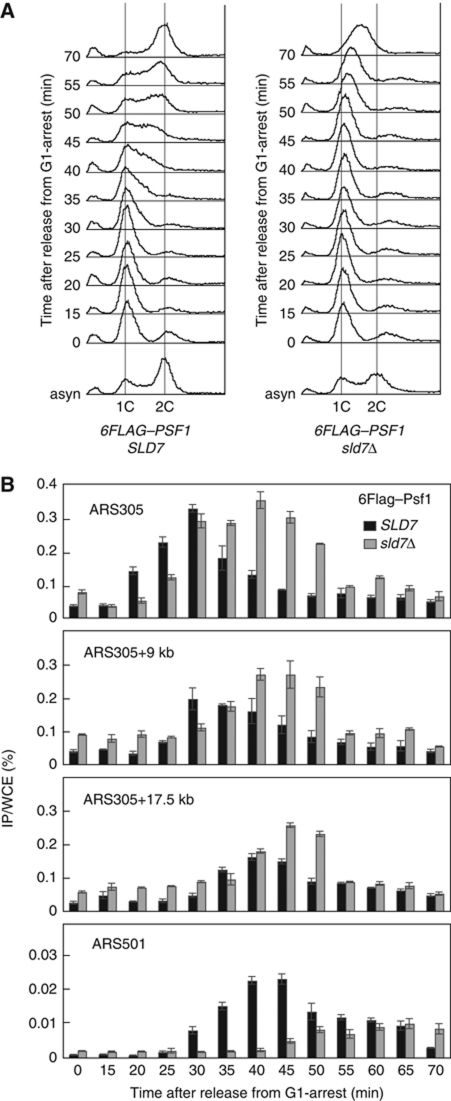
Therefore, we examined the chromatin association of Psf1, a subunit of the GINS complex, using a ChIP assay. In the wild-type cells, 6Flag–Psf1 associated with ARS305 in early S phase (its association level peaked at 30 min), then dissociated quickly from the origins. This was followed by its association with the neighbouring regions when replication started, indicating that Psf1 moves with the replication fork (Figure 6). In sld7Δ cells, the association of Psf1 with ARS305 occurred slightly later but reached the same level as that observed in the wild-type cells at 30 min. This high-level association continued to 45 min. As a consequence, the Psf1 displacement from the origin and its reassociation with the neighbouring sequences were substantially delayed (Figure 6B). This delay was more evident in its association with ARS501, a late-firing origin.
Figure 6.
Chromatin association of GINS in the presence or absence of Sld7. YYT2 (6FLAG–PSF1) or YTT9 (6FLAG–PSF1 sld7Δ) cells were arrested in G1 phase with α-factor and released at 25°C. Aliquots of the cells were sampled at 5 min intervals and subjected to flow cytometric analysis (A) and ChIP assay (B). The black and grey bars indicate the YYT2 (SLD7) and YTT9 (sld7Δ) cells, respectively.
