Abstract
There are major concerns that specific agonal conditions, including coma and hypoxia, might affect ribonucleic acid (RNA) integrity in postmortem brain studies. We report that agonal factors significantly affect RNA integrity and have a major impact on gene expression profiles in microarrays. In contrast to agonal factors, gender, age, and postmortem factors have less effect on gene expression profiles. The Average Correlation Index is proposed as a method for evaluating RNA integrity on the basis of similarity of microarray profiles. Reducing the variance due to agonal factors is critical in investigating small but validated gene expression differences in messenger RNA levels between psychiatric patients and control subjects.
Keywords: Adenine, Uracil-rich elements, bipolar disorder, freezer interval, iron-responsive element, major depressive disorder, postmortem interval
Microarray studies using postmortem brains have become particularly valuable in profiling gene expression patterns related to psychiatric disorders of unknown etiology (Bahn et al 2001; Bunney et al 2003; Hakak et al 2001; Mirnics et al 2001; Van Deerlin et al 2002). In studies using postmortem brain tissue, the variability of ribonucleic acid (RNA) integrity has been a major concern. There have been two major factors that affect RNA integrity in gene expression studies using postmortem brain: 1) agonal factors, defined as specific agonal conditions at the time of death and agonal duration; and 2) postmortem factors, defined as the condition of the postmortem brain tissue after death, including the delay between death and the time the tissue is frozen (postmortem interval [PMI]) and the duration the brain tissue is stored in a freezer (freezer interval [FI]). Prior studies have indicated that agonal factors markedly affect the integrity of RNA, whereas postmortem factors have relatively small effects on RNA integrity (Bahn et al 2001; Barton et al 1993; Gilmore et al 1993; Harrison et al 1995; Johnson et al 1986; Kingsbury et al 1995; Leonard et al 1993; Perrett et al 1988; Ravid et al 1992; Van Deerlin et al 2002). However, because messenger (m)RNAs have been shown to vary in their vulnerability to agonal factors (Barton et al 1993; Burke et al 1991; Harrison et al 1994, 1995), attention should be drawn to the limitation of previous postmortem studies in this area, in which investigators have only analyzed restricted numbers of transcripts. Expression data for thousands of genes obtained by microarray technology facilitate a comprehensive evaluation of the integrity of RNA samples.
In this article, we discuss issues concerning evaluation of agonal and postmortem factors and RNA integrity in microarray study of postmortem brains. The hypothesis raised in the present study was that agonal factors adversely affect integrity of mRNAs and influence microarray expression profiles more than the postmortem factor or other biological factors, such as gender, age, or diagnosis of psychiatric disorder. The hypothesis was tested with 40 well-characterized postmortem brains. We show that agonal factors and RNA integrity can be evaluated by correlation of expression profiles between microarrays. A tool, the Average Correlation Index (ACI), is presented to evaluate the agonal factor and RNA integrity on the basis of the similarity of gene expression profiles for each microarray among a total set of microarray data.
Evaluation of Agonal and Postmortem Factors
Specific agonal conditions, including coma, hypoxia, pyrexia, seizures, dehydration, hypoglycemia, multiple organ failure, head injury, and ingestion of neurotoxic substances at time of death have been evaluated with regard to influences on brain acidosis and RNA integrity in postmortem brain tissues (Barton et al 1993; Harrison et al 1991a, 1991b, 1994b, 1995; Hynd et al 2003; Johnston et al 1997; Miller et al 1990; Morrison-Bogorad et al 1995; Roberge and Krenzelok 2001; Yokota et al 2000). The combination of these specific agonal conditions and the duration of the agonal phase, which might adversely affect RNA integrity in postmortem brain tissue (Barton et al 1993; Hardy et al 1985; Harrison et al 1991a; Johnston et al 1997; Wester et al 1985), are referred to as agonal factors. At the University of California, Irvine Brain Repository (UCIBR), an extensive review of multiple resources was conducted, including the medical examiner’s conclusions, coroner’s investigation, medical records, toxicology results, and interviews of the decedents’ next-of-kin, to obtain information concerning agonal and postmortem factors before the microarray experiments. Each variable for the specific agonal condition (coma, hypoxia, pyrexia, seizures, dehydration, hypoglycemia, multiple organ failure, head injury, and ingestion of neurotoxic substances) was scored as absent (0) or present (1). Agonal duration was separately rated from 1 to 4 according to an agonal duration rating scale by Hardy et al (1985). The Hardy et al (1985) 4-point scale was further refined in this study. The four categories were 1) violent and fast death: deaths due to accident, blunt force trauma, or suicide; terminal phase estimated at less than 10 min; 2) fast death of natural causes: sudden unexpected deaths of people who had been reasonably healthy, after a terminal phase estimated at less than 1 hour. The modal cause of death in this group was myocardial infarction; 3) intermediate death: death after a terminal phase of between 1 and 24 hours; and 4) slow death: death after long illness, with a terminal phase longer than 1 day. Typically, patients in category 4 died of cancer or chronic pulmonary disease. Agonal duration classifications were subsequently recoded to denote the absence or presence of prolonged death. As such, violent deaths and fast deaths of natural causes were considered rapid (0), and intermediate and slow deaths were considered prolonged (1). The scores for these specific agonal conditions and agonal duration were summed to provide an agonal factor score (AFS) for each decedent.
