Abstract
Mitochondrial defects in gene expression have been implicated in the pathophysiology of bipolar disorder and schizophrenia. We have now contrasted control brains with low pH versus high pH and showed that 28% of genes in mitochondrial-related pathways meet criteria for differential expression. A majority of genes in the mitochondrial, chaperone and proteasome pathways of nuclear DNA-encoded gene expression were decreased with decreased brain pH, whereas a majority of genes in the apoptotic and reactive oxygen stress pathways showed an increased gene expression with a decreased brain pH. There was a significant increase in mitochondrial DNA copy number and mitochondrial DNA gene expression with increased agonal duration. To minimize effects of agonal-pH state on mood disorder comparisons, two classic approaches were used, removing all subjects with low pH and agonal factors from analysis, or grouping low and high pH as a separate variable. Three groups of potential candidate genes emerged that may be mood disorder related: (a) genes that showed no sensitivity to pH but were differentially expressed in bipolar disorder or major depressive disorder; (b) genes that were altered by agonal-pH in one direction but altered in mood disorder in the opposite direction to agonal-pH and (c) genes with agonal-pH sensitivity that displayed the same direction of changes in mood disorder. Genes from these categories such as NR4A1 and HSPA2 were confirmed with Q-PCR. The interpretation of postmortem brain studies involving broad mitochondrial gene expression and related pathway alterations must be monitored against the strong effect of agonal-pH state. Genes with the least sensitivity to agonal-pH could present a starting point for candidate gene search in neuropsychiatric disorders.
Keywords: mitochondria, proteasome, apoptosis, chaperone, bipolar disorder I, recurrent major depressive disorder
The pathophysiology of depression and mania is largely unknown leaving open a question of which cellular pathway(s) to investigate. Gene expression screening is a powerful method for probing alterations in neural functions in mood disorder. Examining gene expression profiles has been extremely useful for establishing different phenotypes in cancer.1–3 Various microarray profiles in mood disorder have been reported that use different microarray platforms, analysis methodology, regional differences, agonal factors and brain pH.4–14 Presumably, a substantial heterogeneity in the pathophysiology of mood disorder coupled with small sample sizes and small fold changes contributes to the lack of consistency among the findings.15
In five microarray studies a broad mitochondrial dysfunction was reported in neuropsychiatric disorders.10,11,14,16–18 One study of bipolar disorder (BPD) focused on a broad set of mitochondrial-related gene expression and found that in general, mitochondrial gene expression was predominantly decreased in BPD compared to controls in the dorsolateral pre-frontal cortex (DLPFC) when using Stanley samples with pH above 6.5.10 In a select subgroup of medication-free BPD patients, there were mitochondrial genes that showed increased expression.10 Thus, medication may influence the direction of gene expression change and the precise genes altered in BPD patients. A subset of genes in the oxidative phosphorylation pathway and proteasome family were found to be decreased in the hippocampus by microarray and also in the prefrontal cortex by Q-PCR in bipolar disorder but not schizophrenia11, whereas convincing evidence for a decrease in mitochondrial and proteasomal gene expression was found in hippocampus dentate gyrus neurons sampled with laser capture microscopy.14 Another microarray and proteomic study using a set of 105 DLPFC samples from the Stanley Array Collection found mitochondrial dysfunction in schizophrenia.16
Not all microarray studies find a broad mitochondrial gene expression pattern in bipolar disorder8,12 or schizophrenia.10,19–21 A few microarray studies in major depressive disorder (MDD) have not reported mitochondrial genes were differentially expressed in cortex.4,9,22
There is an emerging hypothesis of mitochondrial dysfunction in bipolar disorder23 and schizophrenia.14,16 Mitochondria are clearly important in cellular functions, involving bioenergetics, cell death and metabolism. Each cell contains multiple mitochondrion, which contain multiple copies of mtDNA that encode 37 genes. A mitochondrial hypothesis of bipolar disorder23 is based on multiple converging lines of data: brain pH, lithium response, known and novel SNPs in nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) encoding mitochondrial genes, and evidence suggesting matrilineal inheritance patterns.23 If pH is altered as part of the pathophysiology of BPD or schizophrenia, it would most likely cause gene expression differences that would be masked by agonal factor that can also induce pH differences. We tested whether gene expressions of nDNA encoding genes with mitochondrial function are altered in mood disorder compared to controls for three brain regions: DLPFC, anterior cingulate cortex (AnCg) and cerebellum (CB). The cerebellum was used as a comparison tissue to measure agonal-pH effects by copy number of mtDNA and by in situ hybridization for a candidate gene, leucine-rich PPR-motif containing (LRPPRC). Leucine-rich PPR-motif containing was chosen as it was dysregulated in the present study and mutations in the gene cause a progressive neurodegenerative disorder, Leigh French Canadian Syndrome24, which involves lactic acidosis25 and alterations in brain pH. Selected nDNA and mtDNA encoding genes were measured with quantitative real time polymerase chain reaction PCR in AnCg and DLPFC. We previously had shown that samples from this cohort with low pH and prolonged death exhibited a decrease in expression of genes involved in energy metabolism and proteolytic activities.26 In the present report, we stringently selected mood disorder samples to increase the signal-to-noise ratio to examine evidence of dysregulation of mitochondrial-related genes in mood disorder.
Methods
Tissue acquisition
Brain tissue was obtained by the University of California, Irvine Brain Repository through a uniform process approved by the Institutional Review Board. An extensive review of multiple sources of information on all subjects included the medical examiner's conclusions, coroner's investigation, medical and psychiatric records, toxicology results and interviews of the decedents’ next-of-kin. These reports were examined for information concerning physical health, medication use, psychopathology, substance use, family psychiatric history and details of death. A neuropathological examination of each brain was conducted to exclude any brains with visible evidence of an infarct, tumor or visible hemorrhage. The agonal duration was rated for each decedent (Table 1) based upon the Hardy, Wester and Johnston rating scales previously published27–29 and refined.30
Table 1.
The demographics of each group for microarray analyses #1, #2, and #3
| Group (N) | PH |
Age |
Gender |
||
|---|---|---|---|---|---|
| Average | s.d. | Average | s.d. | M/F | |
| Microarray analysis #1 (N = 40) | |||||
| BPD (9) | 6.92 | 0.19 | 53.89 | 17.69 | 6M/3F |
| Control (20) | 6.67 | 0.29 | 52.60 | 16.46 | 14M/6F |
| MDD (11) | 6.89 | 0.25 | 50.73 | 14.79 | 8M/3F |
| P-value (two tailed t-test BPD, Control) | 0.03 | 0.85 | |||
| P-value (two tailed t-test MDD, Control) | 0.05 | 0.76 | |||
| Microarray analysis #2 (N = 40) | |||||
| pH > 6.87 | |||||
| BPD (6) | 7.03 | 0.11 | 49.67 | 20.55 | 6M/0F |
| Control (7) | 6.98 | 0.06 | 52.20 | 19.93 | 6M/1F |
| MDD (7) | 7.03 | 0.11 | 48.71 | 15.91 | 5M/2F |
| P-value (two tailed t-test BPD, Control) | 0.38 | 0.84 | |||
| P-value (two tailed t-test MDD, Control) | 0.37 | 0.74 | |||
| pH < 6.87 | |||||
| BPD (3) | 6.71 | 0.12 | 62.33 | 6.03 | 0M/3F |
| Control (13) | 6.53 | 0.25 | 55.54 | 14.76 | 8M/5F |
| MDD (4) | 6.64 | 0.22 | 54.25 | 14.03 | 3M/1F |
| P-value (two tailed t-test BPD, Control) | 0.25 | 0.46 | |||
| P-value (two tailed t-test MDD, Control) | 0.42 | 0.88 | |||
| Microarray analysis #3 (N = 30) | |||||
| BPD-High (6) | 6.97 | 0.10 | 47.5 | 18.7 | 5M/1F |
| Control-High (7) | 6.95 | 0.07 | 50.0 | 16.7 | 6M/1F |
| MDD-High (7) | 7.03 | 0.11 | 48.7 | 15.9 | 5M/2F |
| Control-Low (10) | 6.48 | 0.26 | 57.3 | 16.5 | 5M/5F |
| P-value (two tailed t-test BPD-High, Control-High) | 0.65 | 0.80 | |||
| P-value (two tailed t-test MDD-High, Control-High) | 0.12 | 0.89 | |||
| P-value (two tailed t-test Control-High, Control-Low) | 0.00 | 0.39 | |||
Three groups of subjects (n = 40) were used in microarray analysis #1. Microarray analysis #2 used six groups based on subject assignment to pH above or below the median pH of 6.87 (n = 40 subjects) and diagnosis. Microarray analysis #3 used subject assignment based upon criteria of pH > 6.87 and zero agonal factor to form 3 groups Bipolar-High, Control-High, Major Depression-High. The fourth group Control-Low contained control subjects with pH below the median pH of 6.87, and one or more agonal factors. The agonal factor was the sum of prolonged agonal duration greater than 1 h (0, absence; 1, presence) and agonal risk factors. A 0 agonal factor reflects absence of either agonal factors and/or prolonged agonal duration. Note a significant difference in pH for BPD vs controls in analyses #1 and #2. In analysis #3 to remove the pH difference subjects below a pH of 6.875 were eliminated in mood disorder groups and controls, in total eliminating 10 subjects. The postmortem interval data are shown in methods, there were no group differences.
There were no control subjects with a positive family history for psychiatric disorders, whereas the patients with mood disorders had a significant history of mood or psychotic DSM-IV disorders in first-degree relatives. The subjects in this study with mood disorders were predominantly male.
Brain pH measurement
A piece of tissue (50–100 mg) from a frozen cerebellar cortical slice was placed in double-distilled water purified through a Nanopure Infinity water system to supply 18.2 MΩ-cm resistivity reagent grade water (Barnstead, Dubuque, IO, USA). The brain solution (10% weight/volume) was homogenized with 1.0 mm glass beads using a Bead-Beater (BioSpec Products, Bartlesville, OK, USA) for 60 s at 4°C, centrifuged at 5000 r.p.m. for 2 min at 4°C and quickly equilibrated to room temperature, and the pH measured with a Corning pH meter (Cypress, CA, USA). The refillable electrode (Beckman P/N 511082, Fullerton, CA, USA) which utilizes a 4 m KCl reference solution was calibrated with three standards of pH 4, 7 and 10 (Fisher Scientific, Tustin, CA, USA).
Total RNA extraction
The anterior cingulate cortex (AnCg; area 24), the dorsolateral prefrontal cortex (DLPFC; area 9 plus 46) and the cerebellar cortex (CB) were all dissected from the left hemisphere. The block-designated anterior cingulate was consistently taken from a site located ~1 cm posterior to the genu of the corpus callosum and extending for ~0.8 cm posterior to that point. Dorsoventrally, the anterior cingulate block included only cortex on the crown of the cingulate gyrus, excluding cortex in the callosal sulcus and in the ventral bank of the cingulate sulcus, and thus avoiding areas 32 and 33. Post hoc analysis of Nissl-stained sections from blocks of this type revealed the agranular cytoarchitecture typical of the anterior limbic area, area 24, as defined by Brodmann31 and Vogt et al.32 All cerebellar samples were taken from the lateral aspect of one cerebellar hemisphere and included lateral portions of the middle lobe, containing lateral parts of the superior and inferior semilunar lobules. The cortical and cerebellar samples were microdissected to remove all but a thin ribbon of underlying white matter and the predominant gray matter sample was used for RNA extractions. Total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA, USA), followed by cleaning up of the total RNA by passing over silica-based mini-spin columns (Qiagen RNeasy Mini Kit, Valencia, CA, USA). The total RNA samples were divided into three aliquots and sent to independent sites (AnCg and CB to University of California, Irvine; DLPFC and CB to University of California, Davis; AnCg and DLPFC to University of Michigan, Ann Arbor, USA) for microarray experiments. Total RNA samples were sent to Stanford and UCI for Q-PCR. Total RNA samples were analyzed on a 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA) for quantification of 28S and 18S ribosomal RNA peaks.