Sld7 reduces the association between Cdc45 and Sld3
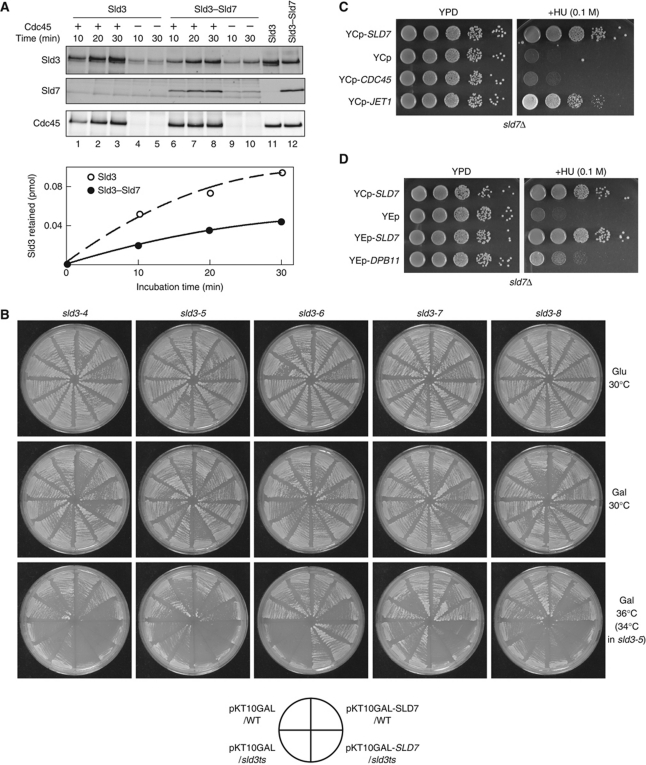
Cdc45, a component of the CMG complex, forms a complex with Sld3 and associates with origins in vivo (Kamimura et al, 2001). However, once DNA replication has started, Cdc45 dissociates from origins, forms a replication fork as a component of the CMG complex, and moves along chromosomal DNA, whereas Sld3 dissociates from origins but does not move with the replication fork (Kanemaki and Labib, 2006). This suggests that the efficient association and dissociation of Cdc45 and Sld3 are important for the proper function of the CMG complex. Therefore, we compared the associations between Cdc45, Sld3, and the Sld3–Sld7 complex using purified proteins. Sld3 and Sld3–Sld7 bound specifically to Cdc45, whereas Cdc45 did not bind to GINS or Sld2 (Supplementary Figure S6). Moreover, Sld3 bound to Cdc45 more efficiently than did Sld3–Sld7 (Figure 7A). This result indicates that Sld7 reduces the association between Cdc45 and Sld3.
Figure 7.
Reduced association between Cdc45 and Sld3 in the presence of Sld7. (A) The Sld3–Flag protein (1 pmol) and the Sld3–Flag–Sld7 complex (1 pmol) were incubated with Cdc45–Flag (0.4 pmol)-bound beads at 4°C for the indicated times. Half of the proteins bound to the beads were subjected to SDS–polyacrylamide gel electrophoresis, followed by western blotting, probed with anti-Sld3 and anti-Cdc45 antibodies. Lanes 11 and 12 contain samples of Sld3 (0.05 pmol), Sld3–Sld7 (0.05 pmol), and Cdc45 (0.2 pmol). (B) Plasmid encoding a Gal-inducible SLD7 gene was introduced into the sld3 temperature-sensitive mutants, sld3-4, -5, -6, -7, and -8. The cells were incubated on glucose or galactose plates lacking uracil at the indicated temperatures for 2 days. (C, D) Suppression of the HU sensitivity of the sld7Δ mutant by the introduction of JET1, an S-CDK bypass allele of the CDC45 gene (C), or DPB11 on a high-copy plasmids (D). YTT1 (sld7Δ) cells were transformed with the indicated plasmids. Tenfold dilutions of the cells were spotted onto plates and incubated at 25°C for 3 days.
The genetic interactions were also consistent with the in vitro result. The overexpression of the SLD7 gene from a galactose-inducible promoter suppressed the phenotype of the sld3-6 mutant, as did the SLD7 gene expressed from a multicopy plasmid (Figure 7B). However, the overexpression of SLD7 inhibited the cell growth of other temperature-sensitive sld3 mutants (sld3-4, -5, -7, and -8), which are suppressed by the multicopy CDC45 gene (Kamimura et al, 2001) at the semipermissive temperatures of these mutants (Figure 7B). We surmise that the increased copies of Sld7 protein further reduced the weak association between Cdc45 and Sld3 in these mutant cells.