Evaluation of Brain Tissue pH
Analyses of postmortem human brain have shown that brain tissue pH is decreased by prolonged death and hypoxia (Hardy et al 1985; Kingsbury et al 1995), whereas brain tissue pH remains stable postmortem (Kingsbury et al 1995). Brain tissue pH has been shown to be unaffected during freezer storage and is remarkably consistent across different brain regions (Johnston et al 1997). Animal studies have suggested that there might be a rapid drop of the brain tissue pH within 10 min after death (Hardy et al 1985; Ravid et al 1992; Swaab and Boer 1972), and after this rapid decline, the brain tissue pH might be stable for 24–36 hours (Johnston et al 1997; Kingsbury et al 1995; Ravid et al 1992; Trotter et al 2002). Thus, brain tissue pH has been evaluated as an indicator for agonal status and as a crude but reliable indicator of RNA integrity (Bahn et al 2001; Harrison et al 1995; Johnston et al 1997; Kingsbury et al 1995). In the UCIBR, brain tissue pH was measured with the following procedures. A piece of tissue (50–100 mg) from a frozen cerebellar cortical slice was mixed with 1.0-mm glass beads (BioSpec Products, Bartlesville, Oklahoma) to form a 10% (wt/vol) solution in distilled deionized water and homogenized with Bead-Beater (BioSpec Products) for 60 sec at 4°C. After centrifugation at 5000 rpm for 2 min at 4°C, the homogenate was warmed quickly to room temperature, and the pH was measured with a pH meter (Corning, Cypress, California) calibrated with three standards (pH 4, 7, and 10). The pH protocol showed reproducibility of the same samples measured on different days within .1 pH units.
Evaluation of RNA Integrity
Integrity of total RNA has been evaluated with quantity of extractable total RNA or quantification of 18S and/or 28S ribosomal RNA, the most abundant transcripts in total RNA involved with the construction of subunits of the ribosome (Bahn et al 2001; Johnson et al 1986; Pardue et al 1994). Total amount of polyadenylated mRNA, which constitutes 3%–5% of total RNA, has been determined by hybridization with a poly-deoxyuridine triphosphate (dUTP) probe (Harrison et al 1991b), and biological activity of mRNA has been evaluated by efficiencies of in vitro translation and peptide synthesis (Johnson et al 1986; Leonard et al 1993; Perrett et al 1988). Also, integrity of specific mRNA has been evaluated by Northern blot hybridization (Johnson et al 1986; Kingsbury et al 1995; Leonard et al 1993; Pardue et al 1994), quantitative in situ hybridization (Gilmore et al 1993; Harrison et al 1991b; Kingsbury et al 1995; Pardue et al 1994), or quantitative reverse transcriptase polymerase chain reaction (Bahn et al 2001; Burke et al 1991; Johnston et al 1997; Leonard et al 1993). Several RNA integrity indicators based on Affymetrix GeneChip (Affymetrix, Santa Clara, California) microarray data are available. The percentage of the total number of probe sets detected as present on the array (percent present call) and the ratio of signal intensities for probe sets designed in 3′ and 5′ end regions of the housekeeping genes glyceraldehyde phosphate dehydrogenase (GAPDH) and β actin (ACTB) can be obtained with MAS 5.0 software (Affymetrix). Also, the coefficient of the RNA degradation slope from 5′ to 3′ ends of the average probe intensities (degradation slope) can be calculated with the R-statistical program AffyRNAdeg (Affy version 2.0), available at http://www.bioconductor.org/. As RNA samples are degraded, it is expected that both the percent (18S+28S) and the percent present call are decreased. The 3′/5′ ratios for GAPDH and ACTB and the “degradation slope” are both found to increase in samples with RNA degradation.”
