Abstract
Transcriptional profiles within discrete human brain regions are likely to reflect structural and functional specialization. Using DNA microarray technology, this study investigates differences in transcriptional profiles of highly divergent brain regions (the cerebellar cortex and the cerebral cortex) as well as differences between two closely related brain structures (the anterior cingulate cortex and the dorsolateral prefrontal cortex). Replication of this study across three independent laboratories, to address false-positive and false-negative results using microarray technology, is also discussed. We find greater than a thousand transcripts to be differentially expressed between cerebellum and cerebral cortex and very few transcripts to be differentially expressed between the two neocortical regions. We further characterized transcripts that were found to be specifically expressed within brain regions being compared and found that ontological classes representing signal transduction machinery, neurogenesis, synaptic transmission, and transcription factors were most highly represented.
Keywords: Microarray, Cerebellum, Prefrontal cortex, Cingulate cortex, Depression, Schizophrenia, Psychiatric disorder
Introduction
1The division of the mammalian brain into structurally and functionally distinct regions implies that gene expression profiles within these regions are likely to vary in concert with structure, function, and brain circuitry. Some neural genes, in fact, are expressed only in very specific regions. For examples, proopiomelanocortin (POMC) is especially expressed in the arcuate nucleus of the hypothalamus (Khachaturian et al., 1984) and, to a lesser degree, in the nucleus of the solitary tract (Bronstein et al., 1992). Other genes, for example, those encoding opioid receptors, have a wider expression pattern (Mansour et al., 1995) but are still expressed in relation to specialized brain circuitry. Many others, such as structural and metabolic genes, are ubiquitously expressed in accord with functions necessary for all cells.
The most detailed studies of brain region-specific expression of particular genes have been carried out in rodents using in situ hybridization histochemistry (ISHH) or other methods of measuring mRNA and protein expression. While most of this work has been conducted in rodents, a significant literature also exists for human and nonhuman primates (Meador-Woodruff, 1994; Tighilet et al., 1998). Inevitably, however, these methods permit examination of expression patterns only of individual genes, and although highly informative about regional specialization and some of the molecular components of brain circuits, description of expression patterns of every gene by this approach will be painstaking. Thus, the use of microarray technology to examine transcriptional profiles within discrete brain regions can quickly define brain region-specific patterns of gene expression and can serve as a powerful first step in advancing the understanding of the specialized functions of brain structures.
DNA microarray technology can begin to describe the transcriptome of specific brain regions by allowing the simultaneous comparison of relative expression levels of thousands of transcripts. Previous studies in rodents have used this technology to identify transcripts enriched in the amygdala (Zirlinger et al., 2001) or specific hippocampal subregions (Zhao et al., 2001), or to analyze transcriptional profiles across defined brain regions in different mouse strains (Sandberg et al., 2000). Each of these studies described brain region-enriched gene expression and laid a foundation for understanding the relationship between molecular composition and functional localization in brain. We followed this line of questioning by using Affymetrix U95A Gene Chips (12,652 probes) to profile gene expression in three brain regions [cerebellum (CB), anterior cingulate cortex (AnCg), and dorsolateral prefrontal cortex (DLPFC)] from postmortem human brains. An important component of this study was that three independent academic institutions independently processed RNA from the same brains for Gene Chip hybridizations. This permitted a three-way replication of results and provided minimization of systematic error to reduce the number of false positives in the results.
The current study was designed to ask both technical and biological questions. First, can we extract quality RNA from postmortem human tissue and generate microarray data that are reproducible across different laboratories? Second, what transcripts are specifically or differentially expressed across three distinct brain structures that might impart specialized function? This article discusses the quality and reproducibility of microarray data using human postmortem brains as the source tissue and presents a comparison of the transcriptional profiles of the three brain regions analyzed. Vawter et al. (2003) describe a comparison of expression profiles from males and females in this data set.
Materials and methods
Dissection of brain tissue and RNA extraction
Human brains were collected by the Brain Donor Program at the University of California, Irvine, Department of Psychiatry and Human Behavior. All subjects used in this study are from a pool of controls that have been established for future studies in which they will be compared with brains from subjects with psychiatric disorders. Table 1 describes the gender, age of death, postmortem interval (PMI, time between death and freezing of the brain), and smoking history (an entry of “History” means the subject had a smoking history but not at the time of death) for each subject. Brains were removed at autopsy and sliced with a knife in the coronal plane into a series of approximately 0.75-cm-thick slabs. For details of the methods see Jones et al. (1992). These slabs were then snap-frozen and stored at −80°C until further handling. Three regions of interest were identified in the slabs using gross anatomical landmarks, and were excised using a fine-toothed saw. They were the lateral part of the cerebellar (CB) hemisphere, the middle part of the superior frontal gyrus (DLPFC, area 9), and the anterior quarter of the cingulate gyrus (AnCg, area 24). The entire dissection was done with the slab sitting on a sheet of dry ice. All cerebellar samples were taken from the lateral aspect of the cerebellar hemisphere, avoiding the white matter core, and thus included predominantly gray matter. Cortical samples were trimmed to include an approximately equal ratio of gray and white matter, the latter being restricted to a region approximately the same thickness as layer VI of the cortex. Samples were taken from the left side for RNA extraction, while corresponding pieces were taken from the right side for ISHH. Specific areas of interest were further dissected at −20°C on a Peltier cooling plate using either a razor or cataract knife.
Table 1.