Oligonucleotide microarrays
The oligonucleotide microarray chip (HGU95Av2) experiments were carried out following the manufacturer's protocol (Affymetrix, Santa Clara, CA, USA) and the microarray procedures from our laboratories are described in recent publications.13,33
Microarray data analysis
Probe level model fit for condensation of affymetrix probe data
A robust probe level model (PLM) fits a linear model to Affymetrix microarray probe data and is freely available at http://www.bioconductor.org. A complete description of the PLM fit algorithm is available at http://www.stat.berkeley.edu/users/bolstad/Dissertation/Bolstad_2004_Dissertation.pdf. The software for the entire process of PLM fit extraction of signal intensity of each gene was implemented in our Consortium wide server and database using the AffyExtensions package (http://oz.berkeley.edu/users/bolstad/AffyExtensions/AffyExtensions.html), which required affy (version 2.0, available at http://www.bioconductor.org/). The PLM fit outputs are coefficients for the diagnosis effect that are estimated on the log2 scale and the associated s.e. The coefficients for mood group were compared to controls by a T statistic for ranking and selecting genes based upon the following formula: T = (Probe level model (PLM) Group Coefficient for Bipolar Disorder–PLM Group coefficient for Control)/((Standard Error Expression for Bipolar Disorder Group Coefficient)z2 + (Standard Error Expression for Control Group Coefficient)2)-1/2. The absolute values of T were rank-ordered, the top 5% of the ranked T were used in gene expression selection for pathway analysis. The PLMfit T statistic that we employ for ranking is comparable to a traditional Student's t-test in estimating the effect size. A fitPLM analysis is useful in the present experimental design involving duplicate chips performed on three groups of subjects across three brain regions. A separate analysis for each brain region analyzed was run. The grouping factors in PLMfit were diagnosis and replicate microarray chip (performed at independent site).
Cohort composition for microarray analysis
A control group analysis was conducted by splitting the controls into subgroups at a median pH of 6.87 and comparing the gene expression profile with PLMfit. This analysis was used to evaluate gene expression results for agonal–pH sensitive genes.
To evaluate overall effects of mood disorders on mitochondrion gene expression profile, three further microarray analyses were conducted (Table 1). The first analysis contained all subjects (n = 40). The second analysis also contained all subjects but controlled for pH by grouping the subjects with low or high pH (above and below pH 6.87). The third analysis was restricted to subjects above pH 6.87 without agonal factors.
In the first analysis, the pH was significantly increased in BPD (P = 0.03) and MDD (P = 0.046) groups compared to controls. Therefore, the second analysis used pH as a categorical factor (low and high) to eliminate pH effects in the analysis of diagnosis main effects and pH main effects (Table 1). The third mood disorder analysis used subjects with zero agonal factor and above median pH cases (Table 1) to evaluate the main diagnosis effect in subjects without agonal-pH-associated factors.
Other reports showed that pH is perhaps most strongly related to gene expression data.10,16 From our own pH measures of 98 brain samples (Vawter, MP, unpublished results), the relationship is minimally weak for PMI within the range of PMI that we are using (r = 0.0684; P = 0.5031). PMI is not universally related to gene expression.26,30,34–36 For extremely long PMI ( > 40 h) and without refrigeration of the brain during prolonged intervals, PMI could be important, although this condition does not apply to autopsied brain samples included in our microarray study. All three groups in the present study were matched for PMI (group mean PMI±s.d.: BP 16.6±7.5; MDD 26.4±6.3; Control 23.0±4.5; P > 0.1 t-test between BPD and MDD groups compared to control group). We did not further measure the effects of PMI on gene expression in the following analyses.
Unigene chip definition file
A custom Affymetrix chip definition was developed owing to the significant increase in EST, cDNA, and genomic sequence information since the design of the Affymetrix chip on the Unigene 95 build. A custom Affymetrix chip definition file based upon a more recent Unigene was developed37 and is available with updates at: http://brainarray.mhri.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp. The custom Unigene chip definition file results in 8846 probesets. In-house comparisons of two custom Unigene chip definition files to the Affymetrix U95Av2 chip definition file did not change the conclusions reached in this paper.
Gene classifications: mitochondrial, proteasome, chaperone, apoptosis and response to reactive oxygen
Five broad gene classifications were chosen a priori based upon reports of mitochondrial, proteasomal and heat-shock protein alterations of gene expression in mood disorders or schizophrenia.8,12,16,17,21 The apoptosis pathway was selected as a possible connection between these pathways and prior reports of apoptotic genes related to lithium treatment.38,39 Genes were extracted from the Top 5, 10, and 25% T ranks from each PLMfit microarray analyses in each brain region. The number of genes expected by chance was compared to the actual number of mitochondrial-related genes extracted. Fisher exact tests were calculated for gene classification enrichment.40 The five broad classes chosen for a priori tests were corrected for multiple testing of the Fisher exact test P-values.
Three Kyoto Encyclopedia of Genes and Genomes (KEGG), GO and GenMapp classifications were collapsed into one classification (oxidative phosphorylation (KEGG 00190, electron transport chain (Gen-MAPP) and mitochondria cellular localization (GO 5739). This resulted in 321 unique Unigene clusters represented by Affymetrix probesets on the HGU95Av2 chip. The proteasome KEGG pathway classification had 32 Unigene clusters represented on the Affymetrix U95Av2 chip. The chaperone classification was derived from the union of four Gene Ontology classes: chaperone activity (GO Molecular Function), chaperonin ATPase activity (GO Molecular Function), chaperonin containing T-complex (GO cellular component), and chaperonin-mediated tubulin folding (GO Biological Process). This resulted in 100 Unigene clusters for the condensed chaperone class represented on the Affymetrix U95Av2 chip.
Apoptotic-related genes were collapsed resulting in 273 probesets from Gene Ontology definitions that contained keywords of apoptosis and not already in the proteasome, chaperone or mitochondria classes. The parent reactive oxygen gene class from Gene Ontology (response to oxidative stress GO:0006979 category) contained 19 unigene clusters.
Functions of genes were annotated using Database for Annotation, Visualization and Integrated Discovery (DAVID) available at http://apps1.niaid.nih.gov/David/upload.asp.41 The EASE software application40 was obtained from the same website at NIH. Some distortion in the percent of transcripts that occur in a functional grouping can be due to the presence of multiple probes for the same gene on Affymetrix arrays. The EASE software corrects for multiple probes that are in same list to reduce a bias toward any gene with multiple probe sets.
Real time quantitative polymerase chain reaction
Real-time quantitative PCR (Q-PCR) with SybrGreen dye was used to replicate the microarray results as previously described42 on total RNA extracted from the DLPFC and anterior cingulate. Real-time quantitative Q-PCR was carried out in an Applied Biosystems 7000 sequence detection system (ABI, Foster City, CA, USA) according to the manufacturer's protocol for SybrGreen PCR using a 25 μl reaction volume and 5 μl of diluted cDNA template. A second site also ran Q-PCR reactions on separately synthesized cDNA on a Bio Rad I-cycler with a SybrGreen protocol. Samples were run twice and averaged or in some assays triplicate measurements were made in one run, and average Ct was calculated for each sample. The delta Ct calculation was used for relative fold change and a significance level of P < 0.05 (one-tailed t-test) was adopted as evidence of microarray validation since the direction was known a priori. For normalization PPIA was selected as a common reference gene for Q-PCR calculations and was stable between brain regions and between diagnostic groups. The primer sequences chosen for PCR of the cDNA for each gene is available upon request from the authors. Genes that showed evidence of any double peaks in the Q-PCR dissociation curves were not analyzed.
Mitochondrial DNA copy number
Genomic DNA samples were obtained from human postmortem brain tissue in the CB following Trizol extraction protocol for RNA and DNA (Invitrogen, Carlsbad, CA, USA) for 40. Primers were designed for both the mitochondrial DNA (mtDNA) and the nuclear DNA (nDNA). The primer sequence for the mtDNA control region was selected in the DLOOP region and did not contain a known single nucleotide polymorphism (SNP) according to NCBI's Entrez system. The nDNA primers were designed within the TATA box-binding protein (TBP-gU) region. Real-time quantitative PCR (Q-PCR) reactions was carried out with known amounts of DNA to obtain copy number for both nDNA and mtDNA using the ABI 7000 sequence detection system.
In situ hybridization
The LRPPRC (leucine-rich PPR-motif containing) gene was tested by quantitative in situ hybridization histochemistry (ISHH).43 Briefly, the right side of each brain was processed for ISHH. To generate subclones for riboprobe synthesis, the unique regions of LRPPRC from a human cDNA brain library (Edge-Biosystems, Gaithersburg, MD, USA) were amplified using PCR forward primer (5′-AAAGAATTCGGCTGTGACAACACTGAAAAC-3′) and reverse primer (5′-AAAGAATTCAGCCAAAGCAATGTTATTGATGA-3′). 35S-labeled sense and antisense riboprobes were prepared and hybridized to sections from the DLPFC, anterior cingulate cortex and CB using previously published method.43 LRPPRC probe sequence matched human LRPPRC (ENSG00000138095), and was complementary to a single exon. Hybridized sections were exposed to Kodak XAR film for 2–4 weeks before development of the film. Sense probes for LRPPRC were hybridized and exposed under similar conditions as the antisense riboprobe, and showed faint signal compared to the more robust antisense probe. The optical density (OD) was adjusted for radioactive standards (nCi/g) and then multiplied by the area that was quantified, which put the relative OD in a range of 20 000–100 000 arbitrary units.
Results
Removal of agonal-pH effects on gene expression
Agonal-pH effects were stringently analyzed to discover gene expression that is codirectional with mood disorder. Codirectional changes are defined as fold changes in control agonal-pH comparisons that are in the same direction as mood disorder comparisons. The direction for control agonal-pH is determined from the fold change of controls with no agonal factors and high pH above 6.87, compared to controls with agonal factors and pH below 6.87. The two control groups had a mean pH difference of 0.5 pH units (P = 0.00036).
In the control analysis of agonal-pH effects, five classifications of gene expression were examined for differential expression (Supplementary Table S1) in two cortical regions in this control analysis. Using a loose criterion of ±1.2-fold change, there were a total of 1609 genes dysregulated in Anterior Cingulate Cortex (AnCg) and 1778 genes dysregulated in DLPFC. The number of genes dysregulated in each classification ranged from 16.5 to 42.2% using a loose criterion of ±1.2-fold change. For example, in the mitochondrial classification, there were 321 genes of which 108 were dysregulated in the cingulate and 103 were dysregulated in the DLPFC with 88 of the genes in common between both cortical regions. This control analysis (Supplementary Table S1) shows that a majority of the differentially expressed mitochondrial, chaperone and proteasome genes are decreased with decreasing pH, whereas the majority of apoptotic and reactive oxygen stresses show increased expression in response to decreased pH, presumably related to hypoxia and prolonged agonal duration.
