Abstract
EMBO J 30 10, 1977–1989 (2011); published online April 05 2011
MicroRNAs are 19–24 nt-long RNAs that post-transcriptionally regulate eukaryotic gene expression. Beside their important roles in patterning and development, miRNAs also orchestrate responses to pathogen infections. In this issue of The EMBO Journal, Schulte et al (2011) investigate the miRNA response of mammalian cells to the intracellular bacterium Salmonella. They find that Salmonella triggers highly specific and robust alterations to the expression of a subset of host miRNAs. In macrophages, these alterations notably include the rapid downregulation of let-7 MIRNA gene family members, following extracellular sensing of bacterial lipopolysaccharides (LPS) via Toll-like receptor 4 (TLR4). The authors identify two interleukins (IL-6 and IL-10) as novel targets of Let-7 and suggest that Salmonella infection rapidly relieves IL-6 and IL-10 from negative control by let-7, thereby potentiating the immune response. Intriguingly, while IL-6 is pro-inflammatory, IL-10 prevents the detrimental consequences of systemic inflammation, suggesting that the observed response is tightly balanced and, hence, biologically relevant.
In eukaryotic RNA silencing, 19–24 nt-long micro (mi)RNAs regulate expression of cellular transcripts exhibiting partially or fully complementary miRNA-binding sites. Beside their roles in development, miRNAs orchestrate various biotic stress responses: in mammals, virus-induced cellular miRNAs contribute to the antiviral immune response, whereas some mammalian DNA viruses encode their own suite of miRNAs to manipulate both viral and cellular mRNA expression (reviewed in Umbach and Cullen, 2009). miRNAs also orchestrate responses to bacteria, which are sensed via receptor-based recognition of pathogen-associated molecular patterns (PAMPs) including lipopolysaccharides (LPS) and flagellin. Seminal work in Arabidopsis showed that a flagellin-derived peptide induces expression of miR-393a, which enhances basal resistance against Pseudomonas synringae notably by repressing the receptor for the hormone auxin, a negative regulator of plant defence (Navarro et al, 2006; Figure 1A). Later on, a role for miRNAs in mammalian bacterial infections was similarly inferred from experiments involving Toll-like receptor (TLR)-mediated sensing of purified PAMP. Hence, LPS recognition by TLR4 and consequent NF-κB signalling induces miR-146a/b and miR-155, both of which control the cytokine pathway (reviewed in Baltimore et al, 2008). It was subsequently found that miR-155-deficient mice are not immunized by attenuated bacterial strains (Rodriguez et al, 2007), and that miR-155 is induced by the human gastric pathogen Helicobacter pylori (Fassi Fehri et al, 2010).
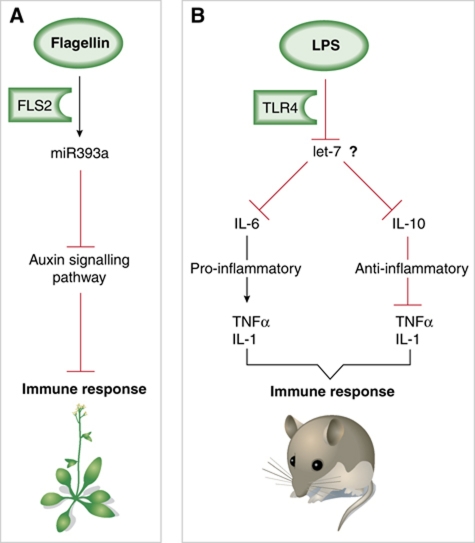
Figure 1.
Plant and mammalian strategies for miRNA-directed, PAMP-triggered immunity. (A) In Arabidopsis, sensing of bacterial flagellin by the cognate receptor FLS2 induces transcription of miR-393a. This miRNA represses a key component of the auxin signalling pathway, which negatively regulates plant defence. Resistance is thus achieved, in this case, by inducing a repressor of a negative regulator of innate immunity. (B) In mouse macrophages, bacterial-derived LPS is sensed by TLR4 to promote rapid downregulation of let-7 MIRNA family members through as yet unidentified mechanisms (?). Downregulation of let-7 promotes enhanced accumulation of its targets, IL-6 and IL-10, with the former displaying pro-inflammatory properties and the latter preventing systemic inflammation. This results in the mounting of a balanced innate immune response that proceeds, therefore, through concomitant repression of a repressor of a positive regulator of defence, and repression of a repressor of a negative regulator of defence.
P. syringae and H. pylori are extracellular, unlike other bacteria that establish intimate connections inside host cells, allowing them to manipulate the transcriptome and signalling pathways. Schulte et al (2011) decided to investigate the role of miRNAs in mounting defences against such an intracellular bacteria, using mouse and human in vitro systems that recapitulate infection by Salmonella enterica, the agent of gastroenteritis. Deep sequencing revealed that Salmonella significantly induces several miRNAs within 4 h of mouse macrophage infection. Perhaps not surprisingly, these miRNAs chiefly included miR-155 and miR-146a, as well as miR-21. Strikingly, several members of the let-7 MIRNA gene family were, by contrast, rapidly downregulated; this was also observed in infected Hela cells, unravelling a generic feature of Salmonella infection in both phagocytic and non-phagocytic cells. Mutant Salmonella defective in cell invasion and/or intracellular replication also triggered let-7 downregulation, suggesting that the process involves sensing of extracellular bacterial antigens. Indeed, these effects on let-7 were recapitulated in mouse macrophages directly treated with purified LPS, but not in cells lacking TLR4, the cognate LPS receptor, or in cells elicited with purified flagellin, sensed by TLR5.