Group composition
| Patient | Gender | Age | Brain pH | PMI (h) | Tobacco use | Laboratory's data used in final analyses |
||
|---|---|---|---|---|---|---|---|---|
| CB | DLPFC | AnCg | ||||||
| 1 | F | 75 | 6.13 | 14 | No | 1,2,3 | 2,3 | |
| 2 | M | 57 | 6.94 | 14 | Unknown | 1,2,3 | 1,2,3 | 2,3 |
| 3 | F | 77 | 6.55 | 16 | Yes | 3 | 1,2,3 | 1,2,3 |
| 4 | M | 55 | 7.14 | 22.3 | No | 2,3 | 1,2,3 | 1,2,3 |
| 5 | F | 53 | 6.98 | 16.5 | History | 2 | 1,2,3 | 1,2,3 |
| 6 | M | 71 | 6.86 | 9.8 | No | 1,2,3 | 1,2,3 | 1,2,3 |
| 7 | M | 72 | 6.84 | 24.5 | No | 1,2,3 | 1,2,3 | 2,3 |
| 8 | M | 88 | 7.03 | 15 | History | 2,3 | 1,2,3 | 1,2,3 |
| 9 | M | 68 | 6.43 | 23 | Yes | 1,2,3 | 1,2,3 | 2,3 |
| 10 | F | 75 | 6.4 | 21 | No | 1,2,3 | 1,2,3 | 2,3 |
| 11 | F | 82 | 6.71 | 28 | Yes | 2,3 | 1,2,3 | 1,2,3 |
| 12 | M | 82 | 6.49 | 30.8 | Yes | 1,2,3 | 2,3 | 2,3 |
| 13 | F | 69 | 6.12 | 21 | No | 1,2,3 | 1,2,3 | |
Total RNA was extracted from each brain region using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and quality verified by electrophoresis on formaldehyde denaturing agarose gels to ensure the integrity of 18S and 28S ribosomal bands. Final purification of total RNA was achieved using RNeasy mini-columns (Qiagen, Valencia, CA, USA) and quantified by spectrophotometric measurement of absorbance at 260 nm. RNA samples from each region were then divided into aliquots that were distributed to the three laboratories.
Gene chip hybridizations and data analysis
Ten micrograms of total RNA was used to prepare biotinylated cRNA for hybridization to Affymetrix U95A arrays, following the standard Affymetrix protocol. Hybridization was allowed to proceed 16 –20 h then Gene Chips were washed and stained according to the Affymetrix fluidics station EukGE wash 2v3 protocol, followed by scanning in an Agilent Gene Array scanner.
Data were initially collected independently at each of the three laboratories with MAS 5.0 software (Affymetrix, Santa Clara, CA, USA) using the default algorithms to generate “signal” values and “present/absent/marginal” calls. Signal intensities were scaled using an arbitrary target value of 150 to represent the 70th percentile of signal from all probes. Data were exported as tab delimited text files to Gene Spring 4.1.2 software (Silicon Genetics, Redwood City, CA, USA) or to Excel (Microsoft Corporation, Bothell, WA, USA). Quality analysis was performed in Excel by calculating the correlation coefficients of the signal intensity values for each chip in comparison to every other chip within its group (same region, same laboratory). A chip was rejected from analysis if its average correlation coefficient was 2 standard deviations from the mean of the correlation coefficients for the entire group. Originally, 13 independent brain samples were processed at laboratories 1 and 3, and 10 of those 13 were processed at laboratory 2. After rejecting low-quality data the following remained: laboratory 1, 7 CB, 7 AnCg, and 11 DLPFC; laboratory 2, 8 CB, 9 AnCg, and 10 DLPFC; laboratory 3, 11 CB, 12 AnCg, and 12 DLPFC. Specific samples passing quality criteria are listed in Table 1.
Using Gene Spring, data were further normalized by chip to the 50th percentile of all values receiving a “present” or “marginal” call from the MAS 5.0 software. Welch t tests were performed in Gene Spring for all 12,652 probe sets, using a P-value cutoff of 0.05 and no multiple testing corrections. Groups that were compared are described under Results. Experiment tree clustering was performed in Gene Spring, using a standard correlation with a separation ratio of 1.0 and a minimum distance of 0.001.
Gene ontologies were obtained from the Genomics Institute of the Novartis Research Foundation (GNF) at http://expression.gnf.org/cgi-bin/index.cgi (Su et al., 2002). Gene Ontology tables were generated using keyword analysis for all U95A probe sets as well as the probe sets within the given results lists, to determine enrichment of gene families within the results.
Cloning of probes and in situ hybridization histochemistry
For probes for ISHH, p21 activated kinase 3 (PAK-3) and checkpoint suppressor 1 (CHES1) fragments were cloned from reverse-transcribed human cDNA using a PCR II TOPO cloning kit (Invitrogen) and sequence verified. The PAK-3 probe was complementary to bases 460 – 697 (AF068864) and CHES1 was complementary to bases 1865–2270 (U68723).
Ten-micrometer thick frozen sections were cut from the samples taken from the right side of each brain using a cryostat. Sections were mounted onto poly-L-lysine-coated slides and frozen at −80°C until used for ISHH. The slide-mounted sections were fixed in 4% paraformaldehyde for 1 h, followed by three washes in 2× SSC (1× SSC is 150 mM sodium chloride, 15 mM sodium citrate). The sections were then placed in a solution containing acetic anhydride (0.25%) in triethanolamine (0.1 M, pH 8) for 10 min at room temperature, rinsed in distilled water, and dehydrated through graded alcohols (50, 75, 85, 95, and 100%). After air-drying, the sections were hybridized with a 35S-labeled cRNA probe. Negative control sections were treated with RNase for 1 h at 37°C immediately following triethanolamine incubation.
Probes were synthesized from linearized templates using 125 μCi [35S]UTP, 125 μCi [35S]CTP, 150 μM each of ATP and GTP, 12.5 mM dithiothreitol, 20 U RNase inhibitor, and 6 U polymerase (T7 for CHES1 and Sp6 for PAK-3). The reactions were incubated for 90 –120 min at 37°C. Then, the probes were separated from unincorporated nucleotides over Micro Biospin 6 columns (Bio-Rad, Hercules, CA). The probes were diluted in hybridization buffer (50% formamide, 10% dextran sulfate, 3× SSC, 50 mM sodium phosphate buffer, pH 7.4, 1× Denhardt's solution, 0.1 mg/ml yeast tRNA, and 10 mM dithiothreitol) to yield approximately 106 dpm/40 μl and pipetted onto the slide-mounted sections. Coverslips were applied and the slides were placed inside a humidified box overnight at 55°C. Following hybridization, the coverslips were removed and the sections rinsed and washed twice in 2× SSC for 5 min each, then incubated for 1 h in RNase (200, μg/ml in Tris buffer containing 0.5 M NaCl, pH 8) at 37°C. The sections were washed in increasingly stringent solutions of SSC (2×, 1×, and 0.5×) for 5 min each, followed by incubation for 1 h in 0.1× SSC at 65°C. After being rinsed in distilled water, the sections were dehydrated through graded alcohols, air-dried, and exposed to a Kodak XAR film (Eastman Kodak, Rochester, NY, USA) for 10 days. The films were then developed using a Kodak X-OMAT processor.