Considering all agonal-pH sensitive genes in the baseline control analysis, across the three brain regions there were 570 genes dysregulated in two or more brain regions that met a fold change criterion of ±1.25 and in the top 5% ranked T-values (Supplementary Table S2). Following over-representation analysis (EASE, NIH) and a Bonferroni correction for multiple testing, there were three major classes that were significant: energy pathways (hydrogen ion transporter, ATP biosynthesis, mitochondrion), ribonucleotide metabolism and synaptic transmission (Supplementary Tables S3A, B). Genes with strong agonal-pH effects have been implicated in neuropsychiatric disorders, especially related to synaptic neurotransmission gene category (AMPH; CRH; DLGAP1; DNM1; DTNA; GABRA1; GABRA5; GABRB3; GABRG2; GAD1; GAD2; GLRB; GPR; GPR51; GRIA1; GRIA3; GRIN2A; HTR2A; KCNQ2; LARGE; MAOA; NPTX1; NQO1; PMP22; SCN2B; SLC1A3; SNAP25; SST; SYN2; SYT1; SYT5). The complete listing of agonal-pH genes is available at http://pritzkerneuropsych.org/data/archive/File022206.aspx. The agonal-pH effect must be considered in expression analysis studies before reaching meaningful conclusions. This baseline control analysis is useful for considering individual genes that can be altered without neuropsychiatric diagnosis, and presumably due to factors related to the agonal-pH measures. The list of genes generated from the control analysis was used to compare against the final microarray analysis #3 (reported below) of mood disorder.
All subjects - microarray analysis #1
In this first analysis of mood disorders, the pH was significantly increased in BPD subjects compared to controls (P < 0.05), but this difference in pH was eliminated in the second and third microarray analysis. Owing to the significant difference in pH in BPD compared to controls (Table 1, Analysis #1), a subset of the mitochondrial pathway (hydrogen-ion transporter) was over-represented following Bonferroni correction (P < 0.004). This data shows that when there are agonalpH factor differences between the mood disorder and control groups, the hydrogen-ion transporter classification is significantly over-represented as also shown in the above baseline agonal-pH control analysis. Table 2 also showed that the number of differentially expressed mitochondrial genes in controls with a high brain pH compared to controls with low brain pH is approximately the same number of mitochondrial genes found in bipolar disorder with high pH compared to controls with high pH.
Table 2.
The frequency of top ranked T-values for nuclear encoded mitochondrial genes in each microarray analysis
| Brain Region | Top Rank%a | Factorial design of microarray analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| Microarray Analysis #1. 40 subjects |
Microarray Analysis #2. 40 subjects pH Category |
Microarray Analysis #3. 30 subjectsb |
||||||
| BPD | MDD | BPD | MDD | BPD-High vs Control-High | MDD-High vs Control-High | Control-High vs Control-Low | ||
| Number of Mitochondrial Genes | ||||||||
| AnCg | 5 | 36 | 35 | 29 | 25 | 29 (6)c | 22 (6)c | 25 |
| 10 | 61 | 61 | 53 | 50 | 48 (17) | 45 (16) | 55 | |
| 25 | 124 | 133 | 105 | 128 | 97 (5) | 106 (65) | 130 | |
| DLPFC | 5 | 37 | 20 | 25 | 20 | 30 (4) | 19 (5) | 24 |
| 10 | 66 | 44 | 53 | 34 | 56 (24) | 44 (16) | 56 | |
| 25 | 120 | 123 | 109 | 97 | 121 (74) | 105 (55) | 131 | |
| Cerebellum | 5 | 37 | 21 | 27 | 13 | 10 (2) | 6 (2) | 33 |
| 10 | 67 | 38 | 58 | 24 | 28 (7) | 23 (7) | 71 | |
| 25 | 134 | 90 | 132 | 69 | 64 (30) | 60 (29) | 135 | |
Top Rank % is the percentile of the top ranked probe selection criteria, e.g. 5% is the top 5% PLMfit T statistic.
Control-High, BPD-High and MDD-High have selection criteria of Agonal factor = 0, pH > 6.87. Control-Low are control subjects with selection criteria of pH <6.87 and with agonal factors.
Analysis #3 shows in parentheses the number of genes in common with the Control-High vs Control-Low analysis. There were a total of 321 mitochondrial Unigene clusters represented on HGU95Av2 chips.
All subjects categorized by low and high pH - microarray analysis #2
The next approach was to assign all control and mood subjects to subgroups with high and low pH (Table 1). The grouping of pH subjects by diagnosis in analysis #2 attenuated the number of differentially expressed nDNA-encoded mitochondrial genes in mood disorder in the cortical regions (Table 2), compared to the first analysis. In analysis #2 of BPD DLPFC, there was a reduction by seven of 36 mitochondrial genes found in the top 5% and the hydrogen-ion transporter was not significantly over-represented category in contrast to analysis #1.
Correlation analyses of mitochondrial-related genes with pH and age
The correlations of pH and age to individual gene expression for 321 nDNA mitochondrial genes were calculated in three different brain regions. There were 171–201 genes that significantly correlated (r = > ±0.361, P = 0.05, df = 28) with pH depending on the individual brain region (Supplementary Table S4). A smaller number of mitochondrial genes (29–66) correlated with age (Supplementary Table S4, S5; Supplementary Figure 1) depending on the brain region. The overall correlation of age and pH was not significant (r = 0.17, P = 0.35). However, when only subjects without agonal factor and pH > 6.87 were used for correlation analyses, the number of significant correlations for pH and mitochondrial gene expression were reduced from 201 genes to four genes (Supplementary Table S4) in AnCg, and from 189 genes to six genes in the DLPFC. Thus, eliminating all subjects with agonal-pH factors substantially decreased the number of significant mitochondrial gene expression correlations by decreasing the range of agonal-pH effects and decreasing the number of subjects. This restricted sample (pH > 6.87 and zero agonal factors) was used in the final microarray analysis #3, and may be more indicative of mood disorder effects that are not dependent on pH-agonal state.
Subjects with a reduced range of agonal-pH factors - microarray analysis #3
We used the same groups with reduced correlations to study which genes might be impacted by mood disorder comparisons. We examined Control-High (sudden death and pH > 6.87) vs BPD-High or MDD-High (sudden death and pH > 6.87) groups for nDNA mitochondrial-encoded gene expression. There was no difference in mean pH amongst the three groups. From the results of these mood disorder comparisons, the genes from the baseline control analysis of agonal-pH effects were contrasted. Three categories of genes emerged: (a) genes that showed no sensitivity to pH but were differentially expressed in bipolar disorder (BPD) or major depressive disorder (MDD); (b) genes that were altered by low pH in one direction and altered in mood disorder in the opposite direction and (c) genes with agonal-pH sensitivity that display codirectional changes in mood disorder. The genes from categories (a) and (b) are shown in Table 3 for microarray results, and genes selected for Q-PCR are shown in Table 4. The codirectional genes for category (c) are shown in Tables S6–S10 for each classification and summarized in Table S11.
Table 3.
Microarray analysis of mitochondrial, chaperone, apoptotic and proteasome genes
| Gene Symbol | Pathway | Anterior Cingulate Cortex |
Dorsolateral Prefronal Cortex |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FC BPD | FC MDD | FC Control | T BPD | T MDD | T Control | FC BPD | FC MDD | FC Control | T BPD | T MDD | T Control | ||
| HSPA2 | Chaperone | 1.36 | 0.68 | 0.69 | 9.00 | –11.58 | –12.22 | 1.21 | 0.69 | 1.01 | 5.95 | –12.25 | 0.36 |
| SPP1 | Apoptosis | 1.28 | 0.84 | 0.36 | 7.67 | –5.88 | –35.42 | 1.32 | 0.86 | 0.70 | 8.49 | –5.10 | –12.35 |
| TM4SF10 | Apoptosis | 1.27 | 0.86 | 0.75 | 7.34 | –4.73 | –10.03 | 1.48 | 0.92 | 0.69 | 10.52 | –2.30 | –10.96 |
| CAT | Oxidative Stress | 1.22 | 0.89 | 0.68 | 6.28 | –3.94 | –13.28 | 1.22 | 0.85 | 0.78 | 6.14 | –4.94 | –8.30 |
| S100B | Apoptosis | 1.21 | 0.94 | 0.68 | 6.39 | –2.25 | –14.39 | 1.19 | 0.89 | 0.75 | 5.16 | –3.57 | –9.70 |
| NR4A1 | Mitochondria | 0.83 | 0.86 | 1.17 | –9.63 | –8.30 | 9.19 | 0.83 | 0.90 | 1.15 | 8.36 | –5.21 | 7.06 |
| BZRAP1 | Mitochondria | 0.82 | 1.02 | 1.16 | –7.05 | 0.68 | 6.01 | 1.00 | 0.96 | 1.21 | 0.10 | –1.13 | 5.47 |
| GSK3B | Apoptosis | 0.76 | 1.03 | 1.20 | –9.16 | 0.94 | 7.11 | 0.93 | 1.00 | 1.05 | –2.43 | 0.02 | 1.76 |
| COX7A1 | Mitochondria | 0.73 | 1.15 | 1.36 | –7.65 | 3.54 | 8.57 | 0.94 | 1.08 | 1.16 | –1.73 | 2.08 | 4.48 |
| UQCRB | Mitochondria | 1.06 | 1.20 | 0.98 | 1.64 | 5.37 | –0.58 | 1.15 | 1.09 | 1.14 | 3.48 | 2.16 | 3.62 |
| DUSP1 | Oxidative Stress | 0.87 | 0.74 | 1.10 | –4.00 | –8.85 | 2.92 | 0.84 | 0.83 | 1.13 | –4.06 | –4.54 | 3.31 |
| DUSP6 | Apoptosis | 0.84 | 0.70 | 1.59 | –4.15 | –8.89 | 12.26 | 0.96 | 0.77 | 1.77 | –0.93 | –5.46 | 13.19 |
| TM4SF10 | Apoptosis | 1.27 | 0.86 | 0.75 | 7.34 | –4.73 | –10.03 | 1.48 | 0.92 | 0.69 | 10.52 | –2.30 | –10.96 |
| ATP6V0E | Lysosyme | 1.15 | 0.88 | 0.59 | 2.89 | –2.87 | –12.33 | 1.36 | 0.90 | 0.63 | 6.10 | –2.23 | –10.14 |
| GLUL | Mitochondria | 1.00 | 0.67 | 0.78 | –0.07 | –7.65 | –5.22 | 1.33 | 0.78 | 0.70 | 4.96 | –4.42 | –7.06 |
| SPP1 | Apoptosis | 1.28 | 0.84 | 0.36 | 7.67 | –5.88 | –35.42 | 1.32 | 0.86 | 0.70 | 8.49 | –5.10 | –12.35 |
| APG-1 | Chaperone | 0.98 | 1.05 | 1.06 | –0.22 | 0.64 | 0.92 | 1.26 | 1.19 | 0.95 | 2.81 | 2.20 | –0.77 |
| HSPA5 | Chaperone | 1.07 | 0.98 | 0.94 | 1.78 | –0.42 | –1.76 | 1.25 | 1.19 | 1.00 | 5.09 | 4.08 | 0.09 |
| GATM | Mitochondria | 1.08 | 0.80 | 0.61 | 2.62 | –7.64 | –18.24 | 1.23 | 0.78 | 0.79 | 7.34 | –8.90 | –9.41 |
| CAT | Oxidative Stress | 1.22 | 0.89 | 0.68 | 6.28 | –3.94 | –13.28 | 1.22 | 0.85 | 0.78 | 6.14 | –4.94 | –8.30 |
| HADHB | Mitochondria | 1.11 | 0.88 | 0.71 | 3.71 | –5.10 | –14.11 | 1.22 | 0.87 | 0.74 | 6.30 | –4.74 | –10.47 |
| CCT3 | Chaperone | 1.15 | 1.07 | 0.96 | 4.98 | 2.43 | –1.67 | 1.22 | 1.04 | 1.03 | 5.97 | 1.34 | 0.96 |
| DAD1 | Apoptosis | 1.13 | 1.00 | 0.88 | 4.36 | 0.15 | –5.31 | 1.21 | 1.00 | 0.93 | 6.31 | 0.05 | –2.86 |
| HSPA2 | Chaperone | 1.36 | 0.68 | 0.69 | 9.00 | –11.58 | –12.22 | 1.21 | 0.69 | 1.01 | 5.95 | –12.25 | 0.36 |
| NR4A1 | Mitochondria | 0.83 | 0.86 | 1.17 | –9.63 | –8.30 | 9.19 | 0.83 | 0.90 | 1.15 | –8.36 | –5.21 | 7.06 |
| NAPG | Mitochondria | 0.94 | 1.07 | 1.69 | –1.32 | 1.53 | 12.83 | 0.83 | 1.04 | 1.83 | –3.06 | 0.60 | 10.97 |
| COX6A1 | Mitochondria | 0.85 | 0.99 | 1.12 | –4.35 | –0.35 | 3.34 | 0.82 | 0.96 | 1.07 | –4.77 | –1.10 | 1.82 |
| MAPK1 | Apoptosis | 1.04 | 1.04 | 1.39 | 0.94 | 0.91 | 8.60 | 0.79 | 1.07 | 1.46 | –3.99 | 1.14 | 7.17 |
| STIP1 | Chaperone | 1.17 | 1.16 | 0.97 | 4.50 | 4.51 | –0.86 | 1.06 | 1.23 | 1.00 | 1.49 | 5.06 | 0.06 |
| DUSP1 | Oxidative Stress | 0.87 | 0.74 | 1.10 | –4.00 | –8.85 | 2.92 | 0.84 | 0.83 | 1.13 | –4.06 | –4.54 | 3.31 |
| SLC25A13 | Mitochondria | 1.06 | 0.91 | 0.97 | 2.33 | –3.94 | –1.55 | 1.03 | 0.83 | 1.03 | 1.11 | –6.61 | 1.10 |
| SST | Apoptosis | 1.16 | 0.88 | 3.34 | 3.92 | –3.70 | 36.03 | 0.85 | 0.78 | 3.71 | 4.96 | –7.89 | 45.04 |
| DUSP6 | Apoptosis | 0.84 | 0.70 | 1.59 | –4.15 | –8.89 | 12.26 | 0.96 | 0.77 | 1.77 | –0.93 | –5.46 | 13.19 |
| USP9Y | 26S proteasome | 1.05 | 0.85 | 1.91 | 0.89 | –3.32 | 13.69 | 1.17 | 0.75 | 2.20 | 2.39 | –4.51 | 13.38 |
| HSPA2 | Chaperone | 1.36 | 0.68 | 0.69 | 9.00 | –11.58 | –12.22 | 1.21 | 0.69 | 1.01 | 5.95 | –12.25 | 0.36 |
| SEMA6A | Apoptosis | 0.84 | 0.74 | 0.68 | –4.41 | –8.25 | –11.00 | 0.92 | 0.62 | 1.10 | –1.70 | –10.66 | 2.29 |
Abbreviations: Bipolar Disorder; FC, Fold change; BPD, MDD, Major Depressive Disorder, T, PLMfit T values.
