Abstract
The rapid separation of isomeric precursor ions of oligosaccharides prior to their analysis by MSn was demonstrated using an ambient pressure ion mobility spectrometer (IMS) interfaced with a quadrupole ion trap. Separations were not limited to specific types of isomers; representative isomers differing solely in the stereochemistry of sugars, in their anomeric configurations, and in their overall branching patterns and linkage positions could be resolved in the millisecond time frame. Physical separation of precursor ions permitted independent mass spectra of individual oligosaccharide isomers to be acquired to at least MS3, the number of stages of dissociation limited only practically by the abundance of specific product ions. IMS-MSn analysis was particularly valuable in the evaluation of isomeric oligosaccharides that yielded identical sets of product ions in MS/MS experiments, revealing pairs of isomers that would otherwise not be known to be present in a mixture if evaluated solely by MS dissociation methods alone. A practical example of IMS-MSn analysis of a set of isomers included within a single HPLC fraction of oligosaccharides released from bovine submaxillary mucin is described.
Keywords: carbohydrate isomers, separation, precursor ions, tandem mass spectrometry, ion mobility spectrometry
INTRODUCTION
Carbohydrates play pivotal roles in a number of biological systems and are involved in embryonic development [1–3], innate and adaptive immune responses [4–6], bacterial, plant and fungal cell wall structures and a wide variety of physiological functions [7–9]. Mass spectrometry has long been an important tool for analysis of carbohydrate molecules [10–12]. Recently, multiple stages of isolation/dissociation (MSn) have been possible in ion traps and Fourier transform ion cyclotron resonance (FTICR) mass spectrometers, which have enabled more detailed structural information to be garnered from the substructures of complex molecules [10–15]. However, the analysis of oligosaccharides from biological sources still precent a central and nontrivial problem: many of the precursor ions are mixtures of isomers [12, 13, 15]. Some of these isomeric precursor ions dissociate to yield sets of product ions all having the same m/z values, even in multiple rounds of isolation/dissociation.
This problem itself prompts two fundamental questions: (1) How can one know if an ion at a given m/z is really one pure isomer? If it is not, one may misassign a structure based on dissociation data from a mixture of molecules. (2) What is a fast and effective means to determine isomeric heterogeneity? In the past, nuclear magnetic resonance spectroscopy (NMR) has been very effective in evaluating isomeric heterogeneity of oligosaccharides, because anomeric H-1 signals invariably appear at different frequencies and intensities when a mixture is present [16]. HPLC has been frequently used to separate fractions, often requiring multiple orthogonal modes such as normal phase, reversed phase, ligand exchange, or other modes to purify structures to homogeneity as evaluated by NMR [16–19]. HPLC separation on a single column in no way ensures that molecules are pure and free of isomers. A strong case can be made for development of any technique used in conjunction with MS that can address the characterization of isomeric species, given that MS is 3~4 orders of magnitude more sensitive than NMR and likely to become even more so in the future.
In recent years, ambient pressure ion mobility spectrometry (APIMS) has been developed as a tool for the rapid resolution of carbohydrate isomers [20, 21], and is of interest in that it separates ions based primarily on a cross-section/charge ratio rather than mass/charge. Compared to conventional chromatographic techniques, IMS allows much faster separation, which occurs in millisecond time periods. APIMS also offers resolving power comparable to GC, and higher than 1-D LC [22]. In addition to speed, IMS is a versatile technique in that changing the nature of the drift gas or adducting species can have marked effects on the separation of isomers [21–23] thus a number of separation conditions can be rapidly screened as compared to running consecutive HPLC columns. Using an APIMS-time-of-flight (TOF) instrument and N2 drift gas, monosaccharide isomers [21], and neutral di- and trisaccharide isomers [20], have been shown to be completely resolved, although in these studies only an m/z was recorded and no dissociation of precursor ions was carried out. Other related IMS methods, high-field asymmetric waveform ion mobility spectrometry [24] and traveling waveform ion mobility spectrometry [25] have been coupled to time-of-flight mass spectrometers and shown to provide some degree of resolution between carbohydrate isomers. IMS can also be readily interfaced with other types of mass spectrometers, such as quadrupoles [26], triple quadrupoles [27], quadrupole ion traps [28], and FTICR [29] mass spectrometers. Previous studies using a two-gate IMS system have shown that collision-induced dissociation (CID) MS/MS spectra could be acquired after selectively introducing ions based on mobility into a quadrupole ion trap [28] or an ICR cell [29], although in these studies no sets of isomeric oligosaccharides were investigated. The key goal of this investigation was to demonstrate whether tandem dissociation of mobility-selected isomeric carbohydrate precursor ions could be achieved.
Differentiation of isomers is a general problem in mass spectrometry, becoming increasingly more formidable in the analysis of ever more complex mixtures of molecules. Oligosaccharides are a paradigm for development of better methods to deal with isomeric mixtures of molecules, having isomers that fall into three main groups: (1) Those isomers where precursor ions dissociate to yield sets of product ions where one or more product ions have different m/z values. This is typically the case for linkage and branch isomers. (2) Those isomers where precursor ions yield product ions all having the same m/z values, even after multiple stages of dissociation, but where the relative abundance of the product ions are significantly different. This is sometimes the case when structures vary solely in their anomeric configurations. (3) Those isomers where precursor ions give rise to product ions all having the same m/z values, even after multiple stages of dissociation, where the relative abundance of product ions at each stage is nearly identical. This is sometimes the case when single asymmetric carbons on a single sugar vary in their stereochemistry. In this study, we have selected examples of all three types to determine if APIMS-MSn can be used to effectively evaluate isomeric mixtures by enabling independent mass spectra to be obtained for individual isomers.
Materials and methods
Materials
Oligosaccharides from bovine submaxillary mucin (Galβ1-3GalNAc-ol, GlcNAcβ1-3[GlcNAcβ1-6]GalNAc-ol, GalNAcβ1-3[GlcNAcβ1-6]GalNAc-ol, Fucα1-2Galβ1-3[GlcNAcβ1-6]GalNAc-ol and Fucα1-2Galβ1-4GlcNAcβ1-3GalNAc-ol) were isolated as previously described [16]. They were released from the protein as oligosaccharide-alditols, and purified by multi-step HPLC fractionation. Structures were determined by nuclear magnetic resonance spectroscopy and tandem mass spectrometry at the University of Colorado. The molecules GlcNAcβ1-6Gal, Glcα1-4Glcα1-4Glcα1-4Glc (maltotetraose) and Glcβ1-4Glcβ1-4Glcβ1-4Glc (cellotetraose) were from Sigma (St. Louis, MO). They were reduced with 1.0 M NaBH4 as previously described [16]. The neutral non-reducing trisaccharides melezitose and raffinose were also purchased from Sigma. All solvents used were HPLC grade and used as supplied without further purification. MeOH was provided by EMD Chemicals Inc. (Gibbstown, NJ). H2O was from J. T. Baker Inc. (Philipsburg, NJ).
Sample Preparation
Beginning with a 200 μM stock solution (1:1 MeOH/H2O) for each oligosaccharide-alditol, mixtures having a pair of isomeric compounds were prepared by mixing equal volumes of two isomers, resulting in 100 μM for each compound. When only a single compound was used in analyses, the oligosaccharide-alditol was diluted in 1:1 MeOH/H2O to 100 μM.
