Abstract
Genes that are differentially expressed between the sexes (sex-biased genes) are among the fastest evolving genes in animal genomes. The majority of sex-biased expression is attributable to genes that are primarily expressed in sex-limited reproductive tissues, and these reproductive genes are often rapidly evolving because of intra- and intersexual selection pressures. Additionally, studies of multiple taxa have revealed that genes with sex-biased expression are also expressed in a limited number of tissues. This is worth noting because narrowly expressed genes are known to evolve faster than broadly expressed genes. Therefore, it is not clear whether sex-biased genes are rapidly evolving because they have sexually dimorphic expression, because they are expressed in sex-limited reproductive tissues, or because they are narrowly expressed. To determine the extend to which other confounding variables can explain the rapid evolution of sex-biased genes, I analyzed the rates of evolution of sex-biased genes in Drosophila melanogaster and Mus musculus in light of tissue-specific measures of expression. I find that genes with sex-biased expression in somatic tissues shared by both sexes are often evolving faster than non–sex-biased genes, but this is best explained by the narrow expression profiles of sex-biased genes. Sex-biased genes in sex-limited tissues in D. melanogaster, however, evolve faster than other narrowly expressed genes. Therefore, the rapid evolution of sex-biased genes is limited only to those genes primarily expressed in sex-limited reproductive tissues.
Keywords: Drosophila, mouse, gene expression, sexual dimorphism, reproduction
Introduction
A substantial fraction of genes are differentially expressed between males and females across a wide array of animal taxa (reviewed in Ellegren and Parsch 2007). These genes are said to have sex-biased expression or, more specifically, female-/male-biased expression. Sex-biased expression has been intensely studied on a genome-wide scale in model organisms, but experimental and analytical approaches differ depending on the taxon under examination. For example, in Drosophila, it is conventional to identify sex-biased genes by comparing transcript quantities in RNA isolated from whole males and females. These experiments have revealed that the expression levels and protein-coding sequences of male-biased genes evolve faster than those of unbiased genes (Ranz et al. 2003, Zhang et al. 2004, Haerty et al. 2007, Zhang et al. 2007), and there is evidence that this rapid evolution is driven by positive selection (Meiklejohn et al. 2003, Pröschel et al. 2006, Baines et al. 2008).
It is important to note, however, that a large fraction of an organism consists of sex-limited tissues (i.e., testis, ovary, and somatic reproductive tissues). In Drosophila, these differences in tissue content between males and females are responsible for most of the sex-biased expression measured in whole flies (Jin et al. 2001, Arbeitman et al. 2002, Arbeitman et al. 2004, Parisi et al. 2003, Parisi et al. 2004). Genes expressed in sex-limited tissues often encode proteins involved in reproduction, and many reproductively related genes are rapidly evolving (Swanson et al. 2001, Swanson et al. 2004, Haerty et al. 2007, Prokupek et al. 2008). This raises the question: Is the rapid evolution of sex-biased genes in Drosophila attributable to expression in reproductive tissues or are sex-biased genes expressed in somatic tissues shared by both sexes (shared somatic tissues) also rapidly evolving?
Sex-biased expression in vertebrates, on the other hand, is measured using individual organs rather than whole organisms (e.g., Yang et al. 2006, Ellegren et al. 2007, Blekhman et al. 2010). Among the many discoveries made in analyzing these data sets, it is worth noting that sex-biased genes tend to have tissue-specific expression profiles (Yang et al. 2006, Mank et al. 2008). Narrowly expressed genes are probably more functionally limited (McShea 2000) and, therefore, less pleiotropic. The correlation between sex-biased expression and tissue specificity has thus been interpreted to mean that the evolution of sex-biased expression in one tissue is impeded by pleiotropic constraints imposed by expression in other tissues (Mank et al. 2008, Mank 2009).