Three categories of genes emerged in mood disorder comparsion to controls: (a) genes that showed no sensitivity to pH but were differentially expressed in bipolar disorder (BPD) or major depressive disorder (MDD); (b) genes that were altered by low pH in one direction and altered in mood disorder in the opposite direction and (c) genes with agonal-pH sensitivity that display codirectional changes in mood disorder. The genes from categories (a) and (b) are shown in this table, the category (c) are in Tables S6–11.
Table 4.
Real-time Q-PCR validation of microarray results for selected nDNA mitochondrial-related candidate genes for mood disorders in two cortical regions. The controls and mood disorder subjects compared by Q-PCR and microarray are restricted to cases without agonal factors and pH > 6.87. Genes were selected from three categories
| Gene symbolabc | Cellular localization | Brain Region | Microarray fold change (BPD vs Control) | QPCR fold change (BPD vs Control) | Microarray fold change (MDD vs Control) | QPCR fold Change (MDD vs control) |
|---|---|---|---|---|---|---|
| HSPA2a | Chaperone, function in apoptosis | DLPFC | 1.21 | 1.81d | 0.69 | 0.77d |
| NR4A1b | Translocates from nucleus and mitochondria | DLPFC | 0.83 | 0.58d | 0.90 | 0.51d |
| ATP5A1c | Complex V, mitochondria | DLPFC | 1.20 | 1.58d | 1.15 | 1.14 |
| ATP6V0Bc | Complex V, lysosomal | DLPFC | 1.18 | 1.01 | 1.07 | 1.48d |
| ATP6VIE1c | Complex V, lysosomal | DLPFC | 1.14 | 1.69d | 1.08 | 0.94 |
| IDH3Bc | Mitochondria Matrix | DLPFC | 1.15 | 1.01 | 1.08 | 1.18d |
| LRPPRCc | Binds to mitochondrial and nuclear RNAs | DLPFC | 1.22 | 1.62d | 1.16 | 1.80d |
| MRPS12c | Ribosomal protein, mitochondrial | DLPFC | 1.09 | 1.14 | 1.11 | 1.27d |
| NDUFV1c | Complex I, mitochondria | DLPFC | 1.12 | 0.99 | 1.10 | 1.43d |
| PSEN1c | Proapoptotic mitochondrial protein | DLPFC | 1.08 | 1.30d | 0.88 | 0.83 |
| HSPA2b | Chaperone, function in apoptosis | AnCg | 1.36 | 2.55d | 0.68 | 0.70 |
| NR4A1b | Translocates from nucleus to mitochondria | AnCg | 0.83 | 0.59d | 0.86 | 0.48d |
| NDUFV1c | Complex I, mitochondria | AnCg | 1.17 | 1.48 | 1.15 | 1.11 |
| IDH3Bc | Mitochondria Matrix | AnCg | 1.14 | 1.00 | 1.10 | 0.99 |
| LRPPRCc | Binds to mitochondrial and nuclear RNAs | AnCg | 1.20 | 1.08 | 1.08 | 1.02 |
| PSEN1c | Proapoptotic mitochondrial protein | AnCg | 1.11 | 1.46d | 0.84 | 1.09 |
| PSMB1c | Proteasome | AnCg | 1.27 | 1.61d | 1.02 | 1.16 |
| PSMD8c | Proteasome | AnCg | 1.15 | 1.35d | 1.10 | 1.23d |
These gene symbols represent categories (a), (b) and (c) genes. The details for the genes (a) and (b) are shown in Table 3. Category (c) genes that show codirectional fold changes in mood disorder and agonal-pH controls are shown in Supplementary Tables S6-S11.
These gene symbols represent categories (a), (b) and (c) genes. The details for the genes (a) and (b) are shown in Table 3. Category (c) genes that show codirectional fold changes in mood disorder and agonal-pH controls are shown in Supplementary Tables S6-S11.
These gene symbols represent categories (a), (b) and (c) genes. The details for the genes (a) and (b) are shown in Table 3. Category (c) genes that show codirectional fold changes in mood disorder and agonal-pH controls are shown in Supplementary Tables S6-S11.
Represents a significant difference between mood disorder and control P< 0.05, one-tailed t-test.
There were nine mitochondrial gene classification differentially expressed for BPD in categories (a and b), that is, not differentially expressed in control analysis, or expressed in the completely opposite direction: NR4A1, BZRAP1, COX7A1, ATP6VOE, GLUL, GATM, HADHB, NAPG and COX6A1. There were ~20 genes dysregulated in BPD or MDD in categories (a and b) in the remaining four classifications (Table 3; Supplementary Figure 2). Seven of the genes that belong to category a or b were altered in two cortical regions. Five nDNA mitochondrial-related genes were differentially expressed in BPD across two cortical regions (HSPA2, SPP1, TM4SF10, CAT, NR4A1). In MDD, both DUSP1 and DUSP6 were decreased in two cortical regions. The genes in Table 3 are not key players in mitochondrial citric acid cycle or electron transport and importantly are in contrast to the mitochondrial genes that have been reported by other groups as dysregulated in neuropsychiatric disorders.10,11,14,44 This is one of the main results of this paper, that is, the usual electron transport genes are dysregulated by pH, but not by mood disorder, and those genes are shown in our control analysis of agonal-pH effects (Supplementary Tables S1, S2).
Category c genes that showed codirectional changes with agonal pH and mood disorders might be viable candidates but more difficult to partition effects to either cause. To purify the potential list of category c genes (Supplementary Tables S6–S11) toward mood disorders, three filters were used: Top 5% T-values from microarray analysis #3 of mood disorder, fold change > 1.2 in mood disorder and non-significant correlation with pH. The resulting genes may contribute to defects in mitochondrial-related functions. Genes in category c can be viewed as a continuum from categories a and b. Several genes in category c (PCCB, TRAI and KARS) are borderline to category a or b, that is, these genes show robust mood disorder effect larger than the agonal-pH effects.
Antidepressant and mood stabilizer effects on mitochondrial related genes
The gene expression alterations in mitochondrial-related genes shown for BPD and MDD prompted the question whether these specific gene expressions might be consistent with antidepressant and mood stabilizer prescriptions, and alcohol use.
Lithium
It was documented from medical records that lithium was discontinued in all BPD subjects ranging from days to months before death. Further, lithium was not detectable in brain tissue assays with a standard clinical assay. Acute lithium treatment per se would not be specifically related to mitochondrial gene expression patterns, although a long-term effect cannot be ruled out. The BPD group was split into two groups according to recent lithium exposure: BPD Lithium prescription at time the of death (BPD-Li) and BPD no lithium prescription at time the of death (BPD-NoLi). In the DLPFC, there were seven mitochondrial-related genes in category a or b that showed a stronger alteration in the BPD-NoLi group compared to the BPD-Li group (Table S12). By definition, these genes also either showed no dysregulation owing to agonal-pH effect in controls or were in the opposite direction compared to the no lithium BPD group. Thus, subjects that were not prescribed lithium showed several dysregulated mitochondrial genes that did not have codirectional agonal effects.
SSRI
MDD subjects were split into two groups: prescribed SSRI at the time of death (MDD-SSRI) and MDD subjects not prescribed SSRI at the time of death (MDD-NoSSRI). For category a and b genes, SSRI status did not affect HSPA2, SLC25A13 and SST. DUSP1 and USP9Y were significantly decreased in MDD-NoSSRI groups but not in MDD-SSRI. These five genes by definition were not found in agonal comparison in controls (Supplementary Table S13).
Alcohol
The effect of alcohol could not be systematically evaluated in both mood groups in the present data set as the MDD patients predominantly used alcohol at the time of death, whereas controls and BPD subjects did not use alcohol at the time of death. The blood alcohol levels when obtained showed a positive result in only two of seven MDD patients, whereas all controls and BPD subjects showed a 0.0% BAL. Thus, acute alcohol intoxication would not be responsible for mitochondrial gene expression differences seen in BPD. However, in MDD, rates of lifetime alcohol abuse in MDD subjects were 67% compared to 0% in controls, and 16% in BPD. Thus chronic alcohol usage in MDD subjects might account for some gene expression effects.
Pathway analysis related to mitochondria, proteasome, chaperone, apoptosis and reactive oxygen gene classifications
Pathway analysis was conducted before and after removal of agonal-pH sensitive genes in controls from the mood disorder list. In general, after removal of agonal-pH genes from each analysis, the over-representation in each significant pathway was no longer significant. A subset of the mitochondrial gene classification (hydrogen ion transporter) was initially over-represented in BPD, but not after removal of agonal-pH genes (Supplementary Table S4).