Sample Introduction by Electrospray Ionization
The ESI emitter was prepared in-house using 360 μm o.d. × 75 μm i.d. fused-silica capillary (Polymicro Technologies, Phoenix, AZ). The emitter was 5 cm long connected with the sample transfer line via a stainless steel zero dead volume union (Upchurch Scientific, Oak Harbor, WA). The ESI voltage (3.0 kV higher than the first ring of IMS tube) was applied at the stainless steel union. The electrospray capillary was positioned at the center of the IMS tube at an upward angle of 20° from the central transverse axis of the IMS tube. For this study, the sample flow in the ESI was 3 μl/min for the APIMS-TOF instrument, and 2 μl/min for the two-gate ambient pressure ion mobility-quadrupole ion trap mass spectrometer (APIMS-ion trap).
Electrospray Ionization Ambient Pressure Ion Mobility (time-of-flight) Mass Spectrometry
Mobility measurements were first performed on the APIMS-TOF. The instrument has been described in detail by Steiner et al [30] and Dwivedi et al [21]. The length of the mobility drift region in this instrument was 17.8 cm. The voltage applied at the first ring electrode of the ion mobility tube was 10.0 kV, resulting in gate voltage of 9.1 kV and an electric field of 511 V/cm across the drift region. Because this was a lab built instrument some danger existed from direct contact with the high voltage. To reduce risk to the operator, the power supply was current limited to 10 μA and high voltage isolators were used for the connections. Nitrogen was heated to 200°C and used as a counter-flowing drift gas with a flow rate of ~1 L/min. A gate pulse width of 200 μs was used in this study. Ion mobility gate pulsing was set at 25 Hz to introduce desolvated ions into the drift region. The extraction frequency for pushing ions into the orthogonally aligned reflectron TOF analyzer was set at 25 kHz. The mass and mobility spectra were displayed by averaging all scans over the acquisition time. It is important to note that the ion mobility separation occurs in the millisecond time frame, while ions are analyzed within microseconds by the TOF mass analyzer. Thus the APIMS-TOF provided mass spectra in real time as the ion mobility spectrum was acquired. This was not the case for the APIMS-ion trap, below, where the ion trap scanned the mass range over a much longer time frame, but enabled mobility selected MSn spectra to be acquired. All samples were therefore run on both instruments.
Electrospray Ionization Ambient Pressure Two-gate Ion Mobility-Quadrupole Ion Trap Mass Spectrometry
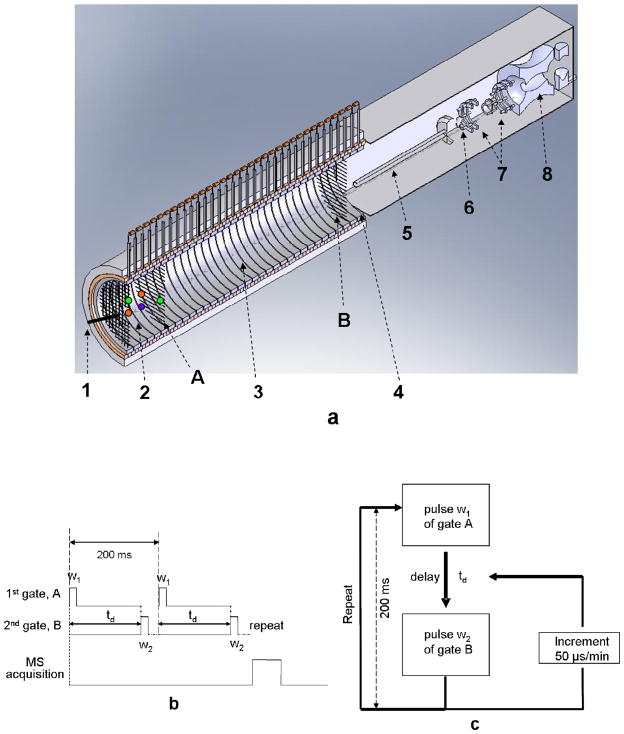
The ionization source of the LCQ Deca quadrupole ion trap mass spectrometer was removed to enable the end of the IMS tube to fit into the MS interface. The IMS instrument was a modified version of the two-gated ion mobility system described by Clowers and Hill [28] coupled to a LCQ Deca quadrupole ion trap mass spectrometer (Thermo Electron, San Joe, CA). Instead of a17.6 cm length for the drift region described previously, 25.4 cm was used in the new design. The desolvation region was 7.5 cm. In addition, the IMS and MS were separated by a distance of 0.8 cm to avoid excessive heat transfer from the heated IMS tube to the MS system. The optimal distance was determined empirically by maximizing the ion abundance in the quadrupole trap. A schematic of the instrument is displayed in Figure 1(a). The instrument consists of the following parts: an in-house built ESI source, an ambient pressure ion mobility tube equipped with two Bradbury-Nielson gates (A and B in Figure 1a) which divide the tubes into three regions: a desolvation region, a drift region, and the 2.6 cm extended region of conducting rings after the second gate for coupling to the mass spectrometer. The Bradbury-Nielson gate was composed of two sets of isolated wires (Alloy 46, California Fine Wire Co., Grover Beach, CA), alternating with an equidistant space of 0.6 mm. The extended part after the second gate allowed the MS capillary inlet to be placed inside the central end of the IMS tube for better ion focusing and transmission. The voltage applied to the first and second gates was 9.2 kV and 0.9 kV, respectively. The resulting electric field strength was 327 V/cm in the ion mobility drift region. The two gate controllers built at Washington State University were two independent MOSFET switches controlled by TTL pulse train signals from an external computer. Under external control, when the pulse was low, one set of wires in the gate received an additional +50 V, while the other set received −50 V. Thus ion flow was stopped. On the other hand, the gate remained open when the pulse was high. The gate control software used in this study for mobility scanning or mobility window selection was written in-house in LabView 6.1 (National Instrument, TX). The external operation modes of the instrument are illustrated in Figure 1(b) and (c). The gate controllers can also be adjusted manually to keep both gates open by applying no additional DC voltage to the gate wires. MS spectra were acquired in the two-gate IM-QIT when both gates remained open.
Figure 1.
Schematics of the electrospray ionization, ambient pressure two-gate ion mobility-quadrupole ion trap mass spectrometer and timing sequence diagrams. a. The instrument setup. 1. electrospray tip; 2. desolvation region; 3. drift region; 4. interface region. 5. heated capillary; 6. skimmer; 7. octapole; 8. quadrupole ion trap. Ions were introduced into the desolvation region by the electrospray ionization process. They entered the drift region once gate A was pulsed open. Gate B was triggered to open by gate A after certain delay of time td to allow ions fly through the drift region. b. Timing sequence of the mobility selected mode. The pulse train signal in high position (gate open) was 5 V. When in low position (gate close), no voltage was applied. Gate A was typically opened for 0.5 ms, and the second gate either opened over a wide window period to initially find approximate regions of precursor ion abundance after a selected delay, or set for a narrow period just wide enough to cover a single ion’s peak width after a drift period td. Multiple ion mobility runs could be used to repetitively fill the trap, and mass spectra that were accumulated using the trap could also be signal averaged. c. Timing sequence of the mobility-scanning mode. A delay time td in milliseconds (i.e. 39 ms) was preset between the pulses of gate A and B. Multiple incremented delays could be performed (i.e. 50 μsec increments/min), keeping the second gate-open time constant (pulse width w2 μsec) to scan drift-time regions of interest (i.e. 39~42 ms). Every ion mobility experiment in this scanning mode was repeated every 200 ms.