We also found that the JET1 mutation partially suppressed the HU sensitivity of the sld7Δ cells (Figure 7C). JET1 is a mutant allele of CDC45 that bypasses the requirement for Sld3 phosphorylation by S-CDK (Tanaka et al, 2007). The phosphorylation of Sld3 is required for the formation of the complex between Sld3 and Dpb11, which is facilitated by Jet1 (Tanaka et al, 2007). We have previously shown that, in addition to JET1, high-copy DPB11 also bypasses the requirement for Sld3 phosphorylation by S-CDK, probably through the restoration of the Sld3–Dpb11 complex (Tanaka et al, 2007). This is also true for the suppression of the HU sensitivity of sld7Δ cells (Figure 7D). These observations are consistent with the increased association between Cdc45 and Sld3 in the absence of Sld7 (see Discussion).
Discussion
A recent genome-wide study of essential yeast genes strongly suggested that all the proteins involved in replication have been identified (Kanemaki et al, 2003). However, we have found a new replication protein, Sld7. This protein is not essential for DNA replication, although cells lacking it exhibit delayed chromosomal DNA replication. Presumably, its dispensability is the reason why Sld7 was not previously identified as a replication protein.
Role of Sld7 in DNA replication
In the absence of Sld7, the Sld3 level decreases (Figure 4B and C). Although we cannot exclude the possibility that Sld7 enhances the transcription and/or translation, it is most likely that the lack of Sld7 destabilizes Sld3. We speculate that Sld7 alters the conformation of Sld3 to increase its resistance to proteases and that the improper conformation of Sld3 in the absence of Sld7 makes it easily accessible to proteases. This Sld7-dependent conformational change may affect the efficiency of the initiation of DNA replication. Although the exact mechanisms underlying this inefficient initiation in sld7Δ cells are not known, we have shown that Sld7 reduces the association between Sld3 and Cdc45 (Figure 7A). Thus, it is conceivable that this reduced association contributes to the efficient displacement of Sld3 (Figure 5) and the CMG complex (Figure 6) from origins, allowing more efficient DNA replication.
Several phenomena observed in sld7Δ cells can be explained by the reduced levels of Sld3 and by the inefficient displacement of Sld3 from replication origins. A genome-wide study estimated the number of TAP-tagged Sld3 and Sld7 protein molecules to be 125 and 1690 per cell, respectively (Ghaemmaghami et al, 2003), and the number of replication origins to be about 247–400 per genome of Saccharomyces cerevisiae, depending on the methods used (Wyrick et al, 2001; Yabuki et al, 2002). In wild-type cells, the amount of Sld3 protein might be sufficient to be distributed evenly across the very early-firing origins during early S phase and to be redistributed to later-firing origins during mid or late S phase (Araki, 2010). However, because of the low levels of Sld3 protein, only a few origins in sld7Δ cells can associate with Sld3. Moreover, the redistribution of Sld3 in sld7Δ cells might be further delayed because the displacement of Sld3 from origins is delayed (Figure 5). Therefore, a very limited number of early-firing origins might fire at the same time in sld7Δ cells, and the redistribution of Sld3 may not be sufficient for the proper firing of origins. A computer simulation based on mathematical modelling predicted that the inefficient displacement of Sld3 from origins compromises the coherent firing of origins, thus causing their incomplete firing (Brümmer et al, 2010). This is consistent with the delay in S phase observed in sld7Δ cells (Figure 1B). In sld7 cells expressing an increased dose of Sld3, higher than that in wild-type cells, Sld3 probably associates with more replication origins than in the wild-type cells, and the redistribution of Sld3 is enough for the proper firing of origins, regardless of the inefficient Sld3 displacement from origins. Consistent with this scenario, high-copy SLD3 suppressed the HU sensitivity of sld7Δ cells (Figure 4A). The displacement of Sld3 from origins might require the establishment of the replication fork, and acceleration of the formation of the replication fork might compensate for the redistribution of Sld3. This might happen through the enhanced interaction between Dpb11 and Sld3, which is caused by JET1 and high-copy DPB11 (Figure 7C and D).