Agonal Factors Decrease Brain Tissue pH and Affect RNA Integrity Indicators
The effects of agonal and postmortem factors on brain tissue pH, RNA integrity indicators, and global gene expression profiles in microarray experiments were analyzed in 40 postmortem brains. All postmortem brain tissues were obtained after informed consent had been received, under an institutional review board–approved protocol, “Organ Donation and Release of Medical Information.” The cohort consisted of 9 subjects with bipolar disorder type I (BPD), 11 with major depressive disorder (MDD), and 20 control subjects between the ages of 18 and 73 years (mean 52.4, SD 16.0); 28 subjects were male and 12 were female. The PMI ranged from 6.5 to 41.3 hours (mean 22.6, SD 7.14), FI ranged from 3 to 112 months (mean 38.1, SD 25.3), and the brain tissue pH ranged from 6.06 to 7.20 (mean 6.79, SD .28). There were 24 subjects who died rapidly without any specific agonal conditions (AFS = 0), whereas the remaining 16 had prolonged deaths and/or the specific agonal conditions (AFS ≥ 1). The details for each subject are shown in Table 1 (AFS = 0 for subjects 1–24; AFS ≥ 1 for subjects 25–40). Total RNA was extracted from anterior cingulate cortex, dorsolateral prefrontal cortex, and cerebellum of the 40 postmortem brains with Trizol (Invitrogen, Carlsbad, California) and purified with silica-based mini-spin columns (Qiagen RNeasy Mini Kit, Valencia, California). Ten micrograms of each purified total RNA sample was applied on an Affymetrix U95Av2 GeneChip. Each microarray experiment was carried out twice independently as experimental duplicates. In total, 240 GeneChips (2 GeneChips × 3 brain regions × 40 cases) were analyzed in this experiment. The procedures for the microarray experiments are described in detail elsewhere (Vawter et al 2003).
Table 1.
Clinical Information Including Agonal and Postmortem Factors for Each of the 40 Subjects
| Agonal Factors |
Postmortem Factor |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject No. | Gender | Age (years) | Diagnosis | PD | Coma | Pyrexia | Hypoxia | MOF | HI | NS | AFS | PMI | FI |
| 1 | M | 69 | BPD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 112 |
| 2 | M | 23 | BPD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 83 |
| 3 | M | 26 | BPD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 70 |
| 4 | F | 56 | BPD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 | 62 |
| 5 | M | 52 | BPD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 9 |
| 6 | M | 59 | BPD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 4 |
| 7 | F | 72 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 89 |
| 8 | M | 19 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 85 |
| 9 | M | 58 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 83 |
| 10 | M | 49 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 | 6 |
| 11 | M | 46 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27 | 16 |
| 12 | M | 49 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27 | 18 |
| 13 | M | 52 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 25 |
| 14 | F | 48 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 20 |
| 15 | M | 39 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 7 |
| 16 | F | 60 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 27 |
| 17 | M | 54 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 22 |
| 18 | M | 70 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27 | 19 |
| 19 | M | 18 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 91 |
| 20 | M | 58 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 83 |
| 21 | M | 55 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 84 |
| 22 | M | 50 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 | 30 |
| 23 | M | 45 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 45 |
| 24 | M | 44 | Control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 6 |
| 25 | F | 63 | BPD | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 22 | 19 |
| 26 | F | 68 | BPD | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 26 | 8 |
| 27 | M | 69 | BPD | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 29 | 4 |
| 28 | M | 53 | MDD | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 17 | 54 |
| 29 | F | 73 | MDD | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 4 | 29 | 13 |
| 30 | M | 72 | Control | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 15 | 13 |
| 31 | M | 19 | Control | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 7 | 90 |
| 32 | M | 25 | Control | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 3 | 14 | 53 |
| 33 | F | 64 | Control | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 27 | 5 |
| 34 | F | 68 | Control | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 25 | 4 |
| 35 | M | 58 | Control | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 15 | 8 |
| 36 | F | 70 | Control | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 3 | 21 | 112 |
| 37 | F | 73 | Control | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 24 | 3 |
| 38 | M | 52 | Control | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 23 | 22 |
| 39 | M | 50 | Control | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 17 | 20 |
| 40 | F | 47 | Control | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 41 | 5 |
The table displays information regarding gender, age, diagnosis, and agonal and postmortem factors. Ratings of the absence (0) or presence (1) of prolonged death, coma, hypoxia, pyrexia, seizures, dehydration, hypoglycemia, multiple organ failure, head injury, and ingestion of neurotoxic substances at time of death are summarized. There are no instances of severe dehydration, seizure or hypoglycemia in the 40 subjects. Variables of prolonged death and specific agonal conditions were summed to provide an agonal factor score (AFS) for each decedent. Twenty-four subjects had rapid deaths with no evidence of the specific agonal conditions (AFS = 0), and 16 subjects had an AFS of at least 1. A line is drawn between the top 24 subjects with an AFS of 0 and the bottom 16 subjects with an AFS of 1 or more.
PD, prolonged death; MOF, multiple organ failure; HI, severe head injury; NS, ingestion of neurotoxic substances; AFS, agonal factor score; PMI, postmortem interval; FI, freezer interval; M, male; F, female; BPD, bipolar disorder; MDD, major depressive disorder.
In this study, the percentage of 18S and 28S ribosomal RNA to total RNA [percent(18S+28S)] was used as an indicator of RNA integrity, as were the 3′/5′ ratios for GAPDH, the percent present call on the Affymetrix microarray provided by MAS 5.0, and the “degradation slope” calculated by AffyRNAdeg. For the quantification of 18S and 28S ribosomal RNA, 1 μg of each RNA sample was applied to a 2100 Bioanalyzer (Agilent, Palo Alto, California). The RNA integrity indicators were averaged across three brain regions.