The following MS conditions were implemented throughout the study: capillary temperature 200°C, capillary voltage 27 V, tube lens voltage 9.0 V, ion injection time 500 ms. In MS2 and MS3 experiments, the normalized collision energy level was set to 40% and 45%, respectively. The isolation width for MS2 and MS3 experiments was 3 amu and 4 amu, respectively. The activation time was set at 30 ms for all MSn experiments. The Xcalibur software from Thermo-Finnigan was used for MS instrument control, data acquisition and processing.
Operation of the Mobility-Selected Mode of the Two-Gate Ion Mobility System
The mobility-selected mode was used to extract ions having a mobility time of interest into the mass spectrometer. The timing sequence for the ion gate control and data acquisition is shown in Figure 1(b). Once the first gate was pulsed open for a period of w1 ms (typically about 0.2 to 0.5 ms), after a delay of td ms, the second gate was opened for w2 ms. This process was repeated while ions of interest were extracted into the ion trap mass spectrometer for detection. A mobility selected time window is defined as td ~ (td+w2), which can be a relatively wide range or a finite mobility drift time window depending on the values for td and pulse width w2. A relatively high w2 value (10–20 ms) was first applied to initially locate precursor ions of interest anywhere within a broad drift-time window. Then a smaller and smaller w2 was selected to rule out time windows that did not contain ions of interest and narrowed the drift time down to a small window, typically a 2-ms range, containing ions of interest. Then an automated step-increment scanning mode was employed using 0.05 ms increments to obtain the mobility spectra of selected ions. Once the mobility spectrum of an ion species was determined by the mobility scanning mode, a finite drift time window based on the mobility spectrum was selected to repetitively introduce ions into the trap to build up ion counts to high enough levels for MSn analysis. In mobility selected MS2 and MS3 experiments in this study, the w1 value used was 0.5 ms.
Operation of the Mobility Scanning Mode of the Two-Gate Ion Mobility System
As shown in Figure 1(c), the timing sequence of the mobility scanning mode used in this study was as follows: The first gate was opened at time 0 for w1 ms. The second gate was opened for w2 ms after a pre-defined time delay td from the rising edge of the first gate. This cycle was repeated at 5 Hz with a 50 μs step added to the td of previous cycle. As td increased to match the actual drift time of an ion species, the ions were extracted into the ion trap and then detected. The ion intensity as a function of td can be plotted as the ion mobility spectrum [28]. Ion mobility spectra from the two-gate APIMS-ion trap system were consequently reconstructed by applying a Gaussian smoothing function using Matlab 7.0 (MathWorks, Natick, MA). Throughout this study, both w1 and w2 in the scanning mode were set at 0.3 ms.
Statistics and Other Experimental Conditions
Each of the eight oligosaccharide alditols and two non-reducing neutral trisaccharides were individually run on the one-gate APIMS-TOF and the two-gate APIMS-ion trap instruments. Five measurements were acquired for each compound on both instruments. One-way ANOVA [31] was performed to compare the reduced mobility values determined from APIMS-TOF and two-gate APIMS-ion trap. Both instruments were operated in the positive mode. Ion mobility experiments were performed under ambient pressure at Pullman, WA (~690–~700 torr).
Calculation of Reduced Mobility and Resolution
Experimental ion mobility (K) can be determined using the following equation:
Where vd is the drifting velocity (cm/s), E is the electric field strength (V/cm), L is the drift length (cm), td is the drift time (s), and V is the electric potential across the drift region (volts). To make valid comparisons between experiments, K has to be normalized for operating temperature and pressure, and is reported as the reduced mobility:
Where T is the temperature in degrees Kelvin, P is the pressure in torr.
The resolution of two mobility peaks in IMS is analogous to the concept in traditional chromatography, and is defined as follow:
Where td and wh denote peak drift time and full width at half maximum (FWHM), respectively.
Results and Discussion
Throughout this study, all isomers were separated in the positive ion mode as sodiated molecules, using nitrogen as the drift gas. Ion mobility experiments performed on the APIMS-TOF and APIMS-ion trap instruments yielded different drift times, but the reduced mobility values Ko for all compounds that were examined were in agreement (Table 1). The P-values reported were more than 0.07, indicating that there was no significant difference in mobility measurement for the two methods.
Table 1.
Comparison of reduced mobility measures (Ko values) for various oligosaccharide isomers by one-gate IMS-tof-MS and two-gate IMS-ion trap-MS.
| Compound | m/z | Ko(1)a | Ko(2)b | P-value (α = 0.05)c |
|---|---|---|---|---|
| Galβ1-3GalNAc-ol | 408 | 1.04 (0.53) | 1.03 (0.40) | 0.42 |
| GlcNAcβ1-6Gal-ol | 408 | 1.01 (0.57) | 1.00 (0.59) | 0.36 |
| Melezitose | 527 | 0.96 (0.57) | 0.95 (0.74) | 0.17 |
| Raffinose | 527 | 0.92 (0.60) | 0.92 (0.77) | 0.35 |
| GlcNAcβ1-3[GlcNAcβ1-6]GalNAc-ol | 652 | 0.78 (0.64) | 0.79 (0.45) | 0.49 |
| GalNAcβ1-3[GlcNAcβ1-6]GalNAc-ol | 652 | 0.72 (0.43) | 0.72 (0.32) | 0.084 |
| Maltotetraitol | 691 | 0.78 (0.31) | 0.78 (0.58) | 0.87 |
| Cellotetraitol | 691 | 0.76 (0.48) | 0.77 (0.51) | 0.082 |
| Fucα1-2Galβ1-4GlcNAcβ1-3GalNAc-ol | 757 | 0.69 (1.48) | 0.70 (0.43) | 0.070 |
| Fucα1-2Galβ1-3[GlcNAcβ1-6]GalNAc-ol | 757 | 0.71 (0.84) | 0.71 (0.74) | 0.39 |
values determined by one-gate IMS-tof-MS;
values determined by two-gate IMS-ion trap-MS. Ko values (cm2/V/s) were reported as mean (%RSD), n = 5.
one-way ANOVA was performed for variance analysis. The two methods were compared for difference at the 95% significance level.
Separation of isomeric species having different mass spectra: the problem of identifying isomeric mixtures and proving direct precursor-product relationships
The MS/MS spectrum of a mixture of two simple isomeric disaccharide-alditols as sodiated molecules is shown in Fig. 2a. It is worth noting that there is no compelling reason, without prior knowledge, to conclude that this MS/MS spectrum is actually derived from a mixture of molecules. There are two main product ions and four minor ones, not an atypical scenario at all for dissociation of a single disaccharide molecule. Thus we delineate two key issues: (1) In an unknown sample, can one know whether a spectrum like Fig. 2a derives from one or more than one isomeric compound without additional information and (2) How can one prove a direct precursor-product relationship between isomeric molecules and their dissociation products?