Both the association and dissociation of the replication proteins from origins might be important. Many replication proteins associate with replication origins during the initiation step of DNA replication. Some of them form the replication machinery on origins and initiate DNA synthesis. In this step, the proteins associate first with origins and then dissociate from them to synthesize the DNA at a distance from origins, so the proteins must switch their affinity for replication origins from an association mode to a dissociation mode. The way in which they alter their affinities is not understood. However, our study of Sld7 provides a clue to this mechanism. Sld3 and Cdc45 form a complex that associates with origins, and dissociate later when replication starts. Because Cdc45 is a component of the CMG complex, which is an active DNA helicase at the replication fork, and Sld3 does not move with the replication fork, the lack of Sld7 seems to compromise the dissociation between Sld3 and Cdc45, which consequently delays the displacement of GINS from the origins. Although Sld3 binds to Sld7 throughout the cell cycle, the reduction in the affinity of Sld3 for Cdc45 caused by Sld7 suggests that Sld3 can alter its affinity for Cdc45 by interacting with other proteins and can alter the affinity modes (associations and dissociations) of protein–protein interactions. Because many proteins assemble and interact with each other at replication origins, the affinities of these complexes for the replication origins may change.
Subunit structure of the Sld3–Sld7 complex
The Sld7 and Sld3 proteins exist as a tight complex. However, why the Sld3 and Sld7 proteins exist as two distinct subunits is unknown. We propose three possible regulatory roles for this subunit structure. First, the subunit structure may maintain the concentration of the Sld3 protein at low levels, even if the level of either protein increases occasionally, because free Sld3 is unstable and is degraded. Although we observed no toxic effect of the simultaneous overproduction of Sld3 and Sld7 (our unpublished result), this regulation may prevent the precocious activation of replication origins. Second, the ratio of the two subunits, Sld3 and Sld7, may regulate the activity of Sld3. It is conceivable that the ratio of the Sld3 and Sld7 proteins varies under different conditions and that changes to the ratio in the complex have an important role in the regulation of Sld3 function. Third, the free subunits may also have a role. For example, the free Sld7 protein levels may be regulated by its interaction with Sld3. Therefore, if the amount of Sld7 that is associated with Sld3 decreases, free Sld7 may disperse inside or outside the nucleus, where it can interact with other factors to perform other functions.
Possible functions of Sld7, other than in DNA replication
The factors that act in DNA replication events also have roles in mitotic events (Shimada and Gasser, 2007; Hemerly et al, 2009). These factors may coordinate the distinct events of the cell cycle. We have demonstrated that Sld7 acts in DNA replication through its interaction with Sld3. At a glance, sld7Δ cells are unlikely to be defective in cell-cycle events except in S phase because the buds of synchronized sld7Δ cells appear at the same time as those of wild-type cells (Figure 1C). However, several lines of evidence suggest that Sld7 has functions other than that in DNA replication. First, sld7Δ cells are sensitive to thiabendazole, an inhibitor of mitosis (our unpublished result). Second, a genome-wide study showed that the Sld7–GFP protein localized to the nucleus and spindle poles (Huh et al, 2003). Third, a mutation in the SLD7 gene causes aberrant mitochondrial morphology (Altmann and Westermann, 2005). Fourth, the IP of Sld3 did not deplete the Sld7 protein completely from extracts (Figure 2F), and high-copy SLD3 did not restore the sensitivity of these cells to thiabendazole to the level in the wild type (our unpublished result), unlike the restoration of the growth of sld7Δ cells. These results suggest that Sld7 interacts with a factor (or factors) other than Sld3 protein to function in situations other than replication. Further analysis is required to test this possibility.
Counterparts of Sld7 in higher eukaryotes
The N-terminal region of the Sld3 protein, which is less conserved between species than the central region (Supplementary Figure S2), binds to Sld7 (Figure 3), indicating that Sld7 protein has diverged more than Sld3. This appears to be the case because homologues of the Sld7 protein are only found in a limited range of yeast species closely related to Saccharomyces, whereas homologues of Sld3 protein have been found in yeast and fungi (Supplementary Figures S1 and S2). This in silico analysis suggests that homologues of Sld7 must exist in the fungi and yeast that have obvious homologues of Sld3, and leads us to expect that functional counterparts of Sld3 and Sld7 could even exist in higher eukaryotes.