Results
HG-U95A Affymetrix Gene Chips were hybridized with biotinylated cRNA prepared from CB, DLPFC, and AnCg from both male and female postmortem brains. Each cRNA sample was prepared and hybridized independently at the three laboratories participating in the study. Some chips were discarded from analysis due to quality measures, as described under Materials and Methods. Those that were used for the final analyses are described in Table 1.
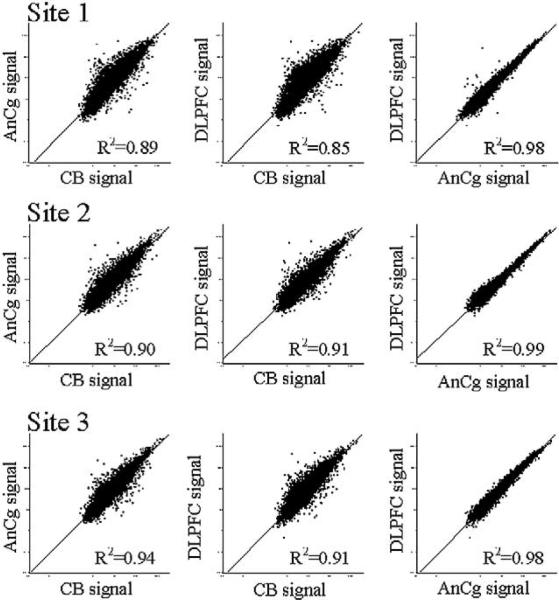
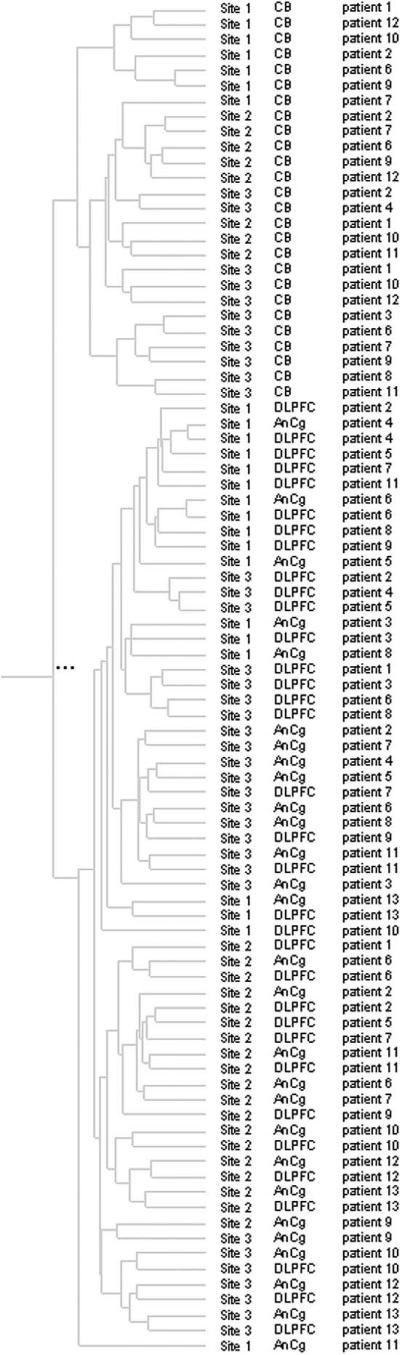
Fig. 1 shows scatterplots of brain region comparisons for each of the three laboratories independently. An average of the “signal” values for all chips representing the given brain region examined at a laboratory is plotted, as indicated, on the axes. These data show average standard correlations of 0.98 for DLPFC versus AnCg, 0.91 for AnCg versus CB, and 0.89 for DLPFC versus CB, showing a higher correlation of expression profiles between the two cerebral cortical regions than between either cortical region and the cerebellar cortex. Fig. 2 shows clustering of the data by individual chips. Only genes that were reliably detected (average detection P value ≤0.05) in at least one brain region were used to cluster the data sets. The cluster tree shows a major branch point with two distinct clusters consisting of all CB data in one and all AnCg and DLPFC data in the other. The second-order separation is based primarily on processing laboratory, with laboratory 2 being most distinct. The third-order separation is less dramatic but based mostly on individuals, with AnCg and DLPFC samples from the same subject tending to cluster together. Separation of AnCg and DLPFC into distinct clusters could not be achieved.
Fig. 1.
Scatterplots of signal intensity values from CB, DLPFC, and AnCg. Signal values are derived from MAS 5.0 for all probe sets scaled to an identical target 70th percentile value. Axes are log scale with arbitrary units. Average signal for all probe sets on the U95A chip are plotted for the various brain regions and sites as indicated in the figure. R2 correlation values are given in the lower right corner of each graph.
Fig. 2.
Cluster analysis of data by individual samples. All data sets used in the final analysis were clustered using GeneSpring's Experiment Tree clustering function using a standard correlation and all genes that had an average detection P value of 0.05 in at least one brain region as the input gene set. The figure lists each sample by site, brain region, and patient number.
Parametric Welch t tests were performed, using all 12,652 probe sets on the array, to identify genes differentially expressed between the three brain regions, using a P value threshold of 0.05. Table 2 lists the number of genes that were found to be different in comparisons made both within laboratories and across all laboratories. This table shows that more than 3000 transcripts were found to be differentially expressed between CB and either of the cerebral cortical regions at each laboratory, and that approximately 1600 of these were reproducible across all three laboratories. The median fold change for CB versus, the cortical regions was 1.86 for transcripts at individual laboratories and 2.20 for those transcripts in common between all laboratories. Comparison of AnCg to DLPFC, however, revealed as few as 559 differentially expressed transcripts at one laboratory with only four of these reproducible across all laboratories, likely because of large number of expected false positives given the large number of observations (12,652 probe sets). Taking the intersection of the lists comparing either of the two cerebral cortical regions with CB shows that 969 transcripts were reproducibly differentially expressed between CB and both cortical regions. All comparisons of either of the cerebral cortical regions to CB showed a highly skewed distribution with many more transcripts enriched in cerebral cortex than were enriched in CB. The reproducible differences between AnCg and CB found 1272 transcripts enriched in AnCg and 359 enriched in CB. Between DLPFC and CB, 1282 transcripts were reproducibly enriched in DLPFC while only 262 were reproducibly enriched in CB. Interestingly, an average of 20% more transcripts were detected in the cortical regions relative to CB using MAS 5.0 default algorithms (data not shown). The comparisons between AnCg and DLPFC yielded only two transcripts reproducibly enriched in each brain region, relative to the other. These include heat shock binding protein 1 (HSBP1) and the purinergic receptor, P2Y1, which were enriched in AnCg relative to DLPFC, and cocaine- and amphetamine-regulated transcript (CART) and an unidentified transcript, KIAA0084, which were enriched in DLPFC relative to AnCg. In all comparisons the majority of transcripts found to be differentially expressed (>75%) were reliably detected (average detection P value <0.05) in at least one brain region.