The tissue specificity of sex-biased genes could also have implications for their evolutionary rates. Pleiotropy may limit adaptive evolution by reducing the probability that mutations are beneficial (Fisher 1930). Narrowly expressed genes evolve faster than broadly expressed genes (Duret and Mouchiroud 2000; Zhang and Li 2004; Liao et al. 2006; Haerty et al. 2007), and, indeed, this rate acceleration may be the result of more adaptive substitutions (Larracuente et al. 2008). Alternatively, narrowly expressed genes may evolve faster because they are under relaxed selective constraints. Additionally, testis-biased genes represent the largest class of tissue-specific genes in Drosophila melanogaster (Chintapalli et al. 2007), further entangling of the relationship between sex bias, expression breadth, and reproductive tissues. Regardless of why narrowly expressed genes are rapidly evolving, the nonindependence of tissue specificity, sex bias, evolutionary rates, and expression in sex-limited tissues means that one must consider all these factors simultaneously to determine which is/are responsible for the rapid evolution of sex-biased genes.
Some work has been carried out to examine the relationship between sex-biased expression and rates of evolution in mammals, but these analyses have focused primarily on genes with testis-biased expression (e.g., Torgerson et al. 2002, Good and Nachman 2005, Khaitovich et al. 2005). These studies confirm the well-established paradigm that genes encoding reproductive proteins are often rapidly evolving (Swanson and Vacquier 2002). Interestingly, in avian brain, female-biased, and not male-biased, genes are rapidly evolving (Mank et al. 2007). Therefore, the selection pressures that drive the rapid evolution of reproductively related sex-biased genes in vertebrates may differ from those acting upon sex-biased genes in shared somatic tissues. On the other hand, these apparent differences might instead reflect different definitions of what a sex-biased gene is in these different tissues.
I analyzed the rates of protein-coding sequence evolution of sex-biased genes in D. melanogaster and Mus musculus (house mouse) to determine whether the rapid evolution of sex-biased genes is limited to only those genes expressed in reproductive tissues. I find some evidence that sex-biased genes in shared somatic tissues evolve faster than unbiased genes, but this rapid evolution is driven by the tissue-specific expression of sex-biased genes. In D. melanogaster, however, there is substantial evidence that sex-biased genes expressed in sex-limited reproductive tissues are rapidly evolving even when I control for breadth of expression. Therefore, the rapid evolution of sex-biased genes appears to be limited to those genes encoding proteins with reproductive functions.
Materials and Methods
Sex-Biased Gene Expression
Processed microarray data comparing gene expression in D. melanogaster males and females were obtained from SEBIDA (http://www.sebida.de; Gnad and Parsch 2006). This database provides the ratio of male-to-female expression level (M/F) and P values for a test of differential expression between males and females estimated using Bayesian inference (Townsend and Hartl 2002). I used a meta-analysis of sex-biased expression that takes sex-specific measurements from whole flies (Parisi et al. 2003, Ranz et al. 2003, Gibson et al. 2004, McIntyre et al. 2006, Ayroles et al. 2009) and gonads (Parisi et al. 2003) and estimates M/F and a P value for each gene (Gnad and Parsch 2006). I also obtained M/F and P values from SEBIDA using the individual experiments that were included in the meta-analysis and a data set collected from gonadectomized flies (Parisi et al. 2003). Adjusted P values that control for the false discovery rate (FDR) (Benjamini and Hochberg 1995) were estimated using the R statistical package (R Development Core Team 2009). If the adjusted P value of a given gene falls below the specified FDR cut off (either 0.01, 0.05, 0.10, or 0.20), then the gene is said to have sex-biased expression. My results are robust to the FDR cut off chosen and data used to estimate M/F (Supplementary Material online) unless otherwise noted.
Microarray measurements of gene expression in four adult tissues (liver, adipose, brain, and muscle) from male and female mice (Yang et al. 2006) were used to estimate M/F in each tissue (Mank et al. 2008). Genes were called as sex-biased in each of the tissues separately if the adjusted P value in a Mann–Whitney test for differences in expression between males and females (Mank et al. 2008) falls below a specified FDR cut off (0.01, 0.05, 0.10, or 0.20). If a gene is female- or male-biased in one tissue, and that gene is either unbiased or has the same sex bias in the other tissues in which expression was measured, the gene was said to be either female- or male-biased (depending on the direction of sex bias). As a second test, genes were called as sex biased if there is a 2-fold difference in expression between females and males (i.e., female-biased if M/F< 0. 5 and male-biased if M/F> 2) (Mank et al. 2008). Additionally, average M/F across the four tissues was used as an estimate of the degree of sex-biased expression. Averages were calculated both for genes with measurable expression in all four tissues and for all genes (ignoring tissues for which expression was not detected).