Three TopBP1-interacting proteins, Treslin/Ticrr (Kumagai et al, 2010; Sansam et al, 2010), GEMC1 (Balestrini et al, 2010), and DUE-B (Chowdhury et al, 2010), have recently been reported as essential replication factors, required for the recruitment of Cdc45 to chromatin. TopBP1 is a possible counterpart of Dpb11. It has eight BRCT domains and its three N-terminal BRCT regions are essential for DNA replication in Xenopus egg extracts (Kumagai et al, 2010). Although a TopBP1–Treslin/Ticrr interaction has been observed in cells treated with roscovitine, an inhibitor of CDK (Sansam et al, 2010), TopBP1 associates with Treslin/Ticrr and GEMC1 in a CDK-dependent manner in Xenopus egg extracts (Balestrini et al, 2010; Kumagai et al, 2010). These characteristics are similar to those of Sld3. Moreover, a recent, sophisticated in silico analysis showed that Sld3 and Treslin/Ticrr have a central conserved region (Sanchez-Pulido et al, 2010), which corresponds to the region that binds to Cdc45 (Figure 3A). Furthermore, Treslin/Ticrr may have a subunit. Although the siRNA-mediated ablation of Treslin/Ticrr inhibited DNA replication in human cells and was rescued by an siRNA-resistant version of Treslin/Ticrr, DNA replication in Treslin/Ticrr-depleted egg extracts could not be restored by the addition of purified recombinant Treslin/Ticrr (Kumagai et al, 2010). The authors argued that either the recombinant Treslin/Ticrr was not active or Treslin/Ticrr forms a multiprotein complex with other unknown replication proteins.
Materials and methods
Microorganisms
The yeast strains used in this study are listed in Supplementary Table 1. To delete the SLD7 gene, its whole open reading frame was replaced with LEU2 using PCR.
Preparation of antibodies
Anti-Sld7 antibodies were raised against purified Sld7. The Sld7 protein was produced in E. coli and separated on sodium dodecyl sulphate (SDS)–polyacrylamide gels, from which the corresponding band was excised. The protein was eluted from the gel and used to immunize rabbits. The antibodies were purified from the sera of the immunized rabbits using Sld7-conjugated resin. The anti-Cdc45 monoclonal antibody was raised against the peptide TDADEVTDEDEEDEDETISNKRGNSS, corresponding to amino acids 189–214 in the protein sequence of Cdc45.
Identification of Sld7-binding protein
YTT4 cells (7 × 107 cells/ml × 2.5 l) expressing Sld7–3Flag–HA were harvested, washed once with distilled water, and disrupted with a mortar grinder (RM100, Retsch) under liquid nitrogen. The cell lysate was suspended in 5 ml of lysis buffer A (50 mM HEPES–KOH (pH 7.5), 300 mM KCl, 0.5% Tween 20, 0.05% NP40, 10% glycerol, 1 × Complete (Roche), 1% protease inhibitor cocktail (Sigma), 50 mM NaF, 2 mM β-glycerophosphate, 0.4 mM Na3VO4, 0.5 mM Na4P2O7, and 1 mM PMSF) and clarified by centrifugation at 12 000 r.p.m. at 4°C for 15 min. The protein extracts were adsorbed to 1 ml of Sepharose 4B (GE Healthcare) at 4°C for 30 min and incubated at 4°C for 2.5 h with 1 ml of anti-Flag M2 affinity gel (Sigma) that had been preincubated with lysis buffer A containing 5 mg/ml bovine serum albumin (BSA). The beads were washed three times with 3 ml of lysis buffer A containing 5 mg/ml BSA. The bound proteins were eluted twice by incubating the beads with 500 μl of Flag elution buffer (lysis buffer A containing 150 μg/ml of 3 × Flag peptide) at 4°C for 1 h. The eluted solution was mixed with 120 μl of anti-HA matrix (Roche) pretreated with anti-Flag M2 affinity gel and incubated at 4°C for 2.5 h. The beads were washed three times with 1 ml of buffer A containing 5 mg/ml BSA, once with 1 ml of buffer A containing 0.1 mg/ml BSA, and once with 1 ml of buffer A. The bound proteins were eluted by incubating the beads twice with 100 μl of HA elution buffer (lysis buffer A containing 1.5 mg/ml 3 × HA peptide [YPYDVPDYAYPYDVPDYAYPYDVPDYA]) at 37°C for 30 min. The proteins were precipitated by the addition of trichloroacetic acid and subsequent centrifugation. The precipitate was boiled for 3 min in 50 μl of 1 × SDS loading buffer. The proteins were separated in an SDS–polyacrylamide gel and stained by SilverQuest (Invitrogen). The protein band was excised from the gel, digested with trypsin, and analysed by mass spectrometry.