Similarly, before removal of agonal effect genes, nine of the 32 proteasome genes were found in the top 5% of differentially expressed genes in BPD in the Anterior Cingulate cortex (Fishers exact test, P = 0.00005, corrected for multiple testing) and predominantly were upregulated. After removal of 10 proteasome genes found in the control comparison of agonal-pH factors, this classification was not significant.
Chaperone genes in the BPD microarray analysis of AnCg showed 12 of 100 chaperone genes in the Top 5% (P = 0.02), but this was not significant after removal of agonal-pH chaperone genes. The top chaperone gene dysregulated was HSPA2, which was increased in BPD, and decreased in agonal-pH control comparisons. For chaperone genes in the top 5% in DLPFC, 8/11 genes were upregulated in BPD and 8/11 genes were downregulated for MDD. This opposite direction of change suggests differences in regulation of chaperone-related processes in mood disorders. Genes related to apoptosis or response to oxidative stress categories were not over-represented in BPD or MDD.
The pathway analysis results shown for the baseline control subjects (Supplementary Tables 1, 2, 3A, 3B) demonstrated that a majority of nDNA encoded genes on the microarray related to mitochondria, proteasome and chaperone were decreased in expression in response to low pH and agonal factors. However, in the apoptosis and reactive oxygen stress pathways, a slight majority of genes were actually increased in expression in response to decreased pH and agonal factors. These over-represented pathways showed different responses in fold change direction to agonal-pH factors.
Validation of candidate nDNA-encoded mitochondrial genes by Q-PCR and ISH
The Q-PCR results showed two proteasome genes, PSMB1 and PSMD8, were increased in BPD AnCg by microarray and were found to be dysregulated by Q-PCR (Table 4). HSPA2 was also increased in BPD AnCg by microarray and Q-PCR. The validation of nDNA-encoded mitochondrial genes by Q-PCR (Table 4) for microarray fold changes ±1.2 was seen in four (ATP5A1, LRPPRC, HSPA2, PSMB1) of five gene comparisons. The Q-PCR validation rate of smaller microarray fold changes < 1.2 was less consistent, although a particular gene was in the Top 5% of dysregulated genes by PLMfit T-values, the algorithm condensed the microarray fold change by about 50% according to the Q-PCR results. The small fold changes are also likely due to careful matching of agonal factors, PMI and pH between groups, thus we are unlikely to find large differences by microarray and Q-PCR. For some genes we found larger Q-PCR increases in gene expression than the microarray fold changes. For example, in Table 4, HSPA2 showed a 1.2 microarray fold change in BPD, and a 1.8-fold change in QPCR. This could suggest deterioration of RNA; however, we have used several housekeeping genes that would also be expected to show such parallel activity to normalize the data. The RNA integrity of the samples were assessed at the same time as running the chips, and microarray chips that did not show low 3′/5′ ratios for two housekeeping genes were eliminated from analysis. If we had left in samples with low pH, then we would perhaps have seen extremely large fold changes by microarray and Q-PCR. Theoretically, RNA slowly degrades during storage; however, we ran each microarray twice in different laboratories, and have averaged the results, thus a loss of RNA integrity is not an explanation for low fold changes observed between mood disorder and controls as the degradation rate would be similar. The fold changes reported in this paper are similar to another report44 using different samples for BPD and different analysis techniques of Affymetrix U95A arrays. These investigators reported significant fold changes (range 1.09–1.39) in the same range of the fold changes, we have reported.44
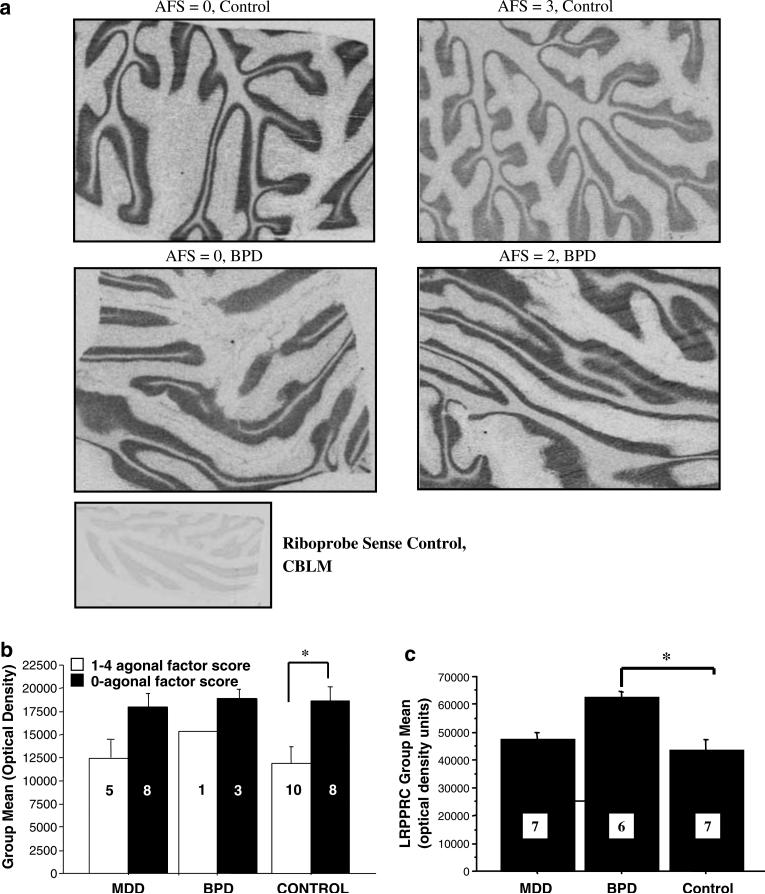
Leucine-rich PPR-motif containing was also co-directionally changed in BPD and agonal-pH by microarray and the changes were validated by Q-PCR. Leucine-rich PPR-motif containing was evaluated with in situ hybridization histochemistry (ISHH). A clear decrease in LRPPRC mRNA expression by 33% in CB was seen in the control agonal factor group compared to the control group with zero agonal factors (P = 0.011, Figure 1a and b). Bipolar disorder cases with zero agonal factors were compared to control cases with zero agonal factors by ISHH for LRPPRC. The LRPPRC ISHH result showed a 19% increase in BPD compared to controls (P = 0.011, Figure 1c). This result supports that LRPPRC was impacted by agonal factors (33%) and less by diagnosis (19%), which underscores the need to account for cases with agonal-pH factors in candidate mitochondrial gene search.
Figure 1.
In situ hybridization for leucine-rich PPR-motif containing (LRPPRC) mRNA in cerebellum. Representative autoradiographs show the pattern of In situ hybridization labeling across three different agonal factor scores. Several poor quality sections were eliminated from analysis (Figures 1b and 1c). (a) ISHH images of LRPPRC expression in control and bipolar disorder (BPD). (b) Controls with agonal factors showed a 36% reduction in LRPPRC compared to controls with zero agonal factors (P = 0.011) in cerebellum and a similar effect was seen across the cortical regions. The optical density (OD) was adjusted for radioactive standards (nCi/g) included on the film and then multiplied by the area that was quantified transforming the relative OD to a range of 20 000–100 000 arbitrary units. The numbers of subjects used for quantitation are shown in each bar. (c) The BPD cases without agonal factors show increased LRPPRC by ISHH compared to controls without agonal factors (P = 0.001) and compared to major depressive disorder (MDD) subjects (P = 0.02) without agonal factors in the DLPFC. The numbers of subjects are shown in each bar.
Mt DNA copy number and gene expressions are dysregulated in agonal-pH comparisons
MtDNA-encoded genes might be dysregulated in mood disorder in conjunction with alterations of specific nDNA-encoded genes in the DLPFC and AnCg. For example, cytochrome c oxidase subunit I (COX1) an mtDNA transcript interacts with LRPPRC.45 COX1 and mtDNA-encoded gene ATP synthase F0 subunit 6 (ATP6) were assayed by real-time Q-PCR (Table 5). MtDNA-encoded transcripts are not included on the Affymetrix U95Av2 arrays, so these transcripts were assayed by real-time Q-PCR. COX1 was decreased in the controls without agonal factors, compared to controls with agonal factor (P < 0.05, t-test) by a fold change of 0.61 in the anterior cingulate and the decrease of 0.54-fold change was also significant in the DLPFC. Similar decreases were seen for agonal factor effects on ATP6 in the AnCg and DLPFC (P < 0.05, t-test for agonal vs no agonal controls). This strongly suggests that agonal-pH factors increase mtDNA-encoded gene expression. This effect is opposite in direction in nDNA-encoded mitochondrial gene expression.
Table 5.
Mitochondrial DNA (mtDNA) encoded genes were analyzed by real-time Q-PCR for differential expression in BPD and MDD compared to controls
| MtDNA Encoded Gene | Gene | Complex | Brain Region |
QPCR fold change |
|
|---|---|---|---|---|---|
| BPD | MDD | ||||
| ATP6/ATP8 | ATP synthase F0 subunit 6 | V | DLPFC | 0.85 | 0.88 |
| ATP synthase F0 subunit 6 | V | AnCg | 0.58a | 0.72a | |
| COX1 | cytochrome c oxidase subunit I | IV | DLPFC | 0.69 | 0.88 |
| COX1 | cytochrome c oxidase subunit I | IV | AnCg | 0.52a | 0.81 |
| COX2 | cytochrome c oxidase subunit II | IV | DLPFC | 0.84 | 1.08 |
| COX2 | cytochrome c oxidase subunit II | IV | AnCg | 0.75b | 0.89 |
| ND1 | NADH dehydrogenase subunit 1 | I | DLPFC | 0.56b | 1.01 |
| ND1 | NADH dehydrogenase subunit 1 | I | AnCg | 0.86 | 0.96 |
| ND2 | NADH dehydrogenase subunit 2 | I | DLPFC | 0.54 | 0.92 |
| ND3 | NADH dehydrogenase subunit 3 | I | DLPFC | 1.07 | 0.86 |
| ND3 | NADH dehydrogenase subunit 3 | I | AnCg | 0.75b | 0.92 |
Two mtDNA encoded genes, ATP6 and COX1, showed a significant decrease by Q-PCR in mood disorders and both genes also showed decreased expression in controls without agonal factors compared to controls with agonal factors. Thus in mtDNA encoded genes tested, it appears that codirectional changes are seen in mood disorder and subjects without agonal factors. For the t-test comparisons and fold changes shown in this table, zero agonal factor cases with pH >6.87 were used in the analysis similar to microarray analysis #3. The data in this table refers to mitochondrial DNA encoded gene expression and not to the microarray nuclear encoded DNA mitochondrial genes shown in Table 3 and Table 4. Thus, mtDNA genes react to agonal factors with an increased gene expression, while nDNA encoded mitochondrial genes show a general decrease in expression in the same subjects.
Significant at P < 0.05 two-tailed.
trend P < 0.1 two-tailed.
The expressions of both mtDNA-encoded ATP6 and COX1 were significantly decreased in the AnCg of BPD-High compared to Controls-High, both groups had no agonal factors, and pH above 6.87. The MDD group showed a significant decrease in COX1 in AnCg. There were also trends for decreased expressions of COX2, ND1 and ND3 in BPD (Table 5). The general trend for mtDNA-encoded genes was a decrease in both BPD and MDD across two cortical regions. Thus, the change in MDD and BPD for COX1 and ATP6 are codirectional with the change induced by agonal factors.