Rates of Evolution
I obtained the ratio of the rate of nonsynonymous to synonymous substitutions (dN/dS) for each D. melanogaster gene estimated five ways: 1) along the branch leading to D. melanogaster after the split with the Drosophila simulans lineage, 2) along all the branches within the D. melanogaster species subgroup, 3) within the D. melanogaster species group, 4) pairwise between D. melanogaster and D. simulans, and 5) between D. melanogaster and Drosophila yakuba (Gnad and Parsch 2006, Larracuente et al. 2008). These results are robust to the estimate of dN/dS unless otherwise noted. Rates of evolution for mouse genes were estimated as the pairwise dN/dS between orthologous genes in the rat genome available from the Ensembl database (Genes 59, NCBIM37).
Tissue-Specific Gene Expression
Gene expression measurements were obtained from FlyAtlas (http://www.flyatlas.org/; Chintapalli et al. 2007) for ten nonoverlapping shared somatic adult tissues from combined D. melanogaster males and females (brain, eye, thoracico-abdominal ganglion, salivary gland, crop, midgut, tubule, hindgut, heart, and fat body) and four adult sex-limited tissues (ovary, testis, accessory gland, and spermatheca). Measurements from spermatheca of virgin and mated flies were averaged and used as a single spermatheca estimate—spermatheca from virgin and mated flies have strongly correlated genome-wide expression values (Supplementary Material online), and analyses using data from either spermatheca of virgin or mated flies yield similar results as those using their average (results not shown).
Counts of the number of expressed sequence tags (ESTs) per gene were obtained from UniGene build #186 for 25 nonoverlapping mouse tissues (adipose, bladder, blood, bone, bone marrow, brain, dorsal root ganglion, eye, vagina, intestine, heart, inner ear, liver, lung, lymph node, mammary gland, muscle, pancreas, prostate, skin, spinal cord, spleen, sympathetic ganglion, testis, and thymus). These tissues are nearly identical to those chosen for a previously published analysis of the relationship between sex-biased expression and tissue specificity (Mank et al. 2008).
Expression measurements for each gene were used to estimate tissue specificity with two methods. In the first method, tissue specificity (τ) was estimated for each gene using the following equation:
| (1) |
In analyzing the D. melanogaster data, Siis the signal intensity in tissue i, Smax is the maximum signal intensity in all tissues, and N is the total number of tissues analyzed (Yanai et al. 2005, Larracuente et al. 2008). All Si<1 were set to 1 so that logSi≥0. For the mouse data, Si is the number of ESTs mapping to a given gene in tissue i, standardized by the total number of mapped ESTs in tissue i (Mank et al. 2008). All genes with fewer than three mapped ESTs across all 25 tissues were removed, and Si<2 were set to 2 for the remaining genes. Larger values of τ indicate more tissue specificity.
The second method was applied only to the D. melanogaster data. If Si≥100 for a particular gene in tissue i, the gene was said to be expressed in that tissue. The number of tissues in which a gene is expressed was used as a measure of the breadth of expression.
Expression Levels
The expression levels of D. melanogaster genes were estimated as the microarray signal intensities measured using RNA isolated from whole flies (males and females combined) obtained from FlyAtlas (Chintapalli et al. 2007). Expression levels of mouse genes were estimated as the average number of ESTs mapping to a gene in the 25 tissues standardized for the length of the gene and the sizes of the libraries.
Statistical Analysis
Spearman's ρ was used to estimate the pairwise correlation between each of the following: τ, expression level, |log(M/F)|, and dN/dS. The 95% confidence interval (CI) of ρ between τ and |log(M/F)| was approximated by bootstrapping the data 1,000 times. Partial correlations between all four variables were estimated from the pairwise correlation coefficients (Schäffer and Strimmer 2005), and I approximated the 95% CI of each partial correlation by bootstrapping the data 1,000 times. All bootstrapping was carried out in the R statistical package (R Development Core Team 2009).
