Abstract
EMBO J 30 10, 1990–2007 (2011); published online April 05 2011
Since the beginning of micro-RNA (miR) research, several attempts have been made to identify the ‘melano-miRs’, which are involved in melanoma progression. Indeed, a small number of miRs have been identified to regulate some genes involved in melanogenesis. However, the miRs that control the pathway to the malignant phenotype are yet undescribed. In this issue of the EMBO Journal, Penna et al (2011) demonstrate that miR-214 is overexpressed in metastatic melanoma cell lines and tumour specimens. miR-214 regulates the expression of two transcription factors AP-2γ (directly) and AP-2α (indirectly). These transcription factors, particularly AP-2α, have been previously shown to play major roles in melanoma metastasis via regulation of genes involved in extravasation, invasion and angiogenesis. As such, this study has identified miR-214 as a driver of melanoma metastasis.
Limited data are available about the changes in miRNA expression levels in malignant melanoma. This investigative effort began in 2008 with the first report by Bemis et al (2008), linking the expression of a single mir, miR-137, with the expression of MITF, a ‘master regulator’ of melanogenesis. MITF, however, has since been shown to be also regulated by miR-182 (Segura et al, 2009). Overexpression of miR-182 was associated with increased survival and invasive properties of melanoma cells via regulation of both MITF and FOX03. miR-182 was found to enhance the invasive capability of melanoma cells in vitro and increase their metastatic potential in vivo in an experimental lung metastasis model. However, these studies never identified the ‘metastatic’ target genes involved in this process. Other miRNAs with known target genes in melanoma are miR-221 and miR-222, which regulate the expression of both p27 and in particular c-KIT, which is downregulated in metastatic melanoma cells. The c-KIT gene is regulated mainly by the AP-2α transcription factor (vide infra). Moreover, treatment with antagomiRs against miR-221/222 inhibited melanoma proliferation and invasion in vitro and melanoma growth in vivo (Felicetti et al, 2008). The first discovered miR to be involved in cancer formation was miRNA let-7. The family of miRs let 7a and let 7b were later reported to play a role in melanoma tumourigenesis. For example, let 7b acts as a negative regulator of melanoma cell proliferation via regulation of cyclin D1, whereas let 7a was demonstrated to regulate the expression of integrin-β3 and the Ras Oncogene (reviewed in Mueller and Bosserhoff, 2009).
All of the above studies focused on single miRs and their individual roles in melanoma progression. Other attempts have been made to identify a cluster of miRs that are either involved in melanogenesis or predictors of survival. For example, Ozsolak et al (2008) searched for miRs that are specific to melanoma progression. To that end, they screened for miRs that are specifically regulated by MITF and identified miR-146a, miR-221/222 and miR-363. Another approach pursued miRNAs silenced by CpG methylation, revealing miR34a to be highly methylated in melanoma cell lines and primary tumours. Although the role of miR-34a in melanoma progression has yet to be elucidated, it was elsewhere reported that miR-34a regulates the expression of MET. While these studies concentrated on the biological functions of these miRs and their contribution to melanogenesis, other studies have attempted to identify miRNA signatures for diagnostic and prognostic purposes. In one such study, Segura et al (2010) analysed the expression of 611 miRs in 59 metastatic specimens with detailed clinical follow-up. They were able to identify a miRNA classifier consisting of six miRs (miR-150, miR-342-3p, miR-455-3p, miR-145, miR-155 and miR-497) that are predictors of post-recurrence survival, with an accuracy of about 80%. A slightly different approach has been taken by Caramuta et al (2010). This group analysed miR expression profiling in lymph node metastases and reported a unique signature consisting of downregulation of miR-191, combined with upregulation of miR-193a, miR-193b, miR-365, miR-338 and let-7. Together, this cluster serves as a predictor of short-term survival of melanoma patients. A broader approach has been more recently taken to characterize the melanoma miRNAome by deep sequencing. However, the identification of an exact, single miR or miRs signature that best predicts metastatic development has yet to be reported or validated. One such miR is described in this issue of the EMBO Journal by Penna et al (2011).
To identify miRs that are specifically affecting melanoma metastasis, Penna et al (2011) have utilized the parental A375P cell line and its metastatic variants that were selected via cycling in nude mice. Among the miRs that were differentially expressed in this model system, miR-214 was found to be overexpressed in the metastatic variants. Overexpression of miR-214 was further validated in metastatic lesions utilizing melanoma tissue array, thus, confirming its upregulation in ‘real’ life. The authors then re-expressed miR-214 in the low-metastatic variants (miR-214 negative) or silenced its expression via antagomiR in the highly metastatic cells, revealing its involvement in migration, invasion and extravasation in vitro. In vivo, miR-214 did not affect the growth of the primary tumours, but did enhance the metastatic potential of cells in an experimental lung metastasis assay, thus, indicating the involvement of miR-214 in the transition point from RGP to VGP of melanoma progression. The novelty of this study, however, is emphasized when the authors identified the downstream target genes for miR-214. These include the transcription factor AP-2γ and the integrin-α3 (ITGA3). Importantly, another family member, the AP-2α transcription factor, was also affected by miR-214. It is not clear, however, at this step whether the downregulation of AP-2α in the metastatic melanoma cells resulted from a direct effect of miR-214 overexpression or by its regulation by AP-2γ. The authors, in this case, analysed the expression of AP-2α after miR-214 overexpression in metastatic melanoma cells endogenously harbouring low levels of AP-2α.