Mitochondrial copy number alterations and agonal duration
The assessment of mtDNA-encoded gene expression for COX1 and other transcripts might be influenced by the number of mitochondria present in brain tissue as a result of agonal-pH factors. The impact of agonal factors on mtDNA copy number was addressed using all 40 subjects’ mtDNA copy numbers that were individually normalized to the nDNA copy numbers. Subjects were divided into rapid and prolonged death groups, that is, no agonal factor compared to agonal factors. The mtDNA copy number data appeared to be non-normally distributed among the 40 cerebellum samples by non-parametric Kolomogorov Smirnov test (P = 0.01), therefore, a non-parametric median test for differences in agonal duration and mtDNA copy number ratio was conducted with a version of the Kruskal–Wallis ANOVA (Statistica vs 5.0, 1999). The median mtDNA copy number was significantly increased in prolonged agonal duration cases (62.7 median mitochondrial copy number) compared to agonal duration less than 1 h, (38.3 median mitochondrial copy number; P = 0.018, Supplementary Figure S3). Agonal-pH factors appear to modulate the number of mtDNA copy number and mtDNA and nDNA gene expression. It is important to control for this effect in microarray, Q-PCR and ISH results.
The Wester, Hardy and Johnston agonal duration scales were each correlated with the mtDNA copy number (Spearman r = 0. 34–0.35, all P-values < 0.032) indicating in all 40 subjects that as agonal duration increased from rapid to prolonged there was an increase in mtDNA copy number. This result demonstrates that elimination of subjects with agonal factors can minimize the impact on mitochondrial findings due to agonal-pH differences. The BPD group without agonal factors compared to controls without agonal factors showed a non-significant increase in mtDNA copy number (P = 0.06). This slight tilting of mtDNA copy number in the bipolar disorder cases might indicate that the bipolar group showed possibly less residual agonal factors than controls. Any residual imbalances could theoretically affect both nDNA and mtDNA gene expression.
DISCUSSION
The pathophysiology of depression and mania is largely unknown leaving open a question of which cellular pathway(s) to investigate.46 This study tested if large and broad alterations of nDNA and mtDNA transcripts occur in mood disorders. The largest fold changes of differentially expressed mitochondrial-related genes were found in comparisons of controls subjects with and without agonal factors. After removing subjects with agonal-pH factors, there were reduced numbers of significant correlations of gene expression with pH, and decreased numbers of significant differentially expressed mitochondrial genes in mood disorders. This suggests that balancing agonal factors will be critical to accurate measurements of dysregulated genes owing solely to mood disorder.
Three categories of differential gene expression in mood disorders were referenced to agonal-pH effects: (a) genes that were not changed by agonal-pH comparisons, (b) genes that showed an opposite change in direction to agonal-pH effects and (c) genes that showed codirectional changes with agonal-pH effects. The mitochondrial and related pathways are not over-represented in these three categories of results. Before filtering genes by agonal-pH-sensitive genes and considering cases without agonal factors, three pathways were significantly over-represented in BPD: mitochondrial, chaperone and proteasome pathways. However, these pathways showed a decrease in gene expression in controls with agonal-pH factors, whereas the apoptotic and reactive oxygen pathways showed prominent increases in gene expression in response to agonal-pH factors. Thus, low pH (prolonged agonal duration) induced opposite changes in the nDNA-encoded pathways selected for analysis in this study. In mtDNA-encoded genes, low pH (prolonged agonal duration) increased expression of several transcripts such as COX1 and ATP6.
Over-representation of mitochondrial genes that were previously reported10,11,14,16 could be possibly due to agonal-pH effects as demonstrated in the present data. Pathways associated with the electron transport chain and proteasome were previously reported to be altered in BPD.11 We show alterations in the same pathways in BPD before applying a stringent criterion. The direction of change for the majority of genes is predominantly codirectional with agonal-pH effects. The simplest explanation is if there is some imbalance in the agonal factors across case-controls, then an agonal-pH effect will likely be detected in the mitochondrial and proteasome pathways.
A recent report of BPD that focused on a broad set of mitochondrial-related gene expression asked whether pH and to a lesser extent medications influence mitochondrial gene expression.10 In that study10 the pH of the BPD group was decreased compared to the control group (pH 6.44 vs 6.60, P < 0.05). The fold change direction of mitochondrial genes was predominantly decreased in BPD compared to controls in the DLPFC, the number of mitochondrial genes showing a decreased expression in BPD was 96 while six genes were increased. Interestingly, Iwamoto et al.10 reported a separate control analysis, in which there were 144 mitochondrial genes (219 probesets) dysregulated between low pH control and high pH control groups. The ratio of downregulated:upregulated was 3:1 comparing low pH to high pH controls. Agreement in fold change consistency between our present agonal-pH control analysis and the Iwamoto et al.10 analysis was high (95.8%, 138/144 genes agreed in direction). In the Iwamoto study, 31% of genes (45) were altered in the control pH analysis and were codirectional with significantly dysregulated genes in either bipolar disorder or schizophrenia. This is consistent with our findings, although we find a larger overlap of codirectional genes with agonal-pH and mood, perhaps owing to using a larger difference in pH among agonal-pH control analysis (delta = 0.47 pH units) than the pH difference in controls used by Iwamoto et al.10 Fold change consistency between DLPFC mitochondrial gene expression in BPD in the Iwamoto et al., report shows that 35 mitochondrial genes agree in direction whereas 75 genes disagree compared to the present study. The genes that show an opposite fold change between both studies might be due to differences in the medication status of the BPD patients, in agonal-pH effects in BPD or controls, or differences due to normalization of results. When a select group of medication-free BPD patients were compared to controls,10 eight of nine genes showed increased expression in agreement with the present study. However, in this subgroup analysis,10 the pH of the drug-free BPD group was significantly increased compared to controls, which could explain the increased gene expression.
The three expression categories described can be a starting place for further genetic, behavioral and in vitro validation studies. The non-codirectional category represents genes that might harbor genetic regulatory variation in mood disorder. In this category, there is no detectable impact of agonal-pH factors on gene expression, and only changes are seen in mood disorder, thus making an imbalance in case–control agonal-pH effects less likely. The potentially stronger effect (agonal-pH) could swamp the signal from mood disorder, however, by considering an opposite fold change direction that effect is plausibly reduced. This leaves medication, patho-physiology, regulatory variations or interactions among these as likely causes of gene expression changes in this category. Examples of genes in this category are HSPA2, NR4A1, DUSP1, DUSP6 and TM4SF10. They are each opposite to agonal-pH effects and dysregulated in two brain regions lending reproducibility to this category. The third category shows the largest number of genes (Supplementary Tables S6-S11) and although agonal-pH subjects were removed from the mood analysis, the changes in genes in this category are codirectional with agonalpH controls. These genes could be involved with mood disorder as the correlations of pH with gene expression are substantially reduced. In this third category, the genes show codirectional agonal-pH effects and mood disorder effects: HSPA8, LRPPRC, CCT2, IMM2, PRDX2 are examples of nDNA-dysregulated genes. LRPPRC was found to be dysregulated in BPD by microarray, QPCR and ISH and to also show agonal-pH effects in controls.
Mitochondrial DNA gene expression in agonal state and mood disorders
The mtDNA-dysregulated genes, ATP6 and COX1, are codirectional in mood disorder with agonal-pH effects in controls. There was decreased expression of COX1 and ATP6 in BPD cases without agonal factors compared to controls without agonal factors. In controls without agonal factors, there was also an increase in nDNA gene expression compared to controls with agonal factors for some nDNA-encoded mitochondrial genes but not all, about 5:1 ratio of increased to decreased genes (Supplementary Table 1). Thus, using the same subjects, in the mtDNA genome, the response to agonal-pH factors is an increase in gene expression and for nDNA mitochondrial genes, the response is a general decrease in expression.
This places the mtDNA-encoded transcripts in the third category of mood dysregulated genes, where it is possible but unlikely that agonal-pH effect entirely accounts for the mood disorder effect. If there is an imbalance in the controls showing more agonal-pH factors, then BPD would show a tendency for a decreased mtDNA expression. The copy number of mtDNA was slightly increased in BPD (P = 0.06); this could indicate there are more agonal factors in the BPD group than in the controls, which should increase mtDNA gene expression. However, mtDNA transcripts are decreased in BPD, thus it is difficult to argue both sides simultaneously. Prolonged agonal duration is associated with agonal factors such as hypoxia, low pH and anaerobic metabolism, which might contribute to increased copy number compared to rapid death cases. Increased mtDNA gene transcription or copy number may lead to a compensatory response by nDNA-encoded gene transcripts. It is unknown whether the increased mitochondria copy number found in this study represent functioning mitochondria in premortem brain resulting from mitochondria biogenesis.
Whatever the explanations for the results, the observed effects presumably are the underlying mitochondrial response to mood and agonal conditions. Genes in categories ‘a, b, and c’ might be involved with both mood and agonal-pH response. A starting point for future inquiry could consider possible explanations of modulatory mechanisms. Transcription factors from nDNA sources modulate transcription in the mitochondrial genome.47,48 Mitochondrial proteins derived from nDNA stabilize mtDNA transcripts within the mitochondria.49 leucine-rich PPR-motif containing LRPPRC interacts to stabilize COX1,45 and LRPPRC (increase) and COX1 (decrease) were altered in BPD, and both genes are coregulated in different tissues.45 When LRPPRC is mutated,24 this leads to Leigh Syndrome French Canadian (LCFS), and the transcripts and enzyme activity of COX1, as well as COX2 and COX3, are deficient owing to an alteration in the localization of LRPPRC within the mitochondria. LCFS individuals with an LRPPRC mutation present with metabolic acidosis, neurodegeneration and elevated lactate levels in CSF50 and the disease is fatal. In LCFS, alterations of LRPPRC and COX1 are associated with alterations in pH, and alterations in pH may trigger alterations in additional pH-sensitive mitochondrial gene expression. Alterations in pH and lactate levels have been reported in bipolar disorder brain neuroimaging studies.51–54 Alterations in Ca2+ release and concentrations have been observed in lymphoblastic cell lines from bipolar patients,55,56 which can implicate calcium homeostatic mechanisms operated by mitochondria or endoplasmic reticulum, or cellular signaling. Taken together, metabolic alterations in BPD possibly involve selective mitochondrial and apoptotic functions consistent with the present gene expression results.
Mitochondrial DNA mutations have been observed in BPD57–60 makes it possible that nDNA transcripts will be altered in response. There is the distinct possibility of mutations in the mtDNA control region (CR), which is the origin of replication of the mitochondrial genome and drives the copy number of mtDNA.61–63 Variations in nDNA genes involving chaperone, apoptotic and oxidative phosphorylation pathways have also been shown in studies of bipolar disorder.51,60,64–68 There might be common regulatory motifs in the promoter regions for some of the oxidative phosphorylation genes as many of these genes appear to be coregulated in multiple studies.47,69 Many proteasome-mitochondrial nDNA genes are transcribed from the same bidirectional promoter,70 increasing the likelihood that genes will be coregulated that share some functional role. Thus, the nDNA and mtDNA genomes share joint functional roles in mitochondrial function, and genetic variants in each may contribute to the present findings in mood disorder.
Candidate pathways
The apoptotic classification shows a high percentage of genes independent of agonal-pH effects, that is, many top-ranked apoptotic genes were not found in agonal-pH control comparisons. There were eight apoptotic genes in the top-ranked mood list, such as GSK3B, and these genes showed directions of change opposite to the agonal-pH direction. These genes are of interest, as others have found apoptotic genes previously to be dysregulated in BPD cortical regions.38,71,72 Alterations of apoptotic gene expression might reflect alterations in cell survival signals in mood disorder. The morphometric postmortem findings in mood disorders are variable within the anterior cingulate cortex.15,73–76 Alterations in neuron and glia possibly coexist in mood disorders with regional variation in the supra- and sub-genual AnCg and frontal cortex of subjects,74,76,77 and alterations in neuropil accompanied by an increase in neuronal packing of smaller diameter cells.74,78 Alterations in mitochondrial-apoptotic-related transcripts might be associated with alterations in brain cell counts in the same brain circuits. There are seven chaperone genes with altered expression in BPD DLPFC that were not agonal-pH-sensitive genes. Further validation of apoptotic and chaperone candidates are warranted based upon the microarray and Q-PCR findings.