Purification of Sld3, Sld7, Sld3–Sld7, and Cdc45
Sld3–Flag was expressed in E. coli BL21-CodonPlus®-RIPL cells. The cells were disrupted by sonication, and the proteins were precipitated with 40% saturated ammonium sulphate. The proteins were incubated with M2 agarose beads at 4°C for 2 h and eluted with 3 × Flag peptide (150 μg/ml). The Sld3–Flag protein was purified further with successive HiTrap Heparin and HiTrap SP-XL column chromatography.
Sld7 was expressed as a glutathione S-transferase (GST)-fused protein connected with a PreScission Protease recognition site in E. coli BL21-CodonPlus-RIPL cells. The cells were disrupted by sonication, the nucleic acids were removed with Polymin P (final concentration, 0.3%), and the proteins were precipitated with 40% saturated ammonium sulphate. The proteins were purified with glutathione Sepharose and then HiTrap Heparin HP. The GST-fused protein was bound again to glutathione Sepharose and cleaved with GST-PreScission Protease. The cleaved Sld7 was purified with MiniS column chromatography.
The Sld3–Sld7 complex was purified from yeast or E. coli. In yeast, 5Flag–Sld3 and Sld7 were coexpressed in BJ2168 cells under the control of the galactose promoter from YEplac112 (Gietz and Sugino, 1988) and pKT10GAL (Kawamura et al, 1996), respectively. The cells were frozen in liquid nitrogen and ground with a mortar grinder. The 5Flag–Sld3–Sld7 complex was purified with anti-Flag M2 agarose affinity gel (Sigma) and then with HiTrap SP-XL. The 5Flag–Sld3–Sld7 complex was purified from E. coli as described for Sld3–Flag protein purified from BL21-CodonPlus-RIPL cells, producing Sld3–Flag and Sld7 simultaneously.
Cdc45–Flag was expressed in E. coli BL21-CodonPlus-RIPL cells. The cells were disrupted by sonication, and the proteins were incubated with M2 agarose beads at 4°C for 2 h and eluted with 3 × Flag peptide (150 μg/ml). The Cdc45–Flag protein was purified further using RESOURCE™ Q column chromatography.