The group of 16 subjects with AFS of 1 or more showed significant decreases in brain tissue pH (p < .001), the percent (18S+28S) (p < .01), and the percent present call (p < .005) compared with the group of 24 subjects with AFS of 0, by Student t test. Also, the group with AFS of 1 or more showed significant increases in the 3′/5′ ratios for GAPDH (p < .005) and the “degradation slope” (p < .001) compared with the group with AFS of 0. The correlations of brain tissue pH with the percent (18S+28S) (r = .54, p < .001) and the percent present call (r = .54, p < .001) were both highly significant. Also, brain tissue pH showed significant negative correlations with the 3′/5′ ratios for GAPDH (r = −.71, p < .001) and the “degradation slope” (r = −.57, p < .001). In contrast to agonal factors, the postmortem interval showed no significant correlation with brain tissue pH (r = .04, p > .1), percent (18S+28S) (r = .05, p > .1), percent present call (r = .05, p > .1), 3′/5′ ratios for GAPDH (r = .02, p > .1), and the “degradation slope” (r = .1, p > .1). The data are consistent with previous reports that agonal factors decrease brain tissue pH and facilitate RNA degradation (Bahn et al 2001; Barton et al 1993; Gilmore et al 1993; Hardy et al 1985; Harrison et al 1995; Johnson et al 1986; Johnston et al 1997; Kingsbury et al 1995; Leonard et al 1993; Perrett et al 1988; Ravid et al 1992; Van Deerlin et al 2002).
Agonal Factors Have a Major Impact on Gene Expression Profiles in Microarrays
To evaluate the effects of agonal and postmortem factors on global gene expression profiles in microarray studies of psychiatric disorders, the 240 GeneChips were analyzed. Signal intensities of 12,625 probe sets on each GeneChip were extracted with MAS 5.0. After the consistency between each of the experimental duplicates was confirmed, signal intensities of the experimental duplicates were averaged. Pearson’s correlation coefficient of the 12,625 probe sets between samples was calculated among 40 subjects, which resulted in 780 correlation coefficients for each brain region. As an example, data based on 40 arrays for anterior cingulate cortex are shown in Figure 1. The matrix in Figure 1A summarizes 780 correlation coefficients for anterior cingulate cortex with three color codes (white, r > .94; gray, .9 < r < .94; and black, r < .9). The matrix is symmetrical around the diagonal line. Almost all of the 24 subjects with an AFS of 0 are highly correlated (r > .94) with each other, regardless of mood disorder diagnoses. The 16 subjects with an AFS of 1 or more show lower correlations (r < .9) with most of the other subjects. The distribution of the correlation coefficient among the 24 subjects with an AFS of 0, and the ones between the 16 subjects with an AFS of 1 or more and all 40 subjects are summarized in Figure 1B. There is a broad dispersion of correlations among subjects with an AFS of 1 or more, as shown in the range of correlations extending to .72. The mean correlation among subjects with an AFS of 0 was .968 (SD .014), whereas the correlation between subjects with an AFS of 1 or more and subjects with an AFS of 0 or subjects with an AFS of 1 or more were lower (mean .886; SD .055). To compare the correlation coefficients (r) between a group comprising only subjects with an AFS of 0 and a group including subjects with an AFS of 1 or more, correlation coefficients were converted to Zr (.5 × loge[(1 +r)/(1 − r)] with Fisher Z transformation to fit a normal distribution. The transformed correlation coefficients (Zr) for each group were compared by Student t test. A group including subjects with an AFS of 1 or more showed significantly lower correlation coefficients than a group comprising only subjects with an AFS of 0 (p < .0001).
Figure 1.
All 24 subjects with an agonal factor score (AFS) of 0 are highly correlated each other. (A) Pearson simple correlations of expression profile composed of 12,625 probe sets were calculated between all 780 pairs among 40 arrays from the anterior cingulate cortex. The 780 correlation coefficients are displayed in gray-scale (white, r > .94; gray, .90 < r < .94; black, r < .9). Almost all of the 24 subjects with an agonal factor score (AFS) of 0 are highly correlated (r > .94) with each other, regardless of mood disorder diagnoses. The 16 subjects with an AFS of 1 or more show lower correlations (r < .9) with most of the other subjects. This simple correlation matrix is used for calculating Average Correlation Index. The matrix is symmetrical around the diagonal line. (B) The distribution of the correlation coefficients calculated for subjects with no agonal factors (AFS = 0, open bars) is different than the distribution for subjects with agonal factors (AFS ≥ 1, filled bars). The individual data for the distribution of 276 correlation pairs among 24 subjects with an AFS of 0 and the correlation of 504 pairs between the 16 subjects with an AFS of 1 or more and all 40 subjects are shown in A. The mean correlation among subjects with an AFS of 0 was .968 (SD .014), whereas the correlation between subjects with an AFS of 1 or more and subjects with an AFS of 0 or subjects with an AFS of 1 or more were lower (mean .886, SD .055). There is a broad dispersion of correlations among subjects with an AFS of 1 or more, as shown in the range of correlation coefficients extending to .72. BPD, bipolar disorder; MDD, major depressive disorder.