Table 2.
Number of genes significantly different between brain regions
| CB × AnCg | CB × DLPFC | AnCg × DLPFC | CB × cortex | |
|---|---|---|---|---|
| Site 1 | 3531 | 3337 | 716 | 2625 |
| Site 2 | 3381 | 3076 | 559 | 2405 |
| Site 3 | 4395 | 5503 | 2697 | 3493 |
| All sites | 1631 | 1544 | 4 | 969 |
We also evaluated the number of transcripts found to be specific to one of the brain regions relative to the other two regions analyzed. Gene transcripts were considered specific to a given brain region if the average of the detection P values of all chips within that brain region were equal to or less than 0.05 and if all detection P values on chips representing the compared brain region(s) were greater than 0.06. (A detection P value greater than 0.06 is called “absent” by MAS 5.0 software under the default criteria.) So our criteria require that transcripts considered to be region specific have an average detection P value meeting MAS 5.0 standards for a “present” call in one brain region and always meet the “absent” call standards in the comparative brain region(s). Overall, approximately 30% of the probe sets on the array were reliably detected in DLPFC and AnCg (3748 and 3623, respectively) while only 22% were reliably detected in CB (2736). Table 3 shows that 15 transcripts were specifically detected in CB and not detected in either of the cerebral cortical regions and that 74 transcripts were detected in both cerebral cortical regions but undetected in CB. No transcripts were specifically detected in only one of the two cerebral cortical regions.
Analysis of the transcripts in Table 3 by functional classification using Gene Ontology (GO) tools (Su et al 2002) identifies functional families enriched in the results set. Table 4 lists a GO analysis of the transcripts found to be specific to CB or cortex as well as their representation in the results set relative to their representation on the U95A Gene Chip. Ontological families that have a higher representation in the results set than on the Gene Chip can be considered enriched in the results and are more likely to be significant as determinants of functional differences between cerebral cortex and CB.
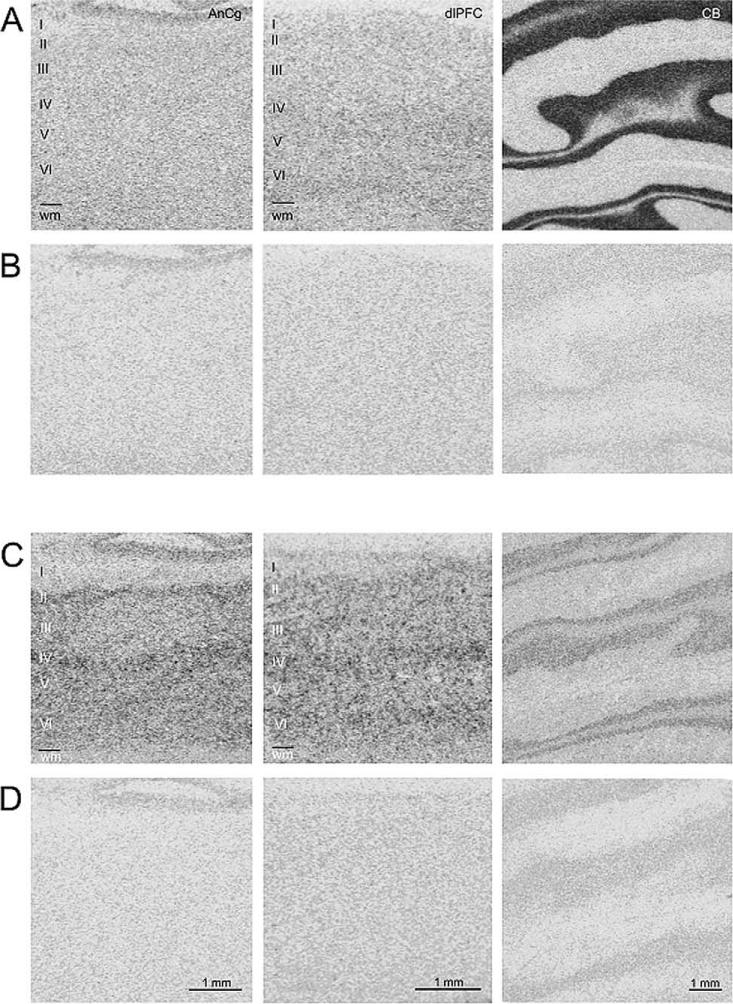
Two transcripts that were specific to either cerebral cortex or CB and that had not been previously reported as such were chosen for evaluation by ISHH. One was specifically detected in CB and the other specifically detected in both cerebral cortical areas. Fig. 3 shows that Checkpoint suppressor 1, detected only in CB by microarray, has a strong specific signal in CB tissue, with rich label in granule cells, and some specific signal in both AnCg and DLPFC, with diffuse labeling throughout the neocortical layers. PAK-3, detected only in cerebral cortex by microarray, shows strong specific signal in both AnCg and DLPFC, with enrichment in layers ii, iv, and vi, and some diffuse specific signal in CB.
Fig. 3.
In situ hybridization histochemistry of PAK-3 and CHES1. The specific signals from 35S-labeled riboprobes relative to RNase-negative controls are shown for representative sections from AnCg, DLPFC, and CB. Rows A and C show specific signal for CHES1 and PAK-3, respectively, across labeled sections. Rows B and D show signal from RNase controls for CHES1 and PAK-3, respectively, of sections adjacent to those immediately above.