SSRI, lithium and alcohol effects on gene expression
Multiple effects of antidepressants and mood stabilizers on mitochondrial functioning,79–82 protea-some83,84 and chaperones are primarily inhibitory. Nicotine increased proteasome gene expression.85 The BPD sample was discontinued from lithium treatment and did not show any measurable lithium levels in brain tissue; although lithium was discontinued at various intervals before death, it could still contribute to gene dysregulation. Several genes showed increased expression in subjects without lithium, and decreased expression in subjects with lithium such as HSPA2, SPP1 and CCT3. Lithium treatment altered energy metabolic and stress responsive genes in rats,86 primarily nine genes were downregulated by at least two-fold.86
In the analysis of SSRI effects on nDNA-encoded mitochondrial gene expression, the comorbidity of alcohol and antidepressants are present in a majority of the MDD subjects, which complicates an interpretation based solely on SSRI treatment. The use of alcohol can clearly complicate gene expression patterns in proteasome and energy production-related pathways18,87 and could play a role in the present MDD subjects who also had a positive history of alcohol abuse. Comorbid alcohol abuse and psychiatric disorders can alter gene expression.18 In alcohol abuser and non-abuser pairs matched for pH, ‘The most remarkable decrease was in the expression of 11 nuclear genes encoding mitochondrial proteins.18’. Notably, the gene expression pattern of six mitochondrial genes in the present study was opposite to that based on alcohol abuse.18 Most bipolar and major depressive patients in our study were not treated with neuroleptic medication, which can alter mitochondrial function.88
In summary, large agonal-pH effects interfere with interpretation of more subtle and selective alterations in microarray gene expression results for brain disorders. With specific criteria, selective alterations in nDNA genes encoding mitochondrial, apoptotic, proteasome and chaperone functions were found in cortical regions in mood disorders. Strong agonal effects were shown for mtDNA copy number and mtDNA-encoded transcripts indicating that interpretation of mitochondrial changes in postmortem studies requires careful evaluation. In the final analysis of the data, the subjects with mood disorders and controls were carefully matched for pH and agonal factors, thereby minimizing the possibility that agonal-pH effects account for findings of selective gene dysregulation. The genes dysregulated in three categories of results represent a starting point for further validation studies of possible genes that may contribute to some of the recurrent features and pathophysiology of mood disorders.
Supplementary Material
Acknowledgments
We appreciate the assistance of Preston Cartagena, PsyD and Richard Stein, PhD for their contributions to postmortem clinical characterization of subjects. We acknowledge Kathleen Burke as well as Jacque Berndt and the investigators and medical examiners at the Orange County Coroners Office for procurement of brain tissue. We also appreciate the technical contributions of Kevin Overman, Sharon Burke, Xiaohong Fan and Phong Nguyen. F Warren Lovell, MD, performed a neuropathological evaluation of the postmortem brains. Tissue specimens were processed and stored at the Human Brain and Spinal Fluid Resource Center, Veteran's Medical Center, Los Angeles under the direction of Wallace W Tourtellotte, MD, PhD This project is supported by the NIMH Conte Center Grant P50 MH60398, Pritzker Family Philanthropic Fund, William Lion Penzner Foundation (UCI), Della Martin Foundation (UCI), NIMH Grant #MH54844 (EGJ), WM Keck Foundation (EGJ) and the NIMH Program Project MH42251 (SJW and HA). The authors are members of a Conte Center supported by the NIMH and members of the Pritzker Neuropsychiatric Disorders Research Consortium, which is supported by the Pritzker Family Philanthropic Fund. A shared intellectual property agreement exists between the Pritzker Family Philanthropic Fund and all the universities involved, in order to encourage the development of appropriate findings for research and clinical applications. The academic and philanthropic entities involved in this Consortium are jointly filing patent applications related to the present findings.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Dyrskjot L. Classification of bladder cancer by microarray expression profiling: towards a general clinical use of microarrays in cancer diagnostics. Expert Rev Mol Diagn. 2003;3(5):635–647. doi: 10.1586/14737159.3.5.635. [DOI] [PubMed] [Google Scholar]
- 2.Ueda M, Ota J, Yamashita Y, Choi YL, Ohki R, Wada T, et al. DNA microarray analysis of stage progression mechanism in myelodys-plastic syndrome. Br J Haematol. 2003;123(2):288–296. doi: 10.1046/j.1365-2141.2003.04601.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 4.Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10(3):309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 5.Huffaker SJ, Ryan M, Sudhakaran P, Webster M, Goedert M, Bahn S. Large scale genechip analysis of post - mortem brains from schizophrenia and bipolar affective disorder patients. Program Number 312.16, in Abstract Viewer/ Itinerary Planner. Society for Neuroscience; Washington, DC: 2003. [Google Scholar]
- 6.Petryshen TL, O'Leary SB, Lehar J, Mootha VK, Raad R, Subramanian A, et al. Identification of altered gene pathways in prefrontal cortex and cerebellum of schizophrenia, bipolar disorder, and depression patients. Program Number 312.12, in Abstract Viewer/Itinerary Planner. Society for Neuroscience; Washington, DC: 2003. [Google Scholar]
- 7.Sequeira A, Gwadry F, Ffrench-Mullen JM, Turecki G. Gene expression changes in suicides with and without major depression. Program Number 640.2, in Abstract Viewer/ Itinerary Planner. Society for Neuroscience; Washington DC: 2003. [Google Scholar]
- 8.Bezchlibnyk YB, Wang JF, McQueen GM, Young LT. Gene expression differences in bipolar disorder revealed by cDNA array analysis of post-mortem frontal cortex. J Neurochem. 2001;79(4):826–834. doi: 10.1046/j.1471-4159.2001.00628.x. [DOI] [PubMed] [Google Scholar]
- 9.Sibille E, Arango V, Galfalvy HC, Pavlidis P, Erraji-Benchekroun L, Ellis SP, et al. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29(2):351–361. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- 10.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14(2):241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 11.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61(3):300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 12.Jurata LW, Bukhman YV, Charles V, Capriglione F, Bullard J, Lemire AL, et al. Comparison of microarray-based mRNA profiling technologies for identification of psychiatric disease and drug signatures. J Neurosci Methods. 2004;138(1–2):173–188. doi: 10.1016/j.jneumeth.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, et al. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA. 2004;101(43):15506–15511. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58(2):85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 15.Harrison PJ. The neuropathology of primary mood disorder. Brain. 2002;125(Part 7):1428–1449. doi: 10.1093/brain/awf149. [DOI] [PubMed] [Google Scholar]
- 16.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JJ, Griffin JL, et al. Mitochondrial dysfunction in Schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9(7):684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 17.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22(7):2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72(6):756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- 19.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myeli-nation-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98(8):4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58(1):11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 21.Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH, III, Donovan DM, et al. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55(5):641–650. doi: 10.1016/s0361-9230(01)00522-6. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9(4):406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- 23.Kato T, Kato N. Mitochondrial dysfunction in bipolar disorder. Bipolar Disord. 2000;2(3 Part 1):180–190. doi: 10.1034/j.1399-5618.2000.020305.x. [DOI] [PubMed] [Google Scholar]
- 24.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100(2):605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson BH, De Meirleir L, Glerum M, Sherwood G, Becker L. Clinical presentation of mitochondrial respiratory chain defects in NADH-coenzyme Q reductase and cytochrome oxidase: clues to pathogenesis of Leigh disease. J Pediatr. 1987;110(2):216–222. doi: 10.1016/s0022-3476(87)80157-9. [DOI] [PubMed] [Google Scholar]
- 26.Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, et al. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13(6):609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- 27.Hardy JA, Wester P, Winblad B, Gezelius C, Bring G, Eriksson A. The patients dying after long terminal phase have acidotic brains; implications for biochemical measurements on autopsy tissue. J Neural Transm. 1985;61(3–4):253–264. doi: 10.1007/BF01251916. [DOI] [PubMed] [Google Scholar]
- 28.Johnston NL, Cervenak J, Shore AD, Torrey EF, Yolken RH, Cerevnak J. Multivariate analysis of RNA levels from postmortem human brains as measured by three different methods of RT-PCR. Stanley Neuropathology Consortium. J Neurosci Methods. 1997;77(1):83–92. doi: 10.1016/s0165-0270(97)00115-5. [DOI] [PubMed] [Google Scholar]
- 29.Wester P, Bateman DE, Dodd PR, Edwardson JA, Hardy JA, Kidd AM, et al. Agonal status affects the metabolic activity of nerve endings isolated from postmortem human brain. Neurochem Pathol. 1985;3(3):169–180. doi: 10.1007/BF02834269. [DOI] [PubMed] [Google Scholar]
- 30.Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55(4):346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Leipzig; Barth: 1909. [Google Scholar]
- 32.Vogt BA, Nimchinsky EA, Vogt LJ, Hof PR. Human cingulate cortex: surface features, flat maps, and cytoarchitecture. J Comp Neurol. 1995;359(3):490–506. doi: 10.1002/cne.903590310. [DOI] [PubMed] [Google Scholar]
- 33.Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29(2):373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan MM, Huffaker SJ, Webster MJ, Wayland M, Freeman T, Bahn S. Application and optimization of microarray technologies for human postmortem brain studies. Biol Psychiatry. 2004;55(4):329–336. doi: 10.1016/j.biopsych.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett. 1995;200(3):151–154. doi: 10.1016/0304-3940(95)12102-a. [DOI] [PubMed] [Google Scholar]
- 36.Miller CL, Diglisic S, Leister F, Webster M, Yolken RH. Evaluating RNA status for RT-PCR in extracts of postmortem human brain tissue. Biotechniques. 2004;36(4):628–633. doi: 10.2144/04364ST03. [DOI] [PubMed] [Google Scholar]
- 37.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manji HK, Duman RS. Impairments of neuroplasticity and cellular resilience in severe mood disorders: implications for the development of novel therapeutics. Psychopharmacol Bull. 2001;35(2):5–49. [PubMed] [Google Scholar]