Results and Discussion
Sex-Biased Genes are Rapidly Evolving
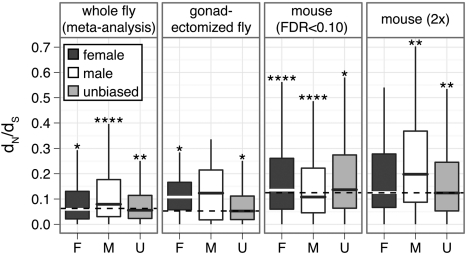
Consistent with previous reports (Zhang et al. 2004, Pröschel et al. 2006, Haerty et al. 2007), my analysis reveals that D. melanogaster male-biased genes evolve faster than unbiased and female-biased genes when expression is measured in whole flies and gonads (fig. 1; Supplementary Material online). Additionally, when gonads are excluded and sex-biased expression is measured (Parisi et al. 2003), female-biased genes evolve faster than other genes (fig. 1). This suggests that rapid evolution is a common feature of sex-biased genes, regardless of whether the genes are expressed in reproductive tissues or shared somatic tissues. In mouse, whether female- or male-biased genes expressed in shared somatic tissues evolve faster depends on the method used to assign genes to sex-biased expression categories (fig. 1). Furthermore, there is some evidence that sex-biased mouse genes as a group evolve faster than unbiased genes (Supplementary Material online).
FIG 1.
Box plots show the distribution of dN/dS for female-biased (dark gray), male-biased (white), and unbiased (light gray) genes, with outliers omitted (boxes extend from the first to third quartile, with the midline indicating the median). Genes were assigned to sex-biased classes using an FDR cut off of 0.10 and the following data sets: meta-analysis of D. melanogaster experiments, gonadectomized flies, and mouse data. The mouse data were also analyzed with a 2-fold cut off to assign genes as sex-baised. The median dN/dS value across all genes within each panel is indicated by a dashed line. Asterisks indicate a significant difference in dN/dS between a subset of genes and the rest of the panel in a Mann–Whitney test (*P<10−2, **P<10−4, ****P<10−8).
Sex-Biased Expression and Tissue Specificity Are Correlated
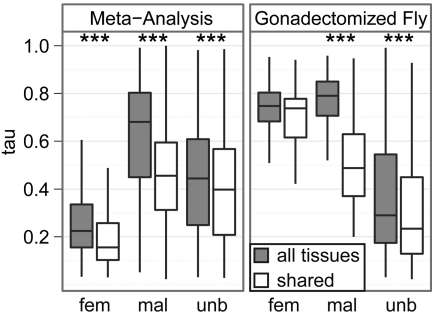
The rapid evolution of sex-biased genes may be affected by expression breadth (Haerty et al. 2007). Indeed, breadth of expression is one of the best predictors of the rate of evolution of protein-coding sequences (Duret and Mouchiroud 2000, Zhang and Li 2004, Liao et al. 2006, Larracuente et al. 2008), and previous work in vertebrates has shown that sex-biased genes have more tissue-specific expression (τ) than unbiased genes (Yang et al. 2006, Mank et al. 2008). As in vertebrates, there is a significant difference in the tissue specificity of female-biased, male-biased, and unbiased D. melanogaster genes (P<<10−10 in a Kruskal–Wallis test of distributions of τ), and male-biased genes have narrower expression profiles than unbiased genes (fig. 2). Unlike in vertebrates, however, female-biased genes have broader expression profiles than unbiased genes (fig. 2).
FIG 2.
Box plots show the distributions of τ for female-biased (fem), male-biased (mal), and unbiased (unb) D. melanogaster genes (outliers have been omitted); larger τ indicates more tissue specificity. Each box extends from the first to the third quartile, with the line in the middle of the box indicating the median. Tissue specificity was calculated using all tissues (gray) or somatic tissues shared by both sexes (white). Sex-biased genes were determined from the meta-analysis of multiple data sets (left) or gonadectomized flies (right). Asterisks indicate significant differences in τ when measured in all tissues compared with when measured in somatic tissues shared by both sexes using a Mann–Whitney test (***P<10−4).