Discussion
We have compared transcriptional profiles from two regions of the cerebral neocortex, DLPFC and AnCg, and the cerebellar hemisphere using human postmortem brains as the source tissue. We independently replicated the study at three different laboratories to reduce false positives due to systematic error. The analysis found approximately 1600 transcripts differentially expressed between CB and either AnCg or DLPFC but only four transcripts differentially expressed between the two cerebral cortical regions. Of the differentially expressed transcripts, 74 were specifically detected in cerebral cortex and not detected in CB, 15 were specifically detected in CB and not in cerebral cortex, while no transcripts were specifically detected in one of the cerebral cortical areas that were not expressed in the other. Furthermore, approximately 20% more transcripts were reliably detected in AnCg and DLPFC than were reliably detected in CB.
It should be emphasized that, because each laboratory started with the same total RNA samples, we are able to address systematic error due to technical variance across the laboratories. We can also address random error due to biological variation because we assayed separately the brain regions of each individual; however, the number of biological replicates was not large (n = 10 –13) and biological replicates were not run within a laboratory. Thus, the interplay between biological variation and technical variation cannot be adequately addressed in the current study but is being evaluated with larger ongoing studies.
Cluster analysis of all individual arrays used in the current study showed a distinct separation of CB and the two cerebral cortical samples into two unique clusters, while AnCg and DLPFC were indistinguishable by this analysis. Second to separation based on CB and cortex differences, the next largest factor was processing laboratory. This is probably due in large part to scanner settings and calibrations. In the current study laboratory 2 had older Affymetrix scanner specifications, which have much higher PMT and voltage settings, while laboratories 1 and 3 had newer Affymetrix scanner specifications with lower PMT and voltage settings. This is the primary reason why between-laboratory comparisons of the same samples were not performed and why reproducibility comparisons were limited to within-laboratory results. All three laboratory's scanners have since been recalibrated and tuned to each other to allow the direct pooling of raw data in ongoing and future studies. Finally, individual patients were the third largest factor in cluster analysis. Data sets from DLPFC and AnCg samples from the same patients tended to cluster as nearest-neighbor pairs more frequently than they clustered with data sets from the same brain region from other subjects. These observations have been replicated in other patient groups in ongoing studies (J. Li, personal communication). Other factors listed in Table 1, such as PMI, age, and gender, were not evident in cluster analysis as significantly contributing to the transcriptional profiles.
An average of approximately 4200 genes were found differentially expressed at each laboratory between divergent samples (CB vs either cerebral cortical region) and approximately 1600 (~38%) of these differences were reproduced at all three laboratories. The large number of genes found reproducibly different between CB and cerebral cortex far exceeds the number of genes expected by random chance with the statistical filters used, and suggests that neocortex and cerebellar cortex have a high percentage of truly differentially expressed genes. This is consistent with expectations based on the different developmental histories, connections, cell types, and functions of these telencephalon-and hindbrain-derived structures. Because of the high degree of true differential expression between CB and cerebral cortex, it is likely that there are a large number of transcripts falling across the continuum of fold-change magnitudes and that the 1600 reproducible changes are skewed toward representation of the higher-magnitude differences. This is supported by the fact that the median fold change in the common data set is of higher magnitude than the median fold change in the individual laboratory's data sets. The large number of genes found differentially expressed at one laboratory that failed to reproduce, then, is likely due to the presence of a large number of transcripts with a moderate fold change magnitude that failed to reproducibly meet our criterion stringency for all laboratories. Therefore in addition to the expected number of false positives at any single laboratory there are probably a large number of false negatives absent from the intersection between laboratories. Both would contribute to the large number of transcripts found differentially expressed at one laboratory but that failed to reproduce across all laboratories.
When comparing similar tissue (AnCg and DLPFC), the three laboratories independently identified 559, 716, and 2697 differentially expressed genes; however, only 4 were reproducible across all three laboratories. The larger number of differences found at laboratory 3 is probably in part due to the fact that a larger number of samples were used for final analysis at this laboratory, providing more statistical power and reducing the number of false negatives. With the exception of the relatively large number of genes found differentially expressed at laboratory 3, the number of genes that did not reproduce across laboratories is within the number of false positives expected by random chance at any single laboratory, since approximately 600 false positives would be expected out of 12,000 genes with a P-value threshold of 0.05 in any single analysis. Furthermore, if the majority of genes each laboratory found differentially expressed between the two cerebral cortical regions were indeed false positives, then the intersection between any two lists should overlap only by approximately 5%, and the intersection of all three lists should overlap only by approximately 0.25%, which is in fact close to what is observed. The lack of many genes found reproducibly differentially expressed between AnCg and DLPFC supports the hypothesis that these two neocortical areas are closely related. This is further evidenced by the inability to separate these two structures by cluster analysis. However, there are likely to be many differences between these two areas that were missed by the current analysis due to the inefficiencies of Gene Chips in reliably detecting relatively low-magnitude changes (Evans et al., 2002). This is supported by the larger number of genes found differentially expressed at the laboratory with the largest number of chips used in the final analysis.
Our observations of reproducing experiments at separate laboratories suggest that when comparing similar samples the majority of results from any single data set analyzed with standard statistical tools are false positives, making confirmation studies laborious. Data sets generated from highly divergent samples also contain a large number of false positives; however, the number of true positives has a much higher representation in the results set and is thus likely to be more efficiently confirmed. Care must also be taken in pooling raw data across laboratories. Even following normalization schemes, data from the current study still clustered by processing laboratory more strongly than by biological sample. This suggests that pooling of data across laboratories should be done only if precautions are taken to measure and minimize technical variation. However, comparison of results following analysis of biological groups within laboratories can provide a powerful means to minimize systematic error in replication studies.