Is the rapid downregulation of let-7 and upregulation of miR-155, miR-146a/b, and miR-21 a mere frontline measure of early infections? To address this issue, the authors took advantage of the fact that TLR4 activity and NF-κB signalling are lost upon pre-exposure of macrophages to extracellular LPS. This desensitization notably prevents runaway systemic inflammation (Biswas and Lopez-Collazo, 2009). Remarkably, let-7 was still efficiently downregulated by Salmonella or its non-invasive/non-replicating variants in desensitized macrophages. Upregulation of miR-21 and miR-146a/b also proceeded normally in those cells, suggesting that the global miRNA response to Salmonella is part of a potent pathogens’ surveillance system that persists independently of successful host invasion and of the onset of the inflammatory response.
While the roles of miR-146a/b and miR-155 in the immune response are well established, gaining insights into the biological relevance of let-7 and its downregulation in this process entailed the identification of possibly novel let-7 targets. Macrophage-mediated regulation of immune responses to systemic bacterial infections relies significantly on interleukin (IL) secretion. Incidentally, previous computer-based predictions had identified potential let-7-binding sites in the 3′-UTRs of the mouse IL-6 and IL-10 transcripts. Moreover, these and additional binding sites uncovered by the authors are conserved in sequence throughout mammalian genomes. Schulte et al (2011) went on testing these predictions by using classical luciferase reporter systems for miRNA activity. In several experimental settings, the 3′-UTR-reporter fusions were repressed in a strict let-7-dependent manner. Furthermore, directly increasing the levels of let-7 in Salmonella-infected mouse macrophages concomitantly reduced the expression of the 3′-UTR-reporter fusions and those of secreted IL-6 and IL-10. Conversely, cells pre-treated with an antisense oligonucleotide that neutralizes let-7 activity displayed increased 3′-UTR-reporter activity and higher levels of secreted IL-6 and IL-10.
Collectively, the results are consistent with let-7 serving as a post-transcriptional brake to IL-6 and IL-10 secretion, a brake that is rapidly relieved upon recognition of Salmonella and perhaps other pathogens. The sophistication of this model lies in the contrasting roles of IL-6 and IL-10 in defence (Klimpel et al, 1995). Indeed, in concert with IL-1 and TNFα, IL-6 promotes inflammation, which may culminate in harmful systemic symptoms including fever or septic shock. IL-10, by contrast, dampens the above pro-inflammatory effects by moderating IL-1 and TNFα production. Thus, let-7 downregulation in response to Salmonella likely allows the mounting of a balanced immune response based on two concomitant, yet opposing pathways involving: (i) repression of a repressor (let-7) of a positive defence effector (IL-6) and (ii) repression of a repressor (let-7) of a negative defence regulator (IL-10) (Figure 1B). In Arabidopsis, flagellin-induced miR-393 resulted in repression of a negative defence regulator (auxin) (Figure 1A). The diversity of these scenarios thus exemplifies the versatility of eukaryotic miRNA networks in mounting defence against pathogens.
While they provide novel directions for the study of mammalian immune responses to bacterial infections, the results of Schulte et al (2011) also raise important questions. First, they show that Hela cells and desensitized mouse macrophages mount a robust miRNA response to Salmonella, notably by dampening let-7 levels, yet both cell types lack a functional TLR4 signalling pathway. This implies that other bacterial stimuli in addition to LPS, and/or alternative LPS sensing systems are involved, and their identification certainly constitutes a major, future endeavour. Second, how is the rapid let-7 downregulation achieved? Of course, transcriptional control comes first to mind, and the authors must have already initiated let-7 promoter gene fusions to address this issue. But miRNAs are also controlled post-transcriptionally, notably at the miRNA precursor level; for instance, the RNA-binding protein Lin-28 specifically hinders processing of mammalian let-7 (Viswanathan et al, 2008). Although the Lin-28 mRNA levels were unaltered 24 h post-infection, effects at the protein level cannot yet be discarded and deserve investigation.
A last, fundamental question is prompted by previous findings made in plants, where phytoviruses and phytobacteria have evolved specific proteins to defeat the effects of RNA silencing. For instance, Pseudomonas virulence factors injected into host cells through the type-III secretion system (T3SS) were shown to suppress transcription, processing or activity of host miRNAs (Navarro et al, 2008). This never-ending molecular arms race characterizes nearly all host–parasite interactions. Thus, given the findings of Schulte et al (2011) and that Salmonella possesses two T3SS required for virulence, the existence of Salmonella-encoded modifiers of the mammalian miRNA pathway arises as a legitimate and fascinating possibility. Owing to their design, none of the experiments described by the authors would have allowed a clear appreciation of the effects of these putative miRNA modifiers. Moreover, some of these factors may oppose each other during specific windows of the infection, and hence, may have confounding effects when studied globally. Individual expression of T3SS virulence factors in various miRNA-reporter systems, as originally carried out in Arabidopsis (Navarro et al, 2008) and now initiated in the author's laboratory (Eulalio et al, 2011), might thus provide a way to address this important issue.
Footnotes
The author declares that he has no conflict of interest.
References
- Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD (2008) MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9: 839–845 [DOI] [PubMed] [Google Scholar]
- Biswas SK, Lopez-Collazo E (2009) Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol 30: 475–487 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Frohlich KS, Mano M, Giacca M, Vogel J (2011) A Candidate approach implicates the secreted Salmonella effector protein SpvB in P-body disassembly. PLoS One 6: e17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T, Churin Y, Gerhard M, Meyer TF (2010) Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One 5: e9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimpel GR, Asuncion M, Haithcoat J, Niesel DW (1995) Cholera toxin and Salmonella typhimurium induce different cytokine profiles in the gastrointestinal tract. Infect Immun 63: 1134–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte LN, Eulalio A, Mollenkopf H-J, Reinhardt R, Vogel J (2011) Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J 30: 1977–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR (2009) The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23: 1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]