Complex formation between Sld3 and Sld7
E. coli BL21-CodonPlus-RIPL cells carrying the pETDuet–(6His)SLD3, pETDuet–SLD7, or pETDuet–(6His)SLD3–SLD7 plasmid were grown in 1 l of LB broth containing ampicillin (200 μg/ml) at 37°C. When the OD600 reached 0.5, 0.1 mM IPTG was added to induce the expression of the proteins, and the cells were incubated at 20°C for 16 h. The cells were harvested and suspended in 5 ml of sonication buffer 300 (50 mM HEPES–KOH (pH 7.5), 300 mM NaCl, 2 mM MgCl2, 0.1 mM DTT, and 10% glycerol), and sonicated five times at 150 W for 30 s. The cell lysate was centrifuged at 15 000 r.p.m. at 4°C for 15 min. Most of the Sld3, Sld7, and Sld3–Sld7 proteins were precipitated. The precipitates were suspended in 1 ml of sonication buffer 500 (50 mM HEPES–KOH (pH 7.5), 500 mM NaCl, 2 mM MgCl2, 0.1 mM DTT, and 10% glycerol) and sonicated five times again at 150 W for 30 s. After incubation on ice for 1 h, the solution was clarified by centrifugation at 15 000 r.p.m. at 4°C for 15 min and the supernatant was then incubated with 50 μl of Ni–NTA agarose beads (Qiagen) at 4°C for 2 h. The beads were washed five times with 1 ml of washing buffer (50 mM HEPES–KOH (pH 7.5), 300 mM NaCl, 2 mM MgCl2, 0.1 mM DTT, 10% glycerol, and 20 mM imidazole), and the proteins were eluted with 200 μl of imidazole elution buffer (50 mM HEPES–KOH (pH 7.5), 300 mM NaCl, 2 mM MgCl2, 0.1 mM DTT, 10% glycerol, 250 mM imidazole, 0.1% Tween 20, and 0.01% Triton X-100). The eluates were boiled for 3 min in 1 × SDS loading buffer.
To mix the two purified proteins, 1.6 pmol of Sld3–Flag bound to M2 anti-Flag-antibody-conjugated Dynabeads M-270 was mixed with 12.5 pmol of Sld7 in 50 mM HEPES–KOH (pH 7.5) containing 0.2 M CH3CO2K, 0.2 M KCl, 2 mM Mg(CH3CO2)2, 1 mg/ml BSA, 2.5 mg/ml casein, 0.1% Tween 20, 0.01% NP40, and 10% glycerol at 30°C for 10 min. After brief washing, the bound proteins were eluted with 20 μl of 200 μg/ml 3 × Flag at 4°C for 15 min.
Complex formation between Sld3 and Cdc45 in vitro
Cdc45–Flag was immobilized on anti-Cdc45-antibody-conjugated rProtein A Sepharose Fast Flow (GE Healthcare). The Cdc45–Sepharose beads (10 μl) containing 0.4 pmol of Cdc45 were incubated with 400 μl of reaction buffer (50 mM HEPES–KOH (pH 7.5) containing 0.2 M CH3CO2K, 0.2 M NaCl, 2 mM Mg(CH3CO2)2, 2 mg/ml BSA, 2.5 mg/ml casein, 0.1% Tween 20, 0.01% NP40, and 10% glycerol) with 1 pmol of Sld3 or Sld3–Sld7 at 4°C for various times. After they were washed, the beads were incubated with 50 mM Tris–HCl (pH 6.8) containing 2% SDS and 10% glycerol at 65°C for 10 min to elute the proteins.
Glycerol gradient sedimentation
The samples were applied to a 15–30% glycerol gradient in 50 mM HEPES–KOH (pH 7.5), 0.15 M NaCl, 1 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and 0.01% Triton X-100 and centrifuged for 16 h at 42 000 r.p.m. at 4°C in an SW50.1 rotor.
Other methods
Proteins were extracted from yeast cells for electrophoretic analysis with the protocol described by Kushnirov (2000). The yeast two-hybrid assay was performed as described previously (Kamimura et al, 2001). The rapid in vivo cross-linking ChIP assay (Kohzaki and Murakami, 2007) was used for the ChIP assay. After western blotting, the proteins were visualized and quantified using fluorescence-conjugated secondary antibodies and the Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Drs K Labib, H Masukata, H Takisawa, and S Tanaka for their critical reading of the manuscript. TT performed all the experiments, except those specified below. MK and YK isolated the sld7 mutation. TU performed the two-hybrid assay, glycerol gradient, and purification of the Sld3–Sld7 complex from yeast. SE purified the Sld3, Sld7, Sld3–Sld7, and Cdc45 proteins and examined the in vitro associations between Sld3 and Sld7 and between Sld3 and Cdc45. SM examined HU sensitivity of sld3 mutants. CO analysed the protein coprecipitated with Sld7 by mass spectrometry. HA organized this project and wrote the paper. This study was partly supported by CREST and by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
The authors declare that they have no conflict of interest.