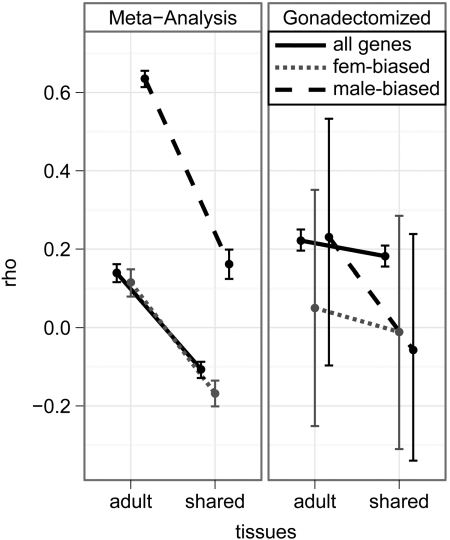
The degree of sex-bias (|log(M/F)|) is also positively correlated with τ for all D. melanogaster genes, female-biased genes, and male-biased genes (fig. 3, left panel, first column of points), as has been previously observed in vertebrates (Mank et al. 2008). This suggests that pleiotropic constrains may prevent the evolution of sex-biased expression in D. melanogaster (Mank et al. 2008, Mank 2009). The correlation is not due to signal saturation in highly expressed genes, stochastic error in lowly expressed genes, X/autosome differences, or sex-biased duplicated genes (Supplementary Material online). Interestingly, the magnitude of the correlation is much greater for male-biased genes than female-biased genes (fig. 3). It is worth noting, however, that this correlation may be conflated by the inclusion of sex-limited tissues in the estimation of M/F and τ.
FIG 3.
Plots show the point estimate of the correlation coefficient (ρ) between |log(M/F| and tissue specificity (τ) and the 95% CI of the estimate for D. melanogaster genes. M/F was estimated using the meta-analysis of multiple data sets (left) or gonadectomized flies (right). Estimates of τ were calculated using all adult tissues (adult) or shared somatic tissues (shared). Correlations were calculated using all genes (black), female-biased genes (gray, short dashes), and male-biased genes (black, long dashes).
Sex-Limited Tissues Drive the Relationship between Sex Bias and Tissue Specificity
If sex-limited tissues are excluded from the estimation of tissue specificity, τ significantly decreases for all genes, female-biased genes, and male-biased genes (fig. 2). Additionally, a large fraction of male-biased genes have extremely sex-biased expression and are primarily expressed in sex-limited tissues (Supplementary Material online). Excluding sex-limited tissues also decreases the correlation between |log(M/F)| and τ, and this leads to ρ<0 for all genes and female-biased genes (fig. 3, first panel). Therefore, a substantial amount of the correlation between sex-biased expression and tissue specificity is driven by sex-biased genes that are primarily expressed in sex-limited tissues.
The effect of sex-limited tissues on the relationship between |log(M/F)| and τ can be seen when gonadectomized flies are used to measure sex-biased expression: both female- and male-biased genes in gonadectomized flies are more narrowly expressed than unbiased genes (fig. 2). Therefore, male-biased genes are narrowly expressed in both sex-limited and shared somatic tissues. On the other hand, there appear to be two types of female-biased genes: genes with female-biased expression driven by ovary expression are broadly expressed, whereas those with female-biased expression in shared somatic tissues are tissue specific. If tissue specificity affects the evolutionary rate of sex-biased genes, the locus of expression of female-biased genes must be taken into account when comparing the evolutionary rates of female- and male-biased genes.
The correlation between |log(M/F)| and τ is also affected by using gonadectomized flies to estimate M/F (fig. 3). As in the analysis using whole flies, ρ≥0 when I use gonadectomized flies to estimate |log(M/F)| and all adult tissues to estimate τ for most FDR cut offs (fig. 3; Supplementary Material online). However, when I analyze all genes in gonadectomized flies, the correlation between |log(M/F)| and τ is not as affected by the exclusion of sex-limited tissues from the estimation of τ (fig. 3). Furthermore, because ρ > 0 when gonads (and most other sex-limited tissues) are excluded, the correlation between sex-biased expression and tissue specificity is not entirely driven by genes with sex-biased expression in sex-limited tissues. This provides further support for the hypothesis that pleiotropic constraints may limit the evolution of sex-biased expression (Mank et al. 2008, Mank 2009).