- 39.Gray NA, Zhou R, Du J, Moore GJ, Manji HK. The use of mood stabilizers as plasticity enhancers in the treatment of neuropsychiatric disorders. J Clin Psychiatry. 2003;64(Suppl 5):3–17. [PubMed] [Google Scholar]
- 40.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 42.Vawter MP, Shannon Weickert C, Ferran E, Matsumoto M, Overman K, Hyde TM, et al. Gene expression of metabolic enzymes and a protease inhibitor in the prefrontal cortex are decreased in schizophrenia. Neurochem Res. 2004;29(6):1245–1255. doi: 10.1023/b:nere.0000023611.99452.47. [DOI] [PubMed] [Google Scholar]
- 43.Neal CR, Jr, Akil H, Watson SJ., Jr Expression of orphanin FQ and the opioid receptor-like (ORL1) receptor in the developing human and rat brain. J Chem Neuroanat. 2001;22(4):219–249. doi: 10.1016/s0891-0618(01)00135-1. [DOI] [PubMed] [Google Scholar]
- 44.Benes FM, Matzilevich D, Burke RE, Walsh J. The expression of proapoptosis genes is increased in bipolar disorder, but not in schizophrenia. Mol Psychiatry. 2006;11(3):241–251. doi: 10.1038/sj.mp.4001758. [DOI] [PubMed] [Google Scholar]
- 45.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J. 2004;382(Part 1):331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vawter MP, Freed WJ, Kleinman JE. Neuropathology of bipolar disorder. Biol Psychiatry. 2000;48(6):486–504. doi: 10.1016/s0006-3223(00)00978-1. [DOI] [PubMed] [Google Scholar]
- 47.Traven A, Wong JM, Xu D, Sopta M, ngles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J Biol Chem. 2001;276(6):4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- 48.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 49.Rotig A, Munnich A. Genetic features of mitochondrial respiratory chain disorders. J Am Soc Nephrol. 2003;14(12):2995–3007. doi: 10.1097/01.asn.0000095481.24091.c9. [DOI] [PubMed] [Google Scholar]
- 50.Morin C, Mitchell G, Larochelle J, Lambert M, Ogier H, Robinson BH, et al. Clinical, metabolic, and genetic aspects of cytochrome C oxidase deficiency in Saguenay-Lac-Saint-Jean. Am J Hum Genet. 1993;53(2):488–496. [PMC free article] [PubMed] [Google Scholar]
- 51.Hamakawa H, Murashita J, Yamada N, Inubushi T, Kato N, Kato T. Reduced intracellular pH in the basal ganglia and whole brain measured by 31P-MRS in bipolar disorder. Psychiatry Clin Neurosci. 2004;58(1):82–88. doi: 10.1111/j.1440-1819.2004.01197.x. [DOI] [PubMed] [Google Scholar]
- 52.Kato T, Shioiri T, Murashita J, Hamakawa H, Inubushi T, Takahashi S. Phosphorus-31 magnetic resonance spectroscopy and ventricular enlargement in bipolar disorder. Psychiatry Res. 1994;55(1):41–50. doi: 10.1016/0925-4927(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 53.Kato T, Takahashi S, Shioiri T, Inubushi T. Alterations in brain phosphorous metabolism in bipolar disorder detected by in vivo 31P and 7Li magnetic resonance spectroscopy. J Affect Disord. 1993;27(1):53–59. doi: 10.1016/0165-0327(93)90097-4. [DOI] [PubMed] [Google Scholar]
- 54.Dager SR, Friedman SD, Parow A, Demopulos C, Stoll AL, Lyoo IK, et al. Brain metabolic alterations in medication-free patients with bipolar disorder. Arch Gen Psychiatry. 2004;61(5):450–458. doi: 10.1001/archpsyc.61.5.450. [DOI] [PubMed] [Google Scholar]
- 55.Wasserman MJ, Corson TW, Sibony D, Cooke RG, Parikh SV, Pennefather PS, et al. Chronic lithium treatment attenuates intracellular calcium mobilization. Neuropsychopharmacology. 2004;29(4):759–769. doi: 10.1038/sj.npp.1300400. [DOI] [PubMed] [Google Scholar]
- 56.Emamghoreishi M, Schlichter L, Li PP, Parikh S, Sen J, Kamble A, et al. High intracellular calcium concentrations in transformed lymphoblasts from subjects with bipolar I disorder. Am J Psychiatry. 1997;154(7):976–982. doi: 10.1176/ajp.154.7.976. [DOI] [PubMed] [Google Scholar]
- 57.Munakata K, Iwamoto K, Bundo M, Kato T. Mitochondrial DNA 3243A > G mutation and increased expression of LARS2 gene in the brains of patients with bipolar disorder and schizophrenia. Biol Psychiatry. 2005;57(5):525–532. doi: 10.1016/j.biopsych.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 58.Kato T, Takahashi Y. Deletion of leukocyte mitochondrial DNA in bipolar disorder. J Affect Disord. 1996;37(2–3):67–73. doi: 10.1016/0165-0327(95)00050-x. [DOI] [PubMed] [Google Scholar]
- 59.Kato T, Stine OC, McMahon FJ, Crowe RR. Increased levels of a mitochondrial DNA deletion in the brain of patients with bipolar disorder. Biol Psychiatry. 1997;42(10):871–875. doi: 10.1016/S0006-3223(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 60.Kato T, Kunugi H, Nanko S, Kato N. Association of bipolar disorder with the 5178 polymorphism in mitochondrial DNA. Am J Med Genet. 2000;96(2):182–186. [PubMed] [Google Scholar]
- 61.Coskun PE, Ruiz-Pesini E, Wallace DC. Control region mtDNA variants: longevity, climatic adaptation, and a forensic conundrum. Proc Natl Acad Sci USA. 2003;100(5):2174–2176. doi: 10.1073/pnas.0630589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci USA. 2004;101(29):10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melov S, Schneider JA, Coskun PE, Bennett DA, Wallace DC. Mitochondrial DNA rearrangements in aging human brain and in situ PCR of mtDNA. Neurobiol Aging. 1999;20(5):565–571. doi: 10.1016/s0197-4580(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 64.Kato T, Kunugi H, Nanko S, Kato N. Mitochondrial DNA polymorphisms in bipolar disorder. J Affect Disord. 2001;62(3):151–164. doi: 10.1016/s0165-0327(99)00173-1. [DOI] [PubMed] [Google Scholar]
- 65.Kato T, Iwamoto K, Washizuka S, Mori K, Tajima O, Akiyama T, et al. No association of mutations and mRNA expression of WFS1/wolframin with bipolar disorder in humans. Neurosci Lett. 2003;338(1):21–24. doi: 10.1016/s0304-3940(02)01334-4. [DOI] [PubMed] [Google Scholar]
- 66.Kakiuchi C, Iwamoto K, Ishiwata M, Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, Kusumi I, et al. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat Genet. 2003;35(2):171–175. doi: 10.1038/ng1235. [DOI] [PubMed] [Google Scholar]
- 67.Washizuka S, Kakiuchi C, Mori K, Kunugi H, Tajima O, Akiyama T, et al. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with bipolar disorder. Am J Med Genet. 2003;120B(1):72–78. doi: 10.1002/ajmg.b.20041. [DOI] [PubMed] [Google Scholar]
- 68.Washizuka S, Kakiuchi C, Mori K, Tajima O, Akiyama T, Kato T. Expression of mitochondria-related genes in lymphoblastoid cells from patients with bipolar disorder. Bipolar Disord. 2005;7(2):146–152. doi: 10.1111/j.1399-5618.2005.00184.x. [DOI] [PubMed] [Google Scholar]
- 69.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36(2):197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 70.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14(1):62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jarskog LF, Gilmore JH, Selinger ES, Lieberman JA. Cortical bcl-2 protein expression and apoptotic regulation in schizophrenia. Biol Psychiatry. 2000;48(7):641–650. doi: 10.1016/s0006-3223(00)00988-4. [DOI] [PubMed] [Google Scholar]
- 72.Li X, Bijur GN, Jope RS. Glycogen synthase kinase-3beta, mood stabilizers, and neuroprotection. Bipolar Disord. 2002;4(2):137–144. doi: 10.1034/j.1399-5618.2002.40201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cotter D, Mackay D, Chana G, Beasley C, Landau S, Everall IP. Reduced neuronal size and glial cell density in area 9 of the dorsolateral prefrontal cortex in subjects with major depressive disorder. Cereb Cortex. 2002;12(4):386–394. doi: 10.1093/cercor/12.4.386. [DOI] [PubMed] [Google Scholar]
- 74.Cotter D, Mackay D, Landau S, Kerwin R, Everall I. Reduced glial cell density and neuronal size in the anterior cingulate cortex in major depressive disorder. Arch Gen Psychiatry. 2001;58(6):545–553. doi: 10.1001/archpsyc.58.6.545. [DOI] [PubMed] [Google Scholar]
- 75.Bouras C, Kovari E, Hof PR, Riederer BM, Giannakopoulos P. Anterior cingulate cortex pathology in schizophrenia and bipolar disorder. Acta Neuropathol (Berlin) 2001;102(4):373–379. doi: 10.1007/s004010100392. [DOI] [PubMed] [Google Scholar]
- 76.Ongur D, Drevets WC, Price JL. Glial reduction in the subgenual prefrontal cortex in mood disorders. Proc Natl Acad Sci USA. 1998;95(22):13290–13295. doi: 10.1073/pnas.95.22.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, Meltzer HY, et al. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999;45(9):1085–1098. doi: 10.1016/s0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 78.Chana G, Landau S, Beasley C, Everall IP, Cotter D. Two-dimensional assessment of cytoarchitecture in the anterior cingulate cortex in major depressive disorder, bipolar disorder, and schizophrenia: evidence for decreased neuronal somal size and increased neuronal density. Biol Psychiatry. 2003;53(12):1086–1098. doi: 10.1016/s0006-3223(03)00114-8. [DOI] [PubMed] [Google Scholar]
- 79.Weinbach EC, Costa JL, Nelson BD, Claggett CE, Hundal T, Bradley D, et al. Effects of tricyclic antidepressant drugs on energy-linked reactions in mitochondria. Biochem Pharmacol. 1986;35(9):1445–1451. doi: 10.1016/0006-2952(86)90108-5. [DOI] [PubMed] [Google Scholar]
- 80.Curti C, Mingatto FE, Polizello AC, Galastri LO, Uyemura SA, Santos AC. Fluoxetine interacts with the lipid bilayer of the inner membrane in isolated rat brain mitochondria, inhibiting electron transport and F1F0- ATPase activity. Mol Cell Biochem. 1999;199(1–2):103–109. doi: 10.1023/a:1006912010550. [DOI] [PubMed] [Google Scholar]
- 81.Eto K, Fukuda T, Araki Y, Inoue B, Ogata M. Effect of tricyclic drugs on mitochondrial membrane. Acta Med Okayama. 1985;39(4):289–295. doi: 10.18926/AMO/31500. [DOI] [PubMed] [Google Scholar]
- 82.Roberton AM, Ferguson LR, Cooper GJ. Biochemical evidence that high concentrations of the antidepressant amoxapine may cause inhibition of mitochondrial electron transport. Toxicol Appl Pharmacol. 1988;93(1):118–126. doi: 10.1016/0041-008x(88)90031-2. [DOI] [PubMed] [Google Scholar]
- 83.Holtz KM, Rice AM, Sartorelli AC. Lithium chloride inactivates the 20S proteasome from WEHI-3B D+ leukemia cells. Biochem Biophys Res Commun. 2003;303(4):1058–1064. doi: 10.1016/s0006-291x(03)00473-x. [DOI] [PubMed] [Google Scholar]
- 84.Rice AM, Sartorelli AC. Inhibition of 20 S and 26 S proteasome activity by lithium chloride: impact on the differentiation of leukemia cells by all-trans retinoic acid. J Biol Chem. 2001;276(46):42722–42727. doi: 10.1074/jbc.M106583200. [DOI] [PubMed] [Google Scholar]
- 85.Kane JK, Konu O, Ma JZ, Li MD. Nicotine coregulates multiple pathways involved in protein modification/degradation in rat brain. Brain Res Mol Brain Res. 2004;132(2):181–191. doi: 10.1016/j.molbrainres.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 86.Bosetti F, Seemann R, Bell JM, Zahorchak R, Friedman E, Rapoport SI, et al. Analysis of gene expression with cDNA microarrays in rat brain after 7 and 42 days of oral lithium administration. Brain Res Bull. 2002;57(2):205–209. doi: 10.1016/s0361-9230(01)00744-4. [DOI] [PubMed] [Google Scholar]
- 87.Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90(5):1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- 88.Ben-Shachar D. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J Neurochem. 2002;83(6):1241–1251. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.