In the current study 89 transcripts were found specifically expressed in either cortex or CB, relative to each other. Approximately 20% of these (18 of 89) were in agreement with previous nonarray studies, providing support for the current findings. These include 4 of the 14 transcripts specifically detected in CB and 14 of the 74 transcripts detected in cortex but not in CB. Fat tumor suppressor 2 (FAT2, MEGF1) (Mitsui et al., 2002), GABAA receptor α6 subunit (GABAA 6) (Mohler et al., 1990), Mab-21-like 1 (Mariani et al., 1998), and M-cadherin (Bahjaoui-Bouhaddi et al., 1997) have all previously been reported as expressed in CB and not in cortex. In contrast, human brain factor 1 (HBF1) (Murphy et al., 1994), neurogranin (NRGN) (Represa et al., 1990), leukocyte antigen-6 (Ly6) (Horie et al., 1998), neuropeptide Y (NPY) (Brene et al., 1989), somatostatin (SST) (Lowe et al., 1987), GABAA receptor α5 subunit (GABAA 5) (O'Hara et al., 1995), Ephrin B3 (Tang et al., 1997), regulator of G-protein signaling 4 (RGS4) (Gold et al., 1997), neuronal SHC-like protein (nSHC) (Nakamura et al., 1998), Mads box transcription enhancer factor 2, polypep-tide C (MEF2C) (Leifer et al., 1993), H-cadherin (Takeuchi et al., 2000), calmodulin-dependent protein kinase II; alpha (CamKII alpha) (McGuinness et al., 1985), Slit-1 (Itoh et al., 1998), and neuronal nicotinic cholinergic receptor alpha, polypeptide 7 (CHRNA7) (Seguela et al., 1993), have previously been reported to have high expression in cortex and little to no expression in CB. Three transcripts, including voltage-gated potassium channel, delayed rectifier, subfamily S (KCNS1) (Salinas et al., 1997), CamKII gamma (Vallano et al., 2000), and 5-hydroxytryptamine receptor 2A (5HT2A) (Eastwood et al., 2001) have been previously detected in CB but we detected these transcripts only in cerebral cortex. This could be due to the inclusion of deep cerebellar nuclei in previous studies whereas our samples were from cerebellar cortex alone. It could also be explained by the sensitivity limits of microarrays that fail to detect many low-abundance transcripts (Evans et al., 2002). However, both the present and previous studies suggest that these three transcripts have a much higher level of expression in cortex than in CB.
Analysis of the specifically expressed 89 transcripts using ontological tools based on GO classification (Su et al., 2002) found that genes involved in signal transduction, neurogenesis, synaptic transmission, and transcription were the most highly represented classes of the region-specific expressers. Furthermore, all of these classes were enriched in this set of 89 genes relative to their representation on the Gene Chip. Given the small number of specifically expressed transcripts in some of the ontological classes it is impossible to accurately estimate the probabilities associated with some of these ratios. Nonetheless, enrichment of these functional families in the results set suggests that they were not the result of random chance and may play a significant role in the maintenance and perhaps development of the functional specialization of cerebral cortex and cerebellum.
Several genes found by the current study to be enriched in cortex have been previously implicated in psychiatric disorders. For example, RGS4 regulation in prefrontal cortex has been implicated in schizophrenia (Mirnics et al., 2001); NPY in bipolar disorder (Caberlotto and Hurd, 1999), cholecystokinin (CCK) in depression (Lofberg et al., 1998), somatostatin in mania (Sharma et al., 1995), schizophrenia (Sharma et al., 1994), and Alzheimer's disease (Minthon et al., 1997), and 5HT2A in major depression and suicide (Turecki et al., 1999). While the expression of some of these well-known genes in frontal cortex may not be surprising, it emphasizes that the microarray analysis is revealing unique features of the transcriptome that might be important for understanding brain pathology. Thus, some of the very gene products that appear unique to cerebral cortex over cerebellum are those that are associated with disorders of higher cortical function, particularly psychosis.
We investigated two transcripts using ISHH in the current study that had not been previously described as enriched in CB or cerebral cortex. CHES1, found in CB but not cerebral cortex by microarray, proved to be highly expressed in CB; however, it also showed diffuse specific labeling throughout neocortex when investigated using ISHH. CHES1 is a member of the forkhead family of transcription factors and has been implicated in delaying progress through the cell cycle in response to UV-induced DNA damage (Pati et al., 1997). Although this function may not be specifically relevant to CB function it does put CHES1 in a role to sense cellular state and control timing through cell progression, which might be involved in cell specialization. PAK-3, detected by microarray in cerebral cortex but not CB, showed strong specific signal in neocortical layers, but also showed weak labeling in CB when investigated by ISHH. PAK-3 is a member of a large family of p21-activated kinases that participate in Rac/Rho/MAPK signal transduction (Bagrodia et al., 1995; Manser et al., 1995). Interestingly, this gene has been implicated in X-linked mental retardation syndrome (Allen et al., 1998) and thus may also play a role in cognitive function.
The fact that we detected signal for these transcripts by ISHH in brain regions that showed no signal in the microarray studies further underlines the sensitivity limits of microarray technology. However, the brain regions predicted by microarray to specifically express these two transcripts did prove to show highly enriched expression. In light of these ISHH results, caution must be given to the interpretation of the large number of transcripts found differentially expressed, but present in all regions analyzed, as there are likely to be a number of false positives in these data as discussed above. However, the specific transcripts named in this article and the gene ontology analyses reported were restricted to those transcripts found by microarray to be specifically regionally expressed, which are likely to be at least highly differentially expressed. This is evidenced by the two transcripts analyzed by ISHH.
The current study found very few differences between DLFPC and AnCg, even though these two cortical regions have quite different functions. DLPFC receives primary input from the mediodorsal nucleus of the thalamus (Jones, 1998) and has been repeatedly shown to be hypoactive in schizophrenia (Weinberger et al., 2001). The AnCg receives primary input from the anterior nuclei of the thalamus, has connections to limbic areas, and is implicated in motor and endocrine outflow (Vogt et al., 1992). The fact that these two areas, although different in function, share a similar transcriptome is undoubtedly a reflection of their possession of the same fundamental cell types, internal circuitry, transmitters, and a similar developmental history. It is therefore the different extrinsic connections of the two areas that provide their functional individuality. There are likely to be small-magnitude differences, however, or differences in low-abundance genes that contribute to significant functional variation but are below the current detection limits of DNA microarray technology.
To conclude, in characterizing the transcripts specifically expressed in functionally distinct brain regions we found the highest representation of gene families to be those that would be expected to provide functional specialization. Many of these transcripts had not been previously described as having specific expression profiles, including two transcripts that we clearly showed, using ISHH, to be regionally enriched. Microarray studies, such as the current one, can provide unique insight into the specific expression profiles of transcripts important to functional specialization and lead to a better understanding of brain structure and function.
Table 3a.