Tissue Specificity Partially Explains the Relationship between Sex-Biased Expression and Rates of Evolution
The nonindependence of tissue specificity and sex bias (Mank et al. 2008, and results described above) means that expression breadth must be considered when comparing the evolutionary rates of sex-biased and unbiased genes. I calculated partial correlations between the degree of sex-biased expression, breadth of expression, and expression level for D. melanogaster and M. musculus genes. Expression level was included in this analysis because it is one of the strongest predictors of the rate of protein-coding sequence evolution (Pál et al. 2001, Rocha and Danchin 2004, Subramanian and Kumar 2004, Wright et al. 2004, Lemos et al. 2005, Larracuente et al. 2008).
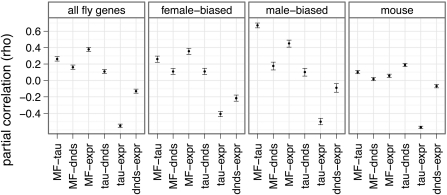
In D. melanogaster, all three factors (|log(M/F)|, τ, and expression level) are significantly correlated with dN/dS in the expected directions: sex biased, narrowly expressed, and lowly expressed genes evolve fastest (fig. 4). These trends hold whether I consider all genes, female-biased genes, or male-biased genes (fig. 4). Additionally, |log(M/F)| and τ remain positively correlated when dN/dS and expression level are taken into account (fig. 4), indicating that their correlation is not an artifact of both being correlated with dN/dS or expression level.
FIG 4.
Point estimates of the partial correlation coefficient (ρ) between |log(M/F)| (MF), τ (tau), dN/dS (dnds), and expression level (expr) are indicated with filled circles, along with their 95% CI (error bars). Correlations were estimated using all D. melanogaster genes, D. melanogaster female-biased genes, D. melanogaster male-biased genes, or data from mouse. For the D. melanogaster analysis, M/F was estimated using the meta-analysis, τ was calculated using all adult tissues, and dN/dS was calculated along the branch leading to D. melanogaster after the split with the D. simulans lineage. For the mouse analysis, genes expressed in at least one of four tissues were used to estimate M/F from the microarray data.
The partial correlations between dN/dS and |log(M/F)| for mouse suggest that the degree of sex-biased expression is not a good predictor of the evolutionary rate of protein-coding genes expressed in shared somatic tissues (fig. 4). Tissue specificity, on the other hand, appears to be the best predictor of the evolutionary rate of mouse genes: only τ is consistently significantly correlated with dN/dS, although expression level is correlated with dN/dS in some of the analyses (fig. 4; Supplementary Material online). Additionally, the correlation between dN/dS and τ is driven by the dN component, and it is not an artifact of rapidly evolving X-linked genes (Supplementary Material online). Finally, |log(M/F)| and τ remain positively correlated when dN/dS and expression level are considered (fig. 4), indicating that this relationship is not an artifact of other correlations.
Expression in Sex-Limited Tissues Drives the Rapid Evolution of Sex-Biased Genes
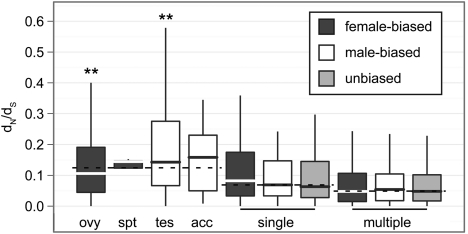
The positive correlation between |log(M/F)| and dN/dS for D. melanogaster genes, even when tissue specificity is taken into account (fig. 4), suggests that sex-biased expression may be an adequate predictor of evolutionary rate in D. melanogaster. However, although the partial correlations take into account expression breadth, they do not consider whether tissue-specific genes are expressed in sex-limited tissues or shared somatic tissues. Indeed, genes expressed in a single sex-limited tissue evolve faster than genes expressed in a single shared somatic tissue (P < 10 − 10 in a Mann–Whitney test), and genes expressed in a single shared somatic tissue evolve faster than genes expressed in multiple tissues (P < 10 − 6) (fig. 5). Additionally, among genes expressed in a single shared somatic tissue or in multiple tissues, there is not a significant difference in the rate of evolution between female-, male-, and unbiased genes (fig. 5); this is true for most sex-bias calls and most dN/dS estimates (Supplementary Material online). However, male-biased genes expressed primarily in male-limited tissues evolve faster than female-biased genes expressed primarily in female-limited tissues (P < 10 − 5 in a Mann–Whitney test). Furthermore, genes that are predominantly expressed in testis evolve significantly faster than other sex-limited or narrowly expressed genes, whereas genes that are predominantly expressed in ovary evolve slower than other genes expressed in sex-limited tissues (fig. 5; Supplementary Material online). Finally, a previous analysis revealed evidence of a “faster-X” effect (Charlesworth et al. 1987) for D. melanogaster male-biased genes (Baines et al. 2008). I find some support for a faster-X effect among D. melanogaster testis-biased genes, but the pattern is dependent on how dN/dS and sex-biased expression are measured (Supplementary Material online).