Figure 2.
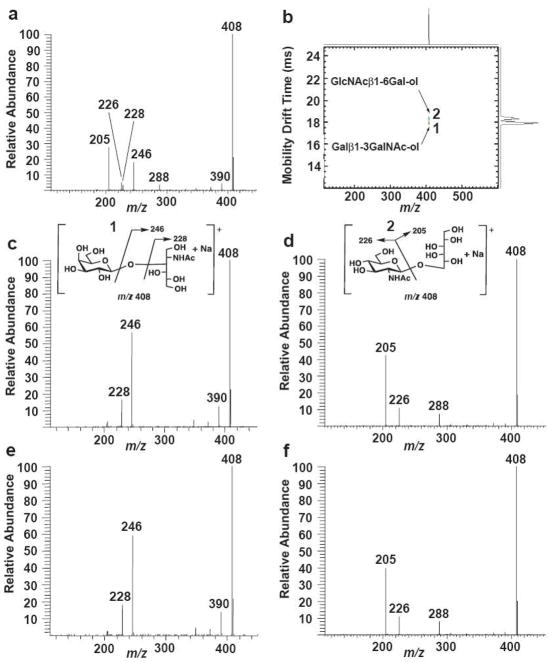
Ion mobility-MS/MS of isomeric disaccharide-alditols illustrating their separation and acquisition of independent MS/MS spectra for the molecules as sodiated molecules. Structural diagrams and postulated dissociation patterns for the ions are shown (m/z 408 precursors). a. The MS/MS spectrum (quadrupole ion trap) of a mixture of the two disaccharide alditols Galβ1-3GalNAc-ol and GlcNAcβ1-6Gal-ol. b. A two-dimensional ion mobility/MS spectrum performed on an APIMS-TOF instrument where mass spectra were acquired in real time during the ion mobility experiment. Note that drift times were different than those obtained for the IMS-ion trap instrument, although the reduced mobility value Ko was constant for each compound. c. The MS/MS spectrum of Galβ1-3GalNAc-ol (1) run individually on the IMS-ion trap instrument, selecting precursor ions within the 39.7–40.4 ms window. d. The MS/MS spectrum of GlcNAcβ1-6Gal-ol (2) run individually on the IMS-ion trap instrument, selecting precursor ions within the 40.7–41.1 ms window. e. The MS/MS spectrum of the 39.7–40.4 ms window on the IMS-ion trap instrument, spraying a mixture of the two isomeric disaccharide-alditols. f. The MS/MS spectrum of the 39.7–40.4 ms window on the IMS-ion trap instrument, spraying a mixture of the two isomeric disaccharide-alditols. All mass spectra were accumulated using identical collision energies. The postulated CID fragmentation mechanisms are similar to those reported by Martensson et al [16]. The CID fragment ions were marked according to Domon and Costello’s nomenclature [32].
Both disaccharide-alditols were run as sodiated molecules individually on the APIMS-TOF instrument using nitrogen as the drift gas (Fig. 2b). They were fully resolved. The two compounds were then run individually on the APIMS-ion trap instrument. By initially keeping the second gate open for a longer time period, then successively narrowing the second gate time, and finally using the mobility scanning mode over a narrow drift time region, the drift time window just covering each precursor ion peak could be determined. The absolute drift times were different in comparing the APIMS–TOF and the APIMS-ion trap although their relative order was the same. Isomer 1 (Galβ1-3GalNAc-ol) and isomer 2 (GlcNAcβ1-6Gal-ol), having drift time windows of 39.7–40.4 ms and 40.7–41.1 ms, respectively, on the APIMS-ion trap, yielded completely different MS/MS spectra as shown in Fig. 2c and 2d, when run individually on the instrument. Moreover, when the two molecules were run as a mixture and these same window times selected, the mass spectra generated were essentially identical to those obtained when the molecules were run individually (Fig. 2e and 2f).
Separation of isomeric species having mass spectra where product ions only vary in relative abundance: isomers varying solely in their anomeric configurations
When oligosaccharides do not have differences in their linkage positions, but vary either in the anomeric configuration of some of those linkages or, sometimes, in the stereochemistry of the sugars, product ions may not be different in m/z between them, although they may vary in their relative abundance. Again, assuming no prior knowledge about the molecules in unknown samples, this is a more subtle and difficult problem to deal with as it can easily lead to the assumption that a mixture of isomers might be one pure molecule. To illustrate this point, two isomeric tetrasaccharide-alditols were examined, molecules that vary only in their anomeric configurations: cellobiitol (Glcβ1-4Glcβ1-4Glcβ1-4Glc-ol) and maltotetraitol (Glcα1-4Glcα1-4Glcα1-4Glc-ol). As indicated in Fig. 3a, the two molecules could be fully separated as sodiated molecules by IMS; an overlay of their IMS spectra run separately on the APIMS-TOF is shown. When the molecules were run individually on the APIMS-ion trap instrument and their MS/MS spectra acquired, it was evident (Fig. 3b and 3c) that the two molecules yielded the same product ions (m/z 529, 509 and 367) from m/z 691 precursor ions. The maltotetraitol had a faster drift time (52.2–52.7 ms window) than cellotetraitol (53.5–53.8 ms) and could be distinguished by MS/MS because the cellotetraitol yielded an m/z 529 product ion having roughly three times the abundance of maltotetraitol, using the same collision energies for dissociation. Again, when the mixture of the two molecules was run, the two compounds were readily distinguished (Fig. 3d and 3e). Their MS/MS spectra were essentially identical to the compounds run individually, and, in combination with the fact that they had different drift times, they could be readily differentiated. It is important to note that these compounds would be extremely difficult to recognize in the presence of the other without prior knowledge of their compositions using standard MSn methods. They could easily have a significant proportion of the other isomer present and mass spectra could be misinterpreted as resulting solely from one or the other, even assuming that their mass spectra are known in advance. In a truly unknown sample, it would be easy to make mistakes in assigning structures to sets of isomers like these, where product ions derive from both precursor ions and normally could not be independently isolated.
Figure 3.
Ion mobility-MS/MS of two isomeric tetrasaccharide-alditols differing only in their anomeric configurations. Structural diagrams and postulated fragmentation patterns are shown for two molecules, Glcα1-4Glcα1-4Glcα1-4Glc-ol (maltotetraitol, 1) and Glcβ1-4Glcβ1-4Glcβ1-4Glc-ol (cellotetraitol, 2). Molecules were separated as sodiated molecules (m/z 691 precursor ions). They yielded the same product ions but in different abundance. a. An overlay of two 2-dimensional ion mobility/MS spectra, performed separately on an IMS-TOF instrument. Note that drift times were different than those observed for the IMS-ion trap instrument. b. The MS/MS spectrum of Glcα1-4Glcα1-4Glcα1-4Glc-ol run individually on the IMS-ion trap instrument, selecting precursor ions within a 52.2–52.7 ms drift time window. c. The MS/MS spectrum of Glcβ1-4Glcβ1-4Glcβ1-4Glc-ol run individually on the IMS-ion trap instrument, selecting precursor ions within a 53.5–53.8 ms drift time window. d. The MS/MS spectrum obtained when the two isomers were analyzed as a mixture on the IMS-ion trap instrument, selecting precursor ions within a 52.2–52.7 ms drift time window. e. The MS/MS spectrum obtained when the two isomers were analyzed as a mixture on the IMS-ion trap instrument, selecting precursor ions within a 53.5–53.8 ms drift time window. Collision energies in the trap were the same for all mass spectra. The postulated CID fragmentation mechanisms are similar to those reported by Martensson et al [16]. The CID fragment ions were marked according to Domon and Costello’s nomenclature [32].