Transcripts specifically detected in cortex and not detected in CB
| Affy ID | Description | Affy ID | Description |
|---|---|---|---|
| 33223-at | KIAA0561 protein | 37951-at | Deleted in liver cancer 1 |
| 37572-at | Cholecystokinin | 34724-at | Glucocorticoid receptor DNA binding factor 1 |
| 41544-at | Serum-inducible kinase | 40388-at | Discs, large (Drosophila) homolog-associated protein 1 |
| 39605-at | Forkhead box G1B | 35469-at | 5-Hydroxytryptamine (serotonin) receptor 2A |
| 40655-at | Huntingtin-associated protein interacting protein (duo) | 38870-at | GDNF family receptor alpha 2 |
| 37516-at | KIAA0749 protein | 32863-at | Calcium/calmodulin-dependent protein kinase IG |
| 33925-at | Neurogranin (protein kinase C substrate, RC3) | 38956-at | N-Acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| 36394-at | Lymphocyte antigen 6 complex, locus H | ||
| 39321-at | GABA-A receptor, alpha 5 | 41355-at | B-cell CLL/lymphoma 11A (zinc finger protein) |
| 36764-at | Calcium channel, voltage-dependent, gamma subunit 3 | 32847-at | Myosin, light polypeptide kinase |
| 38604-at | Neuropeptide Y | 39575-at | Hypothetical protein MOT8 |
| 33785-at | Brain-specific angiogenesis inhibitor 2 | 31813-at | Calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha |
| 37849-at | Slit homolog 1 (Drosophila) | ||
| 41708-at | KIAA1034 protein | 38203-at | K+ intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| 38938-at | T-box, brain, 1 | ||
| 40528-at | LIM homeobox protein 2 | 32105-f-at | Calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma |
| 36073-at | necdin homolog (mouse) | ||
| 34582-at | Solute carrier family 1, member 2 | 1519-at | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) |
| 36261-at | B/K protein | ||
| 32368-at | Protocadherin 8 | 35946-at | NEL-like 1 (chicken) |
| 40375-at | Early growth response 3 | 38280-s-at | Homo sapiens mRNA; cDNA DKFZp761J0523 (from clone DKFZp761J0523) |
| 36707-s-at | Serine/threonine kinase 9 | ||
| 37049-g-at | Translocase of outer mitochondrial membrane 34 | 39849-at | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| 37568-at | Human clone 23560 mRNA sequence | ||
| 38420-at | Collagen, type V, alpha 2 | 41036-at | Hypothetical protein FLJ12242 |
| 31690-at | Homo sapiens, clone MGC:13241 IMAGE:4026312, mRNA | 40913-at | ESTs |
| 40544-g-at | Achaete-scute complex-like 1 (Drosophila) | ||
| 33372-at | RAB31, member RAS oncogene family | 41083-at | Homo sapiens, clone IMAGE:3908182, mRNA, partial cds |
| 33074-g-at | p21 (CDKN1A)-activated kinase 3 | ||
| 40782-at | Short-chain dehydrogenase/reductase 1 | 38275-at | Similar to hypothetical 34.0-kDa protein ZK795.3 in chromosome IV |
| 37782-at | Somatostatin | ||
| 469-at | Ephrin-B3 | 35663-at | Neuronal pentraxin II |
| 39242-at | Synaptotagmin V | 33743-at | KIAA0534 protein |
| 34272-at | Regulator of G-protein signaling 4 | 37785-at | GTP-binding protein |
| 34382-at | Doublecortex; lissencephaly, X-linked (doublecortin) | 39566-at | Cholinergic receptor, nicotinic, alpha polypeptide 7 |
| 36765-at | Chromosome 20 open reading frame 28 | 35350-at | B-cell RAG-associated protein |
| 1511-at | Neuronal Shc | 35022-at | SRY (sex-determining region Y)-box 5 |
| 37712-g-at | MADS box transcription enhancer factor 2, polypeptide C | 39227-at | Protein tyrosine phosphatase, receptor type, T |
| 31815-r-at | Low-density lipoprotein receptor-related protein 3 | ||
| 36065-at | LIM domain binding 2 | 32837-at | 1-Acylglycerol-3-phosphate O-acyltransferase 2 |
| 483-g-at | Cadherin 13, H-cadherin (heart) | 39580-at | KIAA0649 gene product |
| 40808-at | Chromogranin A (parathyroid secretory protein 1) | 118-at | Inositol 1,4,5-trisphosphate 3-kinase A |
| 38338-at | Related RAS viral (r-ras) oncogene homolog |
Table 3b.
Transcripts specifically detected in CB and not detected in cortex
| Affy ID | Description |
|---|---|
| 1535-at | Checkpoint suppressor 1 |
| 38202-at | FAT tumor suppressor homolog 2 (Drosophila) |
| 36389-at | Class MHC-restricted T cell-associated molecule |
| 34025-at | GABA-A receptor, alpha 6 |
| 39297-at | mab-21-like 1 (C. elegans) |
| 514-at | Cas-Br-M (murine) ectropic retroviral transforming sequence b |
| 36271-at | AB028947:Homo sapiens mRNA for KIAA1024 protein |GenBank==AB028947 |
| 35579-at | SLAC2-B |
| 32406-at | KIAA0889 protein |
| 1296-at | Cadherin 15, M-cadherin (myotubule) |
| 819-at | Tissue inhibitor of metalloproteinase 4 |
| 37478-at | Secretagogin, EF-hand calcium binding protein |
| 36396-at | Homo sapiens mRNA; cDNA DKFZp586N2020 (from clone DKFZp586N2020) |
| 34331-at | EphB1 |
| 38951-at | Collagen, type XIII, alpha 1 |
Table 4a.