FIG 5.
Box plots show the distribution of dN/dS for female-biased (dark gray), male-biased (white), and unbiased (light gray) D. melanogaster genes, with outliers omitted (boxes extend from the first to third quartile, with the midline indicating the median). Genes were divided into those that are expressed in a single sex-limited tissue (ovy = ovary, spt = spermatheca, tes = testis, and acc = accessory gland), those that are expressed in a single shared somatic tissue (single), and those that are expressed in multiple tissues (multiple). The median dN/dS value across all genes within a group of genes is indicated by a dashed line. Asterisks indicate a significant difference in dN/dS between a subset of genes and the rest of the group in a Mann–Whitney test (**P < 10 − 4).
Why are genes that are expressed primarily in male-limited tissues rapidly evolving? D. melanogaster male reproductive genes run the gamut from highly conserved to rapidly evolving, and the most rapidly evolving genes are expressed in the accessory gland (Dorus et al. 2006). These accessory gland proteins (Acps) often influence male reproductive success (e.g., Ravi Ram and Wolfner 2007), and natural selection may drive their rapid evolution if they play a role in male–male or male–female competition (Swanson and Vacquier 2002). Accessory-gland-limited genes are not evolving significantly faster than other narrowly expressed genes in my data (fig. 5; Supplementary Material online), but this is probably a methodological artifact of the most rapidly evolving Acps being excluded from this data set. To be included in the analysis of dN/dS, genes had to have a single ortholog in all outgroup species (Larracuente et al. 2008), but Acps have a high rate of turnover as a result of gene duplication and loss (Begun and Lindfors 2005, Haerty et al. 2007). Additionally, the rapid evolution of Acps may decrease the likelihood of ortholog detection. Genes expressed in the lower female reproductive tract are also rapidly evolving (Swanson et al. 2004, Prokupek et al. 2008), and my failure to detect the rapid evolution of spermatheca-biased genes (fig. 5) is likely the result of the aforementioned methodological artifacts as well as small sample sizes.
A previous analysis of the D. melanogaster sperm proteome found that testis-expressed components are among the slowest evolving genes encoding sperm proteins (Dorus et al. 2006). Interestingly, I observe a robust signal of rapid evolution for testis-biased genes (fig. 5; Supplementary Material online). The apparent discrepancy between the two analyses is likely attributable which genes were included: I included genes with testis-biased expression, whereas Dorus et al. (2006) examined genes whose protein products were observable in a mass spectrometry analysis of sperm proteins. Additionally, I found an overrepresentation of genes involved in adenosine triphosphate (ATP) biosynthesis among genes with testis-biased expression (Supplementary Material online). Much of the D. melanogaster sperm proteome is encoded by genes involved in energy metabolism, but these genes are thought to contribute to the highly conserved components of the male ejaculate (Dorus et al. 2006). Although some of the testis-biased genes involved in ATP biosynthesis in my data are highly conserved, I also observe a subset of these genes with a high rate of evolution (Supplementary Material online). Interestingly, recently duplicated testis-specific nuclearly encoded mitochondrial genes are also rapidly evolving (Galach et al. 2010). Further work is required to determine what drives the rapid evolution of testis-biased genes involved in energy metabolism.
Conclusions
It is well established that sex-biased genes are rapidly evolving (Ellegren and Parsch 2007), but the definition of sex-biased genes varies across studies. I showed that sex-biased genes expressed in D. melanogaster sex-limited reproductive tissues evolve faster than other genes with similar expression breadth. Sex-biased genes expressed in D. melanogaster shared somatic tissues are also rapidly evolving, but this can be explained by their narrow expression profiles. Additionally, there is no evidence for a relationship between sex-biased expression and rate of evolution for genes expressed in M. musculus shared somatic tissues. This is in contrast to testis-biased and reproductively related mouse genes for which there is substantial evidence for rapid evolution (e.g., Mouse Genome Sequencing Consortium 2002, Torgerson et al. 2002, Good and Nachman 2005). Therefore, the selection pressures driving the rapid evolution of genes with biased expression in sex-limited tissues do not affect genes with sex-biased expression in shared somatic tissues. Future studies that examine the evolution of so-called “sex-biased” genes should explicitly consider what aspect of the genes' expression or function is sex biased.