Because gender and age affect gene expression in the brain (Lee et al 2000; Vawter et al 2003), these factors should also be taken into account in microarray studies of psychiatric disorders. For example, Y-chromosomal genes, including DBY, SMCY, UTY, and RPS4Y, were expressed only in male tissues, consistent with prior findings (Vawter et al 2003); however, gender and age seem to have less influence on overall gene expression profiles, compared with agonal factors. Among the 24 subjects with an AFS of 0, correlation coefficients between subjects were consistently high regardless of gender, age, and diagnoses of BPD or MDD. For example, 20 male subjects are highly correlated with four female subjects (averaged r = .968; SD = .014). Four subjects younger than 30 years were highly correlated with four subjects older than 60 years (averaged r = .967; SD = .013). Furthermore, nine control subjects were highly correlated with six subjects with BPD (averaged r = .967; SD = .015) and nine subjects with MDD (averaged r = .967; SD = .015). These patterns are stable in the three brain regions. The data suggest that agonal factors introduce larger divergences of global gene expression profiles than do gender, age, or diagnoses of psychiatric disorders.
Average Correlation Index
To test the hypothesis that these wide divergences of correlation coefficients between arrays might reflect agonal factors and RNA integrity, similarity between arrays was summarized as the ACI for each of the brain regions. The ACI was calculated for each of the 40 subjects with the following steps. Step 1: Pearson’s simple correlation of expression profile, composed of 12,625 probe sets, was calculated between all 780 pairs among 40 arrays, as shown as gray-scale grids in Figure 1A. Step 2: in the correlation matrix, the average of the 39 correlations between one chip and the other 39 arrays was calculated for each array. Step 3: an array with the lowest averaged correlation of the 40 was determined, and this lowest averaged correlation was referred to as the ACI for this array. The array with the lowest ACI was removed from the group of 40 for the remaining calculations. Step 4: for each remaining array, the average correlation with the other 38 arrays was recalculated. Step 5: the lowest ACI of the 39 was determined, this array was removed from the group for the remaining calculations, and the averages of the remaining group were recalculated. Step 6: these iterative calculations (steps 2–5) for ACI were carried out until ACIs were obtained for all 40 arrays. All of these steps were performed with Excel software (Microsoft, Redmond, Washington).
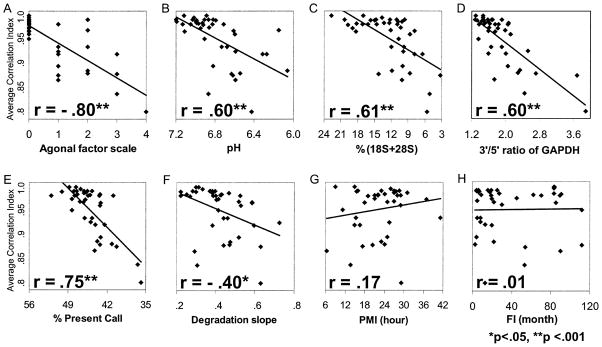
The ACIs for the three brain regions of the 40 brains were calculated. The correlation coefficients between ACI and each of the following variables were calculated: AFS, brain tissue pH, RNA integrity indicators [percent(18S+28S), 3′/5′ ratio of GAPDH, percent present call, and degradation slope] and postmortem factors (PMI, FI). As an example, Figure 2 displays the results for anterior cingulate cortex. The ACI was significantly correlated with AFS and all of the RNA integrity indicators analyzed, whereas ACI showed no significant correlation with PMI and FI. These observations were similar in all three brain regions analyzed. Figure 3 displays the average ACI across three brain regions plotted for presence (more than 1) or absence (0) of AFS for each subject. All subjects with no agonal factors had average ACIs greater than .95, whereas of 16 subjects with an agonal factor, 15 had average ACIs of less than .95. These observations were also stable among alternative methods for data analyses. For example, different condensation programs, such as Robust Multi-Array analysis in the Affy package version 2.0 (Irizarry et al 2003), or Spearman rank correlation.
Figure 2.
Agonal factors and ribonucleic acid (RNA) degradation decrease Average Correlation Index (ACI). The ACI was calculated for the anterior cingulate cortex for 40 subjects and plotted on the y axis in the eight dot plots (A–H). The x axis shows the corresponding (A) agonal factor score (AFS); (B) brain tissue pH; (C) percentage of 18S and 28S ribosomal RNA to total RNA [%(18S+28S)]; (D) ratio of signal intensities for probe sets designed in 3′ and 5′ end region of glyceraldehyde phosphate dehydrogenase (GAPDH) gene (3′/5′ ratio of GAPDH; (E) the percentage of the total number of probe sets detected as present on the array (% Present Call); (F) coefficient of the RNA degradation slope from 5′ to 3′ ends of the averaged probe intensities calculated with the R-statistical program AffyRNAdeg (Degradation slope); (G) postmortem interval (PMI); and (H) freezer interval (FI). Average Correlation Index was significantly correlated with AFS and all of the RNA integrity indicators analyzed, whereas ACI showed no correlation with PMI and FI.