The two examples in Figs. 2 and 3 above illustrate a few key points worthy of note concerning the value of APIMS-ion trap MSn in analysis of carbohydrate isomers: (1) If an oligosaccharide at a given m/z from some biological sample is thought to be pure, a demonstration that the sample gives more than one peak using APIMS, where the MS/MS spectrum is different for each peak, strongly indicates that the sample contains isomers. There may be additional peaks when complex biological samples are used, and there may be more than one isomer in each peak, where additional isomers might happen to coincidentally co-migrate under one set of conditions in the APIMS. However, the presence of two (or more) peaks yielding different tandem mass spectra means with high probability that the precursor ion is not a single isomer. (2) This requires no prior knowledge about the origin of the sample. (3) As we have shown previously, APIMS can be performed with different gases which affect both the relative drift times and in some cases even the relative order of the drift times of compounds21–23. Because ion mobility is an inherent physical property of a compound, changes in MS/MS spectra of peaks under different APIMS conditions indicates isomeric heterogeneity, even when multiple isomers are examined in a mixture, as long as dissociation conditions are identical. Moreover, if a peak remains Gaussian under several conditions and yields the same MS/MS spectrum, one gains confidence in the probability that it is just one isomer and that direct precursor product-relationships can be established for that molecule. It does not prove the above point, as some molecules might consistently comigrate; it merely increases the probability each time a different set of conditions is used such as a different IMS gas or adducting ion. All of the above, of course, warrants the highest IMS resolution possible.
Separation of isomeric species having near-identical mass spectra: the problem of very closely-related isomers
Another layer of complexity is found in analysis of oligosaccharides by mass spectrometry: those isomers that yield nearly identical MS/MS spectra and nearly identical MSn spectra. For example, two isomers that vary only in the stereochemistry at a single location (epimers at one carbon in one sugar) are shown in Fig. 4. Remarkably, these two isomers were very well resolved using nitrogen as the drift gas, as shown using the APIMS-TOF instrument (Fig. 4a). However, their MS/MS spectra were virtually identical as the single stereochemical difference between them does not have a major effect on their dissociation patterns (Fig. 4b and 4c). Selecting either of these well-separated peaks, at drift mobility windows of 51.6–52.3 ms or 56.3–57.2 ms, yielded MS/MS spectra on the APIMS-ion trap instrument that, at the same collision energies, were impossible to distinguish between on a reproducible basis, giving two major product ions at m/z 449 (base peak) and 431. The other ions m/z 634, 532, 413 and 246 were less than 5% in relative abundance and both isomers gave rise to them. There could be any number of biological systems where very minor differences in structure fail to enable two molecules to be differentiated by MS dissociation methods alone. Both these structures, for example, were previously purified from the same source (bovine submaxillary mucin) and clearly shown to have different NMR spectra16. Note that it would be essentially impossible to know which of these structures actually gives rise to spectra as shown in Fig. 4b and 4c, let alone to decide whether the two structures might both be present as a mixture. Again, without any prior knowledge, such closely-related structures may be demonstrated in a mixture if they separate using APIMS-ion trap mass spectrometry.
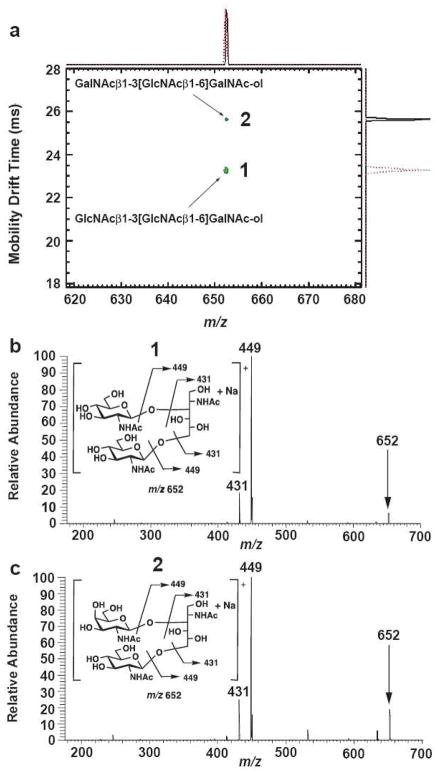
Figure 4.
Ion mobility-MS/MS of two isomeric trisaccharide-alditols varying in the stereochemistry at a single asymmetric carbon, having near-identical dissociation spectra. Structural diagrams and postulated dissociation patterns are shown for the two molecules, GlcNAcβ1-3[GlcNAcβ1-6]GalNAc-ol (1) and GalNAcβ1-3[GlcNAcβ1-6]GalNAc-ol (2) as sodiated molecules (m/z 652 precursor ions are indicated with arrows). Both are closely-related O-linked core structures isolated from bovine submaxillary mucin. a. An overlay of two 2-dimensional ion mobility/MS spectra, performed separately on an IMS-TOF instrument. Note that drift times were different than those observed for the IMS-ion trap instrument. Although very similar in structure, the two molecules were well resolved using ambient pressure IMS. b. The MS/MS spectrum of GlcNAcβ1-3[GlcNAcβ1-6]GalNAc-ol spraying a mixture of both isomers on the IMS-ion trap instrument, selecting precursor ions within the 51.6–52.3 ms window. c. The MS/MS spectrum of GalNAcβ1-3[GlcNAcβ1-6]GalNAc-ol spraying a mixture of both isomers on the IMS-ion trap instrument, selecting precursor ions within the 56.3–57.2 ms window. Note the close similarity of the spectra, both compounds yielding identical ions with only minor differences in abundance. The postulated CID fragmentation mechanisms are similar to those reported by Martensson et al [16]. The CID fragment ions were marked according to Domon and Costello’s nomenclature [32].