Supplementary Material
Supplementary materials are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work benefitted from discussions with Tim Connallon, Tony Greenberg, Amanda Larracuente, Ran Blekhman, Andy Clark, other members of the Clark lab, Erin Kelleher, and Nathan Clark. Judith Mank kindly provided her previous analysis of the mouse microarray data. The comments of Lauren McIntyre and two anonymous reviewers greatly improved the quality of this manuscript. R.P.M. is supported by the National Institute of Health fellowship F32GM087611.
References
- Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–2021. doi: 10.1242/dev.01077. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, et al. (11 co-authors) Systems genetics of complex traits in. Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Sawyer SA, Hartl DL, Parsch J. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in. Drosophila. Mol Biol Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA. Rapid evolution of genomic Acp complement in the melanogaster subgroup of. Drosophila. Mol Biol Evol. 2005;22:2010–2021. doi: 10.1093/molbev/msi201. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, Karr TL. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–1445. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–70. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Hultin-Rosenberg L, Brunström B, Dencker L, Kultima K, Scholtz B. Faced with inequality: chicken does not have general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford: Clarendon Press; 1930. [Google Scholar]
- Galach M, Chandrasekaran C, Betrán E. Analysis of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol. 2010;2:835–850. doi: 10.1093/gbe/evq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, Nuzhdin S, Wayne M. Extensive sex-specific nonadditivity of gene expression in. Drosophila melanogaster. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- Good JM, Nachman MW. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol Biol Evol. 2005;22:1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- Haerty W, Jagadeeshan S, Kulathinal RJ, et al. (11 co-authors) Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in. Drosophila melanogaster. Nat Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Mank JE. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am Nat. 2009;173:141–150. doi: 10.1086/595754. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Axelsson E, Ellegren H. Rapid evolution of female-biased, but not male-biased, genes expressed in the avian brain. Mol Biol Evol. 2007;24:2698–2706. doi: 10.1093/molbev/msm208. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Zwahlen M, Ellegren H. Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am Nat. 2008;171:35–43. doi: 10.1086/523954. [DOI] [PubMed] [Google Scholar]
- McIntyre LM, Bono LM, Genissel A, Westerman R, Junk D, Telonis-Scott M, Harshman L, Wayne ML, Kopp A, Nuzhdin SV. Sex-specific expression of alternative transcripts in. Drosophila. Genome Biol. 2006;7:R79. doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShea DW. Functional complexity in organisms: parts as proxies. Biol Philos. 2000;15:641–668. [Google Scholar]
- Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. Rapid evolution of male-biased gene expression in. Drosophila. Proc Natl Acad Sci U S A. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, et al. (12 co-authors) A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokupek A, Hoffmann F, Eyun Si Moriyama E, Zhou M, Harshman L. An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution. 2008;62:2936–2947. doi: 10.1111/j.1558-5646.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R:a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. 2009 [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Ravi Ram K, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comput Biol. 2007;47:427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- Schäffer J, Strimmer K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat Appl Genet Mol Biol. 2005;4:32. doi: 10.2202/1544-6115.1175. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci U S A. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Kulathinal RJ, Singh RS. Mammalian sperm proteins are rapidly evolving: evidence for positive selection in functionally diverse genes. Mol Biol Evol. 2002;19:1973–1980. doi: 10.1093/oxfordjournals.molbev.a004021. [DOI] [PubMed] [Google Scholar]
- Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA levels across multiple strains or treatments. Genome Biol. 2002 doi: 10.1186/gb-2002-3-12-research0071. 3:research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Yau CBK, Looseley M, Meyers BC. Effects of gene expression on molecular evolution in Arabidopsis thaliana and. Arabidopsis lyrata. Mol Biol Evol. 2004;21:1719–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, et al. (12 co-authors) Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus. Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in. Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.