Figure 3.

Average Correlation Index (ACI) of the microarray chips is a sensitive and specific indicator of agonal factors. The consistency of the ACI across three brain regions (anterior cingulate cortex, dorsolateral prefrontal cortex, and cerebellum) was examined for each subject. The ACI was averaged for each subject across the three brain regions and the result plotted by presence of agonal factors (agonal factor score [AFS] ≥ 1) or absence of agonal factors (AFS = 0). The vertical discrimination line is set to an ACI of .95. All subjects without an agonal factor were above this cut-off (n = 24), whereas 15 of 16 subjects with an agonal factor were below this ACI discrimination threshold. The ACI averaged across three brain regions was 100% sensitive to cases without agonal factors, with 93.7% specificity.
Interestingly, AFS and brain tissue pH showed higher correlations with ACI than did the other RNA integrity indicators in three brain regions, which suggests that ACI can be a reliable indicator of the effects of agonal factors and brain acidosis on RNA integrity.
Taken together, agonal factors affect gene expression profile divergence more than do postmortem factors and other biological factors, including age, gender, and diagnoses of mood disorders. The large divergences in expression profiles influenced by agonal factors are associated with RNA degradation, which suggests that the divergence might be due to the differential vulnerability of mRNAs to degradation during the agonal phase. The postmortem factors seem to have less effect on integrities of mRNAs. After excluding subjects with specific agonal conditions, such as coma and hypoxia, or prolonged death, correlation coefficients between gene expression profiles of remaining subjects were consistently high. It might be feasible to detect relatively smaller effect size between psychiatric disorder and control groups by excluding subjects with the specific agonal conditions.
Why Does Gene Expression Divergence Seem to Be Affected More by Agonal Factors Than by Postmortem Factors?
When ribonucleases are active, the half-life of most mRNAs is on the order of a few hours (Hargrove and Schmidt 1989). Ribonucleic acid seems to become more stable after death, with possible inactivation of its normal processes for degradation (Barton et al 1993). It might be that catabolism of RNA is energy dependent and ceases rapidly after death, owing to a precipitous fall in adenosine triphosphate concentration (Morrison and Griffin 1981); or that the postmortem activity of endogenous ribonuclease inhibitors persists longer than that of the ribonuclease themselves; or that once mRNA stops being translated it is less susceptible to ribonuclease digestion (Barton et al 1993). On the other hand, agonal factors induce brain acidosis (Hardy et al 1985; Kingsbury et al 1995), which might influence both gene transcription and stability of mRNAs. The activation of acid ribonuclease as a result of brain acidosis during the terminal phase might be relevant to the effect of agonal factors on RNA integrity (Albrecht and Yanagihara 1979).
Stability of mRNA varies greatly among eukaryotic mRNAs (Hargrove and Schmidt 1989). Messenger RNA can be degraded by a variety of pathways, and decay is directed by different cis-elements within mRNAs, such as Adenine–Uracil-rich elements and iron-responsive element (Hollams et al 2002). Half-lives of mRNA can increase or decrease in response to a variety of stimuli, including environmental factors such as hypoxia, mitogens and growth factors, hormones, and second messengers that are released by signaling cascades (Hollams et al 2002). These differential vulnerabilities among mRNAs during the agonal phase might cause the divergence in gene expression profiles.
Variable sensitivity among cell types to agonal stress and autolysis might also affect mRNA expression profiles. Degradation of mRNA might be facilitated in cell types more vulnerable to agonal stressors. As an example, granule cells are particularly sensitive to autolysis (Netsky 1968). Also, it was shown that heat shock protein 70 mRNA was degraded in neurons several times more rapidly than in glia (Pardue et al 1994).
Conclusion
Agonal factors impact RNA integrity and gene expression profiles; therefore, the similarity between microarray expression profiles is decreased by agonal factors. The postmortem factors contribute less to the RNA integrity and the expression profile similarity compared with agonal factors. The ACI is a reliable tool for measuring RNA integrity based on microarray expression profiling. When the influences of agonal factors are minimized, microarray profiling of human postmortem brain can be a viable method for detecting relatively small but reliable and validated differences in mRNA levels between psychiatric disorder and matched control groups.
Acknowledgments
This project was supported by National Institute of Mental Health (NIMH) Conte Center Grant P50 MH60398, the Pritzker Neuropsychiatric Disorders Research Consortium, the William Lion Penzner Foundation (UCI), the Della Martin Foundation (UCI), NIMH Grant #MH54844 (EGJ), the W.M. Keck Foundation (EGJ), and NIMH Program Project MH42251 (SJW and HA).