Separation of isomeric species where at least one of them has a very simple dissociation pattern: the value of MS3 and potentially higher order MS to identify some isomers
In some cases, very simple MS/MS spectra due to the presence of one major product ion can make it very difficult to identify the presence of two isomers in a mixture without further dissociation of product ions. For example, in Fig. 5a, the separation of two fucosylated tetrasaccharide-alditol isomers isolated from bovine submaxillary mucin16 is shown as an overlay of two 2-dimensional IMS-MS spectra acquired using the APIMS-TOF instrument. The two structures were baseline resolved. When the separation was performed on the APIMS-ion trap instrument, isomer 1, Fucα1-2Galβ1-3[GlcNAcβ1-6]GalNAc-ol, had a drift time window between 58.3–59.1 ms and isomer 2, Fucα1-2Galβ1-4GlcNAcβ1-3GalNAc-ol, completely separated with a drift time of 59.4–59.9 ms. The mass spectrum of structure 1 (Fig. 5b) showed a very simple MS/MS spectrum, having a very abundant m/z 611 product ion and a very low abundance m/z 449 product ion. Yet these same ions were seen in the MS/MS spectrum of isomer 2 illustrating another issue in differentiating isomers from a mixture: the need for higher order MS analysis. Even with prior knowledge of their MS/MS spectra it would be very difficult to detect the presence of a moderate amount of isomer 1 in a mixture with isomer 2 using MS/MS with selection of the common precursor ion m/z 757. Differentiating them would require that MS3 be used on selected product ions. However, without prior knowledge, an APIMS-ion trap physical separation clearly revealed two isomers, which was verified by the differences in their MS/MS spectra. The MS/MS dissociation spectrum of isomer 1 was, however, structurally rather uninformative. It was therefore demonstrated that the mobility-selected mode could be used to accumulate enough of the m/z 611 product ion following MS/MS to acquire its MS3 spectrum (Fig 5d). This spectrum was far more structurally informative, yielding 4 characteristic product ions for isomer 1 (m/z 491, 449, 431 and 408). For the other tetrasaccharide-alditol isomer, dissociation of the product ion m/z 611 yielded a single major product ion of MS3 at m/z 388 (Fig. 5e). Thus the APIMS-ion trap could be used to at least MS3 to extract more detailed structural information about individual molecules within specific drift time windows, which should have value when some carbohydrate molecules such as structure 1 in Fig. 5b give rise to very simple mass spectra.
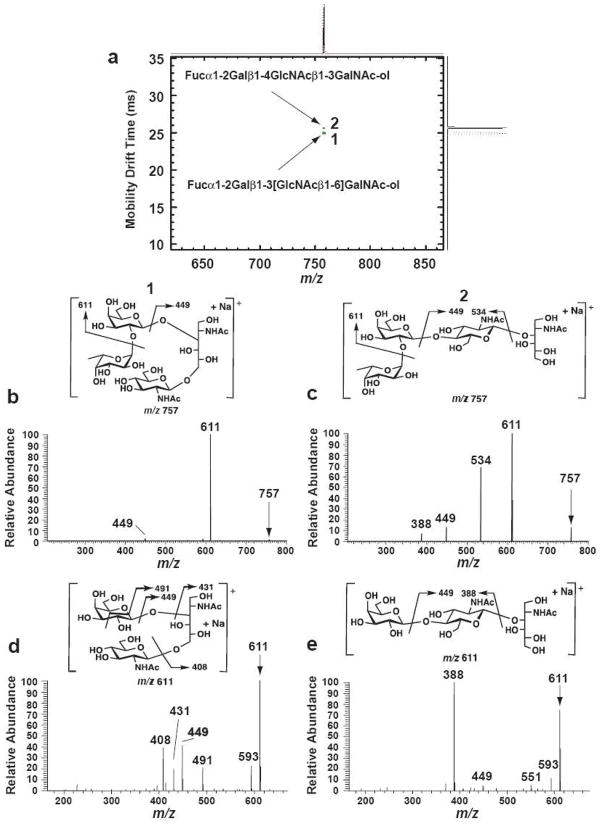
Figure 5.
Ion mobility separation of two isomeric tetrasaccharide-alditols from bovine submaxillary mucin, demonstrating their complete physical separation and independent mass spectra obtained to MS3 using the APIMS-ion trap instrument. Structural diagrams and postulated fragmentation patterns are shown for the two molecules, the branched Fucα1-2Galβ1-3[GlcNAcβ1-6]GalNAc-ol (1) and the linear Fucα1-2Galβ1-4GlcNAcβ1-3GalNAc-ol (2). Isomers were separated as sodiated molecules that yielded very different dissociation patterns, both in MS2 (m/z 757 precursor ion selected) and MS3 (m/z 611 product ion selected, derived by neutral loss of fucose from both m/z 757 precursors). a. An overlay of two 2-dimensional ion mobility/MS spectra, performed separately on an IMS-TOF instrument. Note that drift times were different than those observed for the IMS-ion trap instrument. b. The MS/MS spectrum of isomer 1 (Fucα1-2Galβ1-3[GlcNAcβ1-6]GalNAc-ol) run on the IMS-ion trap instrument, selecting those precursor ions within a 58.3–59.1 ms drift time window. c. The MS/MS spectrum of isomer 2 (Fucα1-2Galβ1-4GlcNAcβ1-3GalNAc-ol), selecting those precursor ions within a 59.4–59.9 ms drift time window. d. The MS3 spectrum of the m/z 611 product ion derived from isomer 1 (58.3–59.1 ms window). e. The MS3 spectrum of the m/z 611 product ion derived from isomer 2 (59.4–59.9 ms window). Structures of the postulated product ions and their dissociation patterns are illustrated in panels d and e. The postulated CID fragmentation mechanisms are similar to those reported by Martensson et al [16]. The CID fragment ions were marked according to Domon and Costello’s nomenclature [32].
Separation of isomeric non-reducing trisaccharides: issues associated with reducing versus non-reducing oligosaccharides
It probably has not escaped the attention of readers that all data up to this point has been presented using oligosaccharides where the reducing sugar has been converted to an alditol via borohydride reduction. Reducing oligosaccharides frequently yield more than one ion mobility peak21, which we have attributed to the fact that their reducing monosaccharides are usually found in multiple configurations that include the α and β pyranose and α and β furanose forms. This is tantamount to actually running four separate isomers as gas phase ions because each form can potentially have a different overall cross-sectional area, hence may show up as an independent IMS peak. This greatly complicates analyses. To date, we have observed that reduction of a number of oligosaccharides to their alditols has always resulted in a single IMS peak. In addition, some natural non-reducing oligosaccharides were examined, as shown in Fig. 6, to verify whether these types of structures yielded single ion mobility peaks and to further demonstrate the value of IMS interfaced with ion trap mass spectrometry. The two isomeric trisaccharides melezitose and raffinose were fully separated using the IMS-TOF instrument (Fig. 6a). The precursor ions (m/z 527) were then selected in separate time windows (43.3–43.9 ms and 44.5–45.2 ms, respectively, for melezitose and raffinose) on the APIMS-ion trap instrument; their MS/MS spectra are shown in Figs. 6b and 6c. Notably, both precursor ions generated an abundant product ion at m/z 365 and a minor product ion at m/z 445. However, melezitose yielded an additional product ion at m/z 347 and 275 (Fig. 6b). Raffinose (structure 2, Fig. 6c) gave a single major dissociation product (m/z 365) that was also present in the MS/MS spectrum of melezitose (structure 1, Fig. 6b). Again, in a similar fashion to the fucosylated tetrasaccharides illustrated in Fig. 5, it would be very difficult to verify the presence of a small amount of raffinose in the presence of melezitose based on MS/MS spectra alone. This scenario, where one isomer yields just one major product ion with little additional structural information, is not uncommon in oligosaccharide MS. It sometimes occurs when cleavages at single sites or neutral losses of single water molecules are highly predominant dissociation events. However, the m/z 365 product ions could be independently isolated from either compound based on their independent ion mobilities and showed clear-cut differences in their MS3 dissociation patterns. That derived from raffinose yielded a major m/z 305 product ion in the next stage of dissociation (Fig 6e) whereas this ion was generated in negligible abundance from the m/z 365 product ion derived from melezitose (Fig. 6d). Thus, admittedly with a limited number of non-reducing structures examined to date, they (1) yielded single IMS peaks and (2) generated independent mobility-selected MS/MS and MS3 spectra.