The expression of the AP-2α transcription factor has been shown to be downregulated in metastatic melanoma cell lines and tumour specimens (reviewed in Bar-Eli, 1999). Loss of AP-2α drives the melanoma metastatic phenotype by regulating a cascade of genes, such as c-KIT, MCAM/MUC18 (one of the target genes identified by Penna et al (2011)), HER-2, VEGF, MMP-2 and the thrombin receptor PAR-1, all of which were shown to play a role in melanoma metastasis (Figure 1). PAR-1 activation, in turn, results in the expression and secretion of proangiogenic factors, such as IL-8, PDGF and VEGF. PAR-1 has also recently been shown to regulate the expression of the tumour suppressor gene, Maspin, in melanoma (Villares et al, 2011). As such, AP-2α plays a pivotal role in melanoma metastasis. However, up until this point, the mechanism(s) for its loss of expression in metastatic melanoma cells have not been known. The study by Penna et al (2011) provides the missing piece of the puzzle: the identification of miR-214 as a post-transcriptional regulator of AP-2α, to drive melanoma metastasis. By affecting the expression of AP-2α and its downstream target genes, miR-214 is largely involved in modulating the melanoma tumour microenvironment to enhance invasion and metastasis. It will be interesting now to determine whether overexpression of miR-214 could be used for prognostic purposes and more importantly, whether its targeting could provide a new treatment modality for melanoma.
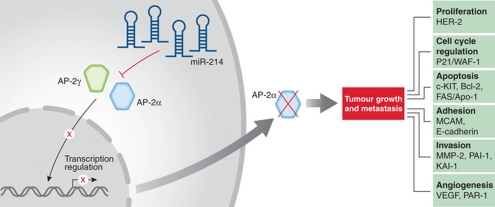
Figure 1.
Proposed model for how miR-214 drives melanoma metastasis. miR-214 post-transcriptionally downregulates the expression of AP-2γ and AP-2α. In turn, AP-2α functions as a master switch regulating a cascade of genes involved in melanoma metastasis, including genes regulating proliferation (HER-2), cell cycle (p21), apoptosis (c-KIT, Bcl-2, Fas/APO-1), adhesion (MCAM/MUC18, E-cadherin), invasion (MMP-2, PAI-1, KAI-1) and angiogenesis (VEGF, PAR-1).
Footnotes
The author declares that he has no conflict of interest.
References
- Bar-Eli M (1999) Role of AP2 in tumor growth and metastasis of human melanoma. Cancer Metastasis Rev 18: 377–385 [DOI] [PubMed] [Google Scholar]
- Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA (2008) MicroRNA-137 targets microphtalmia-associated transcription factor in melanoma cell lines. Cancer Res 68: 1362–1368 [DOI] [PubMed] [Google Scholar]
- Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO (2010) MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol 130: 2062–2070 [DOI] [PubMed] [Google Scholar]
- Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Care A (2008) The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res 68: 2745–2754 [DOI] [PubMed] [Google Scholar]
- Mueller DW, Bosserhoff AK (2009) Role of miRNAs in the progression of malignant melanoma. Br J Cancer 101: 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE (2008) Chromatin structure analyses identify miRNA promoters. Genes Dev 22: 3172–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Polisano L, Haimovic A, Osella S, DePitta C, Pinatel E, Provero P, Bernengo MG, Osman I, Taverna D (2011) microRNA-214 contributes to melanoma tumor progression through suppression of TFAP2C. EMBO J 30: 1990–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Belitskaya-Levy I, Rose AE (2010) Melanoma microRNA signature predicts post-recurrence survival. Clin Cancer Res 16: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menedez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E (2009) Aberrant miR-182 expression promote melanoma metastasis by repressing FOX03 and microphtalmia-associated transcription factor. Proc Natl Acad Sci USA 106: 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares GJ, Zigler M, Dobroff AS, Wang H, Song R, Melnikova VO, Huang L, Braeuer RR, Bar-Eli M (2011) Protease activated receptor-1 inhibits the Maspin tumor-suppressor gene to determine the melanoma metastatic phenotype. Pro Natl Acad Sci USA 108: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]