We thank Preston Cartagena, Psy.D., and Richard Stein, Ph.D., of the UCI Brain Repository for their contributions to postmortem clinical characterization of subjects; Kathleen Burke for procurement of brain tissue; and Chief Deputy Coroner Jacque Berndt and the staff of the Orange County Coroners Office. We also thank Karen Lopez, Sharon Burke, and Phong Nguyen for their technical contributions. Neuropathologic evaluation of the postmortem brains was performed by F. Warren Lovell, M.D. Tissue specimens were processed and stored at the Human Brain and Spinal Fluid Resource Center, Veteran’s Medical Center, Los Angeles, California, under the direction of Wallace W. Tourtellotte, M.D., Ph.D.
Footnotes
Contributions to this work were also made by other members of the Pritzker Neuropsychiatric Disorders Research Consortium and the National Institute of Mental Health Conte Center.
References
- Albrecht J, Yanagihara T. Effect of anoxia and ischemia on ribonuclease activity in brain. J Neurochem. 1979;32:1131–1133. doi: 10.1111/j.1471-4159.1979.tb04607.x. [DOI] [PubMed] [Google Scholar]
- Bahn S, Augood SJ, Ryan M, Standaert DG, Starkey M, Emson PC. Gene expression profiling in the post-mortem human brain—no cause for dismay. J Chem Neuroanat. 2001;22:79–94. doi: 10.1016/s0891-0618(01)00099-0. [DOI] [PubMed] [Google Scholar]
- Barton AJ, Pearson RC, Najlerahim A, Harrison PJ. Pre- and postmortem influences on brain RNA. J Neurochem. 1993;61:1–11. doi: 10.1111/j.1471-4159.1993.tb03532.x. [DOI] [PubMed] [Google Scholar]
- Bunney WE, Bunney BG, Vawter MP, Tomita H, Li J, Evans SJ, et al. Microarray technology: A review of new strategies to discover candidate vulnerability genes in psychiatric disorders. Am J Psychiatry. 2003;160:657–666. doi: 10.1176/appi.ajp.160.4.657. [DOI] [PubMed] [Google Scholar]
- Burke WJ, O’Malley KL, Chung HD, Harmon SK, Miller JP, Berg L. Effect of pre- and postmortem variables on specific mRNA levels in human brain. Brain Res Mol Brain Res. 1991;11:37–41. doi: 10.1016/0169-328x(91)90018-s. [DOI] [PubMed] [Google Scholar]
- Gilmore JH, Lawler CP, Eaton AM, Mailman RB. Postmortem stability of dopamine D1 receptor mRNA and D1 receptors. Brain Res Mol Brain Res. 1993;18:290–296. doi: 10.1016/0169-328x(93)90092-4. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Wester P, Winblad B, Gezelius C, Bring G, Eriksson A. The patients dying after long terminal phase have acidotic brains: Implications for biochemical measurements on autopsy tissue. J Neural Transm. 1985;61:253–264. doi: 10.1007/BF01251916. [DOI] [PubMed] [Google Scholar]
- Hargrove JL, Schmidt FH. The role of mRNA and protein stability in gene expression. FASEB J. 1989;3:2360–2370. doi: 10.1096/fasebj.3.12.2676679. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Barton AJ, Najlerahim A, McDonald B, Pearson RC. Regional and neuronal reductions of polyadenylated messenger RNA in Alzheimer’s disease. Psychol Med. 1991a;21:855–866. doi: 10.1017/s0033291700029858. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Barton AJ, Procter AW, Bowen DM, Pearson RC. The effects of Alzheimer’s disease, other dementias, and premortem course on beta-amyloid precursor protein messenger RNA in frontal cortex. J Neurochem. 1994;62:635–644. doi: 10.1046/j.1471-4159.1994.62020635.x. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: Selective mRNA vulnerability and comparison with their encoded proteins. Neurosci Lett. 1995;200:151–154. doi: 10.1016/0304-3940(95)12102-a. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Procter AW, Barton AJ, Lowe SL, Najlerahim A, Bertolucci PH, et al. Terminal coma affects messenger RNA detection in post mortem human temporal cortex. Brain Res Mol Brain Res. 1991b;9:161–164. doi: 10.1016/0169-328x(91)90143-l. [DOI] [PubMed] [Google Scholar]
- Hollams EM, Giles KM, Thomson AM, Leedman PJ. MRNA stability and the control of gene expression: Implications for human disease. Neurochem Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. J Neurochem. 2003;85:543–562. doi: 10.1046/j.1471-4159.2003.01747.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Morgan DG, Finch CE. Extensive postmortem stability of RNA from rat and human brain. J Neurosci Res. 1986;16:267–280. doi: 10.1002/jnr.490160123. [DOI] [PubMed] [Google Scholar]