Figure 6.
Ion mobility separation of two trisaccharides, illustrating the value of ion mobility interfaced with ion trap mass spectrometry performed to MS3 for the differentiation of natural non-reducing isomers within a mixture. Structural diagrams and postulated fragmentation patterns are shown for the two precursor molecules as sodiated molecules having m/z 527, the branched molecule melezitose (1) and the linear molecule raffinose (2). a. An overlay of two 2-dimensional ion mobility/MS spectra, performed separately on an IMS-TOF instrument. Note that drift times were different than those observed for the IMS-ion trap instrument. b. The MS/MS spectrum of isomer 1 (melezitose) run on the IMS-ion trap instrument, selecting those precursor ions within a 43.3–43.9 ms drift time window. c. The MS/MS spectrum of isomer 2 (raffinose), selecting those precursor ions within a 44.5–45.2 ms drift time window. d. The MS3 spectrum of the m/z 365 product ion derived from isomer 1 (43.3–43.9 ms window). e. The MS3 spectrum of the m/z 365 product ion derived from isomer 2 (44.5–45.2 ms window). The postulated CID fragmentation mechanisms are similar to those reported by Martensson et al [16]. The CID fragment ions were marked according to Domon and Costello’s nomenclature [32].
Examination of the isomeric heterogeneity of a single HPLC fraction from bovine submaxillary mucin by ion mobility spectrometry-tandem ion trap mass spectrometry
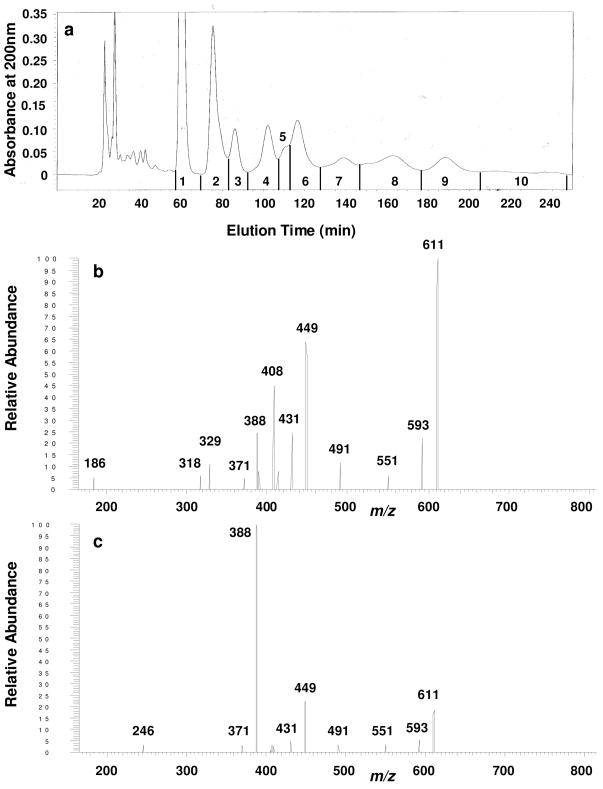
While HPLC is a valuable first step in characterizing oligosaccharide mixtures, it does not have infinite resolution thus it is possible that each HPLC fraction from any complex biological sample might contain isomeric structures as well as structures having different masses. One of the goals of building the IMS-ion trap was to use it for rapid and flexible evaluation of the isomeric heterogeneity of HPLC fractions. The HPLC separation of neutral reduced oligosaccharide-alditols released from bovine submaxillary mucin is shown in Fig. 7a, with fractions indicated. Fraction 5 was concentrated and further examined on the APIMS-ion trap. As indicated in Fig. 7b and 7c, m/z 661 precursor ions were selected from two separate narrow drift time windows by ion mobility and were subsequently isolated and dissociated in the ion trap under identical conditions. They yielded MS/MS spectra that were markedly different, strongly indicating that at least two isomers were present in this HPLC fraction and that APIMS interfaced with an ion trap provides a valuable independent assessment of isomeric components found within HPLC fractions
Figure 7.
Ion mobility-MSMS of an HPLC fraction of neutral oligosaccharide-alditols isolated from bovine submaxillary mucin, showing that IMS-MSn can independently and rapidly evaluate isomers within HPLC fractions derived from complex biological samples. a. The HPLC profile, using a semi-preparative column of Glycopak N [16], eluted with 74/26 acetonitrile/water, 5.0 mL/min, with detection at 200 nm. Fractions as indicated were concentrated. b. Ambient pressure ion mobility-MS/MS spectrum of oligosaccharide-alditols from fraction 5 from the HPLC column shown in panel a. The MS/MS spectrum was obtained from m/z 611 precursor ions (sodium adducts) isolated within a 51.1–51.6 ms drift time window. c. The MS/MS spectrum obtained from m/z 611 precursor ions (sodium adducts) isolated within a 50.2–50.7 ms drift time window, from the same mixture shown in panel b. Dissociation conditions in the trap were identical for precursor ions isolated from each independent drift time window.
Limitations of the technique as the instrument is currently configured and other considerations
Two limitations of the instrument as it was operated are worth mentioning, with the potential for modifications and improvements in the future. First, the sensitivity of the method for acquisition of MSn spectra was much less than direct MSn using a quadrupole ion trap. This is in part due to the loss of some ions during ion mobility and in part due to a suboptimal interface to the ion trap at this point, but sensitivity can probably be significantly improved in the future. In addition, linear ion traps can effectively store more ions thus would be favored to interface with APIMS in the future. Second, it was valuable to have access to an IMS-TOF instrument separately from the IMS-trap. While the IMS-TOF cannot provide MSn spectra, it can rapidly assess whether isomers physically resolve prior to their independent analyses on the IMS-ion trap. In addition, the TOF can provide higher mass accuracy.
One other point is worth mentioning. The value of ion mobility spectrometry as an analytical tool is based on the premise that a number of precursor ions of the same compound migrate down the drift tube as single Gaussian peak distributions, at least for most small molecules. Every effort has been made in the design of instruments over the years to avoid clustering of ions, the principal cause of multiple peaks for single compounds. While the vast majority of small molecules behave as single peaks, some ions exist in different configurations prior to electrospray, and some ions undergo conformational changes that are slow on the IMS time scale hence yield more than one peak. In our analyses of oligosaccharide-alditols to date, only one peak has been observed per molecule. Admittedly, with larger oligosaccharide structures it might be possible that sodium ions could coordinate at more than one location, potentially giving rise to more than one cross-sectional area thus more than one peak. More observations of many more oligosaccharide-alditols by IMS will be required to see whether there are exceptions.