References
- Altmann K, Westermann B (2005) Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell 16: 5410–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H (2010) Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol 22: 766–771 [DOI] [PubMed] [Google Scholar]
- Araki H, Leem SH, Phongdara A, Sugino A (1995) Dpb11, which interacts with DNA polymerase II (ε) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA 92: 11791–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V (2010) GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol 12: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Ann Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Brümmer A, Salazar C, Zinzalla V, Alberghina L, Höfer T (2010) Mathematical modelling of DNA replication reveals a trade-off between coherence of origin activation and robustness against rereplication. PLoS Comput Biol 6: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Liu G, Kemp M, Chen X, Katrangi N, Myers S, Ghosh M, Yao J, Gao Y, Bubulya P, Leffak M (2010) The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol Cell Biol 30: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann PR, Sclafani RA (2006) Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae. Genetics 174: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS (2004) Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci 29: 579–585 [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B (2009) Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR (2010) Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 18: 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H (2001) Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 20: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Labib K (2006) Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 25: 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K (2003) Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–724 [DOI] [PubMed] [Google Scholar]
- Kawamura M, Kominami K, Takeuchi J, Toh-e A (1996) A multicopy suppressor of nin1-1 of the yeast Saccharomyces cerevisiae is a counterpart of the Drosophila melanogaster diphenol oxidase A2 gene, Dox-A2. Mol Gen Genet 251: 146–152 [DOI] [PubMed] [Google Scholar]
- Kohzaki H, Murakami Y (2007) Faster and easier chromatin immunoprecipitation assay with high sensitivity. Proteomics 7: 10–14 [DOI] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H (2003) A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG (2010) Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Labib K (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Gambus A (2007) A key role for the GINS complex at DNA replication forks. Trends Cell Biol 17: 271–278 [DOI] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL (2008) Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell 32: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H (2002) S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H (2000) Dpb11 controls the association between DNA polymerases α and ε and the autonomously replicating sequence region of budding yeast. Mol Cell Biol 20: 2809–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H (2010) CDK-dependent complex formation between replication proteins, Dpb11, Sld2, Polε and GINS in budding yeast. Genes Dev 24: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Pursell ZF, Kunkel TA (2008) DNA polymerase epsilon: a polymerase of unusual size (and complexity). Prog Nucleic Acid Res Mol Biol 82: 101–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Diffley JFX, Ponting CP (2010) Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol 20: R509–R510 [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA (2010) A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev 24: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Labib K (2006) Regulating initiation events in yeasts. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 295–311. New York: Cold Spring Harbor Press [Google Scholar]
- Shimada K, Gasser SM (2007) The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell 128: 85–99 [DOI] [PubMed] [Google Scholar]
- Sivaprasad U, Dutta A, Bell SP (2006) Assembly of pre-replication complexes. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 63–88. New York: Cold Spring Harbor Press [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H (2010) Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma 119: 565–574 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Valens M, Bohn C, Daignan-Fornier B, Dang VD, Bolotin-Fukuhara M (1997) The sequence of a 54.7 kb fragment of yeast chromosome XV reveals the presence of two tRNAs and 24 new open reading frames. Yeast 13: 379–390 [DOI] [PubMed] [Google Scholar]
- Walter JC, Araki H (2006) Activation of pre-replication complexes. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 89–104. New York: Cold Spring Harbor Press [Google Scholar]
- Wang H, Elledge SJ (1999) DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 3824–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.