- Johnston NL, Cervenak J, Shore AD, Torrey EF, Yolken RH, Cerevnak J. Multivariate analysis of RNA levels from postmortem human brains as measured by three different methods of RT-PCR. Stanley Neuropathology Consortium. J Neurosci Methods. 1997;77:83–92. doi: 10.1016/s0165-0270(97)00115-5. [DOI] [PubMed] [Google Scholar]
- Kingsbury AE, Foster OJ, Nisbet AP, Cairns N, Bray L, Eve DJ, et al. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Brain Res Mol Brain Res. 1995;28:311–318. doi: 10.1016/0169-328x(94)00219-5. [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Leonard S, Logel J, Luthman D, Casanova M, Kirch D, Freedman R. Biological stability of mRNA isolated from human postmortem brain collections. Biol Psychiatry. 1993;33:456–466. doi: 10.1016/0006-3223(93)90174-c. [DOI] [PubMed] [Google Scholar]
- Miller MT, Sleigh J, Rawlinson F, de Jongh R, Godwin Y. Amoxapine overdose: Recovery after severe metabolic acidosis (pH 6.69) and status epilepticus. Anaesth Intensive Care. 1990;18:246–248. doi: 10.1177/0310057X9001800217. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Lewis DA, Levitt P. Analysis of complex brain disorders with gene expression microarrays: Schizophrenia as a disease of the synapse. Trends Neurosci. 2001;24:479–486. doi: 10.1016/s0166-2236(00)01862-2. [DOI] [PubMed] [Google Scholar]
- Morrison MR, Griffin WS. The isolation and in vitro translation of undegraded messenger RNAs from human postmortem brain. Anal Biochem. 1981;113:318–324. doi: 10.1016/0003-2697(81)90083-x. [DOI] [PubMed] [Google Scholar]
- Morrison-Bogorad M, Zimmerman AL, Pardue S. Heat-shock 70 messenger RNA levels in human brain: Correlation with agonal fever. J Neurochem. 1995;64:235–246. doi: 10.1046/j.1471-4159.1995.64010235.x. [DOI] [PubMed] [Google Scholar]
- Netsky M. Degradation of the Cerebellum and Its Pathways. New York: McGraw Hill; 1968. [Google Scholar]
- Pardue S, Zimmerman AL, Morrison-Bogorad M. Selective postmortem degradation of inducible heat shock protein 70 (hsp70) mRNAs in rat brain. Cell Mol Neurobiol. 1994;14:341–357. doi: 10.1007/BF02088715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett CW, Marchbanks RM, Whatley SA. Characterisation of messenger RNA extracted post-mortem from the brains of schizophrenic, depressed and control subjects. J Neurol Neurosurg Psychiatry. 1988;51:325–331. doi: 10.1136/jnnp.51.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid R, Van Zwieten EJ, Swaab DF. Brain banking and the human hypothalamus—factors to match for, pitfalls and potentials. Prog Brain Res. 1992;93:83–95. doi: 10.1016/s0079-6123(08)64565-3. [DOI] [PubMed] [Google Scholar]
- Roberge RJ, Krenzelok EP. Prolonged coma and loss of brainstem reflexes following amitriptyline overdose. Vet Hum Toxicol. 2001;43:42–44. [PubMed] [Google Scholar]
- Swaab DF, Boer K. The presence of biologically labile compounds during ischemia and their relationship to the EEG in rat cerebral cortex and hypothalamus. J Neurochem. 1972;19:2843–2853. doi: 10.1111/j.1471-4159.1972.tb03822.x. [DOI] [PubMed] [Google Scholar]
- Trotter SA, Brill LB, 2nd, Bennett JP., Jr Stability of gene expression in postmortem brain revealed by cDNA gene array analysis. Brain Res. 2002;942:120–123. doi: 10.1016/s0006-8993(02)02644-6. [DOI] [PubMed] [Google Scholar]
- Van Deerlin VMD, Ginsberg SD, Lee MYL, Trojanowski JQ. The use of fixed human postmortem brain tissue to study expression in neurodegenerative diseases: Application of microdissection and mRNA amplification. In: Gregg JP, editor. Microarrays for the Neurosciences: An Essential Guide (Cellular and Molecular Neuroscience) Cambridge, Massachusetts: MIT Press; 2002. pp. 201–235. [Google Scholar]
- Vawter M, Evans SJ, Choudary PV, Tomita H, Woodruff JM, Molnar M, et al. Gender specific gene expression in postmortem human brain: Localization to sex chromosomes. [Accessed on December 5, 2003];Neuropsychopharmacology. 2003 doi: 10.1038/sj.npp.1300337. Advance online publication October 29, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester P, Bateman DE, Dodd PR, Edwardson JA, Hardy JA, Kidd AM, et al. Agonal status affects the metabolic activity of nerve endings isolated from postmortem human brain. Neurochem Pathol. 1985;3:169–180. doi: 10.1007/BF02834269. [DOI] [PubMed] [Google Scholar]
- Yokota H, Yamamoto Y, Naoe Y, Fuse A, Sato H, Unemoto K, et al. Measurements of cortical cellular pH by intracranial tonometer in severe head injury. Crit Care Med. 2000;28:3275–3280. doi: 10.1097/00003246-200009000-00025. [DOI] [PubMed] [Google Scholar]