Conclusions
The examples provided here demonstrate that ambient pressure ion mobility spectrometry interfaced with an ion trap will be a valuable tool for analysis of oligosaccharide isomers and probably many other types of isomers as well. The high resolution that could be achieved with the ambient pressure ion mobility portion of the instrument enabled complete physical separation of isomers prior to their independent MSn analyses, demonstrated to MS3. Isomeric oligosaccharides of representative classes were selected to illustrate the usefulness of the technique in addressing a range of problems that MSn alone was unable to effectively solve; this included linkage and branch isomers, anomeric isomers and very closely related isomers varying at a single stereochemical position. The front-end APIMS was especially valuable in evaluating isomeric heterogeneity when the MS/MS spectra of two isomers contained identical product ions, where the spectra either varied only in the relative abundance of these ions or when the relative abundance was nearly identical for all ions. With the mindset that one might expect to find unknown molecules as yet in any complex biological sample, APIMS-ion trap MSn has the key advantage in that it can be used to rapidly reveal the presence of isomers without any prior assumptions or knowledge about their structures, and can provide dissociation spectra for independent isomers that physically separate on the millisecond time scale.
Acknowledgments
This work was supported by a grant from NIH (#5R21RR02004602). Partial support of the preliminary studies was also from a NSF grant (CHE0137986). Thermo Finnigan provided the LCQ Deca quadrupole ion trap mass spectrometer that made this study possible.
References
- 1.Jacquinot PM, Leger D, Wieruszeski JM, Spik G. Glycobiology. 1994;4:617–624. doi: 10.1093/glycob/4.5.617. [DOI] [PubMed] [Google Scholar]
- 2.Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Angata K, Huckaby V, Ranscht B, Terskikh A, Marth JD, Fukuda M. Mol Cell Biol. 2007;27:6659–6668. doi: 10.1128/MCB.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sverremark E, Fernandez C. Immunology. 1997;92:153–159. doi: 10.1046/j.1365-2567.1997.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 6.Ge CH, Stanley P. Pro Natl Acad Sci. 2008;105:1539–1544. doi: 10.1073/pnas.0702846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Tan L, Lamport DT, Kieliszewski MJ. Phytochemistry. 2008;69:1631–1640. doi: 10.1016/j.phytochem.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Ulloa-Aguirre A, Timossi C, Barrios-de-Tomasi J, Maldonado A, Nayudu P. Biology of Reproduction. 2003;69:379–389. doi: 10.1095/biolreprod.103.016915. [DOI] [PubMed] [Google Scholar]
- 9.Platt I, Rao LG, El-Sohemy A. Experimental Biology and Medicine. 2007;232:246–252. [PubMed] [Google Scholar]
- 10.Kochetkov NK, Chizhov OS. Biochim Biophys Acta. 1964;83:134–136. doi: 10.1016/0926-6526(64)90064-3. [DOI] [PubMed] [Google Scholar]
- 11.Vink J, Ridder JJ, Kamerling JP, Vliegenthart JF. Biochem Biophys Res Commun. 1971;42:1050–1056. doi: 10.1016/0006-291x(71)90010-6. [DOI] [PubMed] [Google Scholar]
- 12.Morelle W, Michalski JC. Curr Pharm Res. 2005;11:2615–2645. doi: 10.2174/1381612054546897. [DOI] [PubMed] [Google Scholar]
- 13.Reinhold VN, Reinhold BB, Costello CE. Anal Chem. 1995;67:1772–1784. doi: 10.1021/ac00107a005. [DOI] [PubMed] [Google Scholar]
- 14.Ashline D, Singh S, Reinhold V. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polfer NC, Valle JJ, Bendiak B. Anal Chem. 2005;78:670–679. doi: 10.1021/ac0519458. [DOI] [PubMed] [Google Scholar]
- 16.Martensson S, Levery SB, Fang TT, Bendiak B. Eur J Biochem. 1998;258:603–622. doi: 10.1046/j.1432-1327.1998.2580603.x. [DOI] [PubMed] [Google Scholar]
- 17.Lamblin G, Boersma A, Lhermitte M, Roussel P, Mutsaers J, Halbeek HV, Vliegenthart J. Eur J Biochem. 1984;143:227–236. doi: 10.1111/j.1432-1033.1984.tb08363.x. [DOI] [PubMed] [Google Scholar]
- 18.Dua VK, Narasinga Rao BN, Wu SS, Dube VE, Bush CA. J Biol Chem. 1986;261:1599–1608. [PubMed] [Google Scholar]
- 19.Bendiak B, Harris-Brandts M, Michnick SW, Carver JP, Cumming DA. Biochemistry. 1989;28:6491–6499. doi: 10.1021/bi00441a050. [DOI] [PubMed] [Google Scholar]
- 20.Clowers BH, Dwivedi P, Steiner WE, Hill HH, Bendiak B. J Am Soc Mass Spectrom. 2005;16:660–669. doi: 10.1016/j.jasms.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Dwivedi P, Bendiak B, Clowers BH, Hill HH. J Am Soc Mass Spectrom. 2007;18:1163–1175. doi: 10.1016/j.jasms.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Asbury GR, Hill HH. J Microcolumn Separations. 2000;12:172–178. [Google Scholar]
- 23.Asbury GR, Hill HH. Anal Chem. 2000;72:580–584. doi: 10.1021/ac9908952. [DOI] [PubMed] [Google Scholar]
- 24.Gabryelski W, Froese KL. J Am Soc Mass Spectrom. 2003;14:265–277. doi: 10.1016/S1044-0305(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 25.Vakhrushev SY, Langridge J, Campuzano I, Hughes C, Peter-Katalinic J. Anal Chem. 2008;80:2506–2513. doi: 10.1021/ac7023443. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, Klasmeier J, Hill HH. Rapid Commun Mass Spectrom. 1999;13:1138–1142. doi: 10.1002/(SICI)1097-0231(19990630)13:12<1138::AID-RCM625>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Adamov A, Viidanoja J, Kärpänoja E, Paakkanen H, Ketola RA, Kostiainen R, Sysoev A, Kotiaho T. Review of Scientific Instruments. 2007;78:044101-1–5. doi: 10.1063/1.2723742. [DOI] [PubMed] [Google Scholar]
- 28.Clowers BH, Hill HH. Anal Chem. 2005;77:5877–5885. doi: 10.1021/ac050700s. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, Bruce JE, Hill HH. Rapid Commun Mass Spectrom. 2007;21:1115–1122. doi: 10.1002/rcm.2928. [DOI] [PubMed] [Google Scholar]
- 30.Steiner W, Clowers B, Fuhrer K, Hill HH. Rapid Commun Mass Spectrom. 2001;15:2221–2226. doi: 10.1002/rcm.495. [DOI] [PubMed] [Google Scholar]
- 31.Martinez WL. Martinez AR Computational Statistics Handbook with MATLAB. Chapman & Hall/CRC; Boca Raton, FL: 2007. [Google Scholar]
- 32.Dmon B, Costello CE. Glycoconjugate J. 1988;5:397–409. [Google Scholar]