Cortex-specific transcript ontology
| Gene ontology | Ratio of enrichment | No. of transcripts |
|---|---|---|
| None | 0.74 | 14 |
| Signal transduction | 2.07 | 14 |
| Neurogenesis | 7.58 | 8 |
| Synaptic transmission | 7.65 | 7 |
| Transcription factor | 2.14 | 6 |
| Integral plasma membrane protein | 0.96 | 6 |
| Protein phosphorylation | 3.11 | 4 |
| Calmodulin binding | 45.45 | 4 |
| Brain development | 16.49 | 3 |
| Receptor protein tyrosine kinase | 11.62 | 3 |
| Peripheral plasma membrane protein | 3.30 | 3 |
| Tumor suppressor | 2.62 | 3 |
| Integral membrane protein | 1.95 | 3 |
| Cell–cell signaling | 1.60 | 3 |
| DNA binding | 1.54 | 3 |
| Extracellular space | 1.48 | 3 |
| Transcription activating factor | 3.82 | 3 |
| Peptide hormone | 10.65 | 2 |
| GTPase activator | 8.12 | 2 |
| Digestion | 7.58 | 2 |
| Potassium transport | 6.20 | 2 |
| Muscle development | 5.78 | 2 |
| Central nervous system development | 5.33 | 2 |
| Protein serine/threonine kinase | 3.02 | 2 |
| Negative control of cell proliferation | 2.09 | 2 |
| Protein binding | 1.38 | 2 |
| Small molecule transport | 1.30 | 2 |
| Transcription from Pol II promoter | 1.28 | 2 |
| Cell adhesion | 0.89 | 2 |
| Oncogenesis | 0.83 | 2 |
| Membrane fraction | 0.76 | 2 |
| Nucleus | 0.42 | 2 |
Table 4b.
CB-specific transcript ontology
| Gene ontology | Ratio of enrichment | No. of transcripts |
|---|---|---|
| None | 1.31 | 5 |
| Plasma membrane | 6.21 | 4 |
| Signal transduction | 2.35 | 3 |
| Cell adhesion | 6.60 | 3 |
| Integral plasma membrane protein | 1.69 | 2 |
Acknowledgments
This work was supported by the Pritzker Consortium for Severe Psychiatric Disorders, Family Philanthropic Foundation, and NIH Conte Center Grant L99MH60398.
References
- Allen KM, Gleeson JG, Bagrodia S, et al. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat. Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- Bagrodia S, Derijard B, Davis RJ, Cerione RA. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J. Biol. Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- Bahjaoui-Bouhaddi M, Padilla F, Nicolet M, Cifuentes-Diaz C, Fell-mann D, Mege RM. Localized deposition of M-cadherin in the glomeruli of the the granular layer during the postnatal development of mouse cerebellum. J. Comp. Neurol. 1997;378:180–195. [PubMed] [Google Scholar]
- Brene S, Lindefors N, Kopp J, Sedvall G, Persson H. Regional distribution of neuropeptide Y mRNA in postmortem human brain. Brain Res. Mol. Brain Res. 1989;6:241–249. doi: 10.1016/0169-328x(89)90070-3. [DOI] [PubMed] [Google Scholar]
- Bronstein DM, Schafer MK, Watson SJ, Akil H. Evidence that beta-endorphin is synthesized in cells in the nucleus tractus solitarius: detection of POMC mRNA. Brain Res. 1992;587:269–275. doi: 10.1016/0006-8993(92)91007-2. [DOI] [PubMed] [Google Scholar]
- Caberlotto L, Hurd YL. Reduced neuropeptide Y mRNA expression in the prefrontal cortex of subjects with bipolar disorder. Neuro-Report. 1999;10:1747–1750. doi: 10.1097/00001756-199906030-00022. [DOI] [PubMed] [Google Scholar]
- Eastwood SL, Burnet PW, Gittins R, Baker K, Harrison PJ. Expression of serotonin 5-HT(2A) receptors in the human cerebellum and alterations in schizophrenia. Synapse. 2001;42:104–114. doi: 10.1002/syn.1106. [DOI] [PubMed] [Google Scholar]
- Evans SJ, Datson NA, Kabbaj M, et al. Evaluation of Affymetrix Gene Chip sensitivity in rat hippocampal tissue using SAGE analysis. Eur. J. Neurosci. 2002 doi: 10.1046/j.1460-9568.2002.02097.x. in press. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J. Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M, Okutomi K, Taniguchi Y, Ohbuchi Y, Suzuki M, Takahashi E. Isolation and characterization of a new member of the human Ly6 gene family (LY6H). Genomics. 1998;53:365–368. doi: 10.1006/geno.1998.5462. [DOI] [PubMed] [Google Scholar]
- Itoh A, Miyabayashi T, Ohno M, Sakano S. Cloning and expressions of three mammalian homologues of Drosophila Slit suggest possible roles for Slit in the formation and maintenance of the nervous system. Brain Res. Mol. Brain Res. 1998;62:175–186. doi: 10.1016/s0169-328x(98)00224-1. [DOI] [PubMed] [Google Scholar]
- Jones EG. A new view of specific and nonspecific thalamocortical connections. Adv. Neurol. 1998;77:49–71. discussion 72–73. [PubMed] [Google Scholar]
- Jones EG, Hendry SH, Liu XB, Hodgins S, Potkin SG, Tourtel-lotte WW. A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J. Neurosci. Methods. 1992;44:133–144. doi: 10.1016/0165-0270(92)90006-y. [DOI] [PubMed] [Google Scholar]
- Khachaturian H, Lewis ME, Haber SN, Akil H, Watson SJ. Proopiomelanocortin peptide immunocytochemistry in rhesus monkey brain. Brain Res. Bull. 1984;13:785–800. doi: 10.1016/0361-9230(84)90237-5. [DOI] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, et al. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc. Natl. Acad. Sci. USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofberg C, Agren H, Harro J, Oreland L. Cholecystokinin in CSF from depressed patients: possible relations to severity of depression and suicidal behaviour. Eur. Neuropsychopharmacol. 1998;8:153–157. doi: 10.1016/s0924-977x(97)00046-1. [DOI] [PubMed] [Google Scholar]
- Lowe WL, Jr., Schaffner AE, Roberts CT, Jr., LeRoith D. Developmental regulation of somatostatin gene expression in the brain is region specific. Mol. Endocrinol. 1987;1:181–187. doi: 10.1210/mend-1-2-181. [DOI] [PubMed] [Google Scholar]
- Manser E, Chong C, Zhao ZS, et al. Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J. Biol. Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Akil H, Watson SJ. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci. 1995;18:22–29. doi: 10.1016/0166-2236(95)93946-u. [DOI] [PubMed] [Google Scholar]
- Mariani M, Corradi A, Baldessari D, et al. Mab21, the mouse homolog of a C. elegans cell-fate specification gene, participates in cerebellar, midbrain and eye development. Mech. Dev. 1998;79:131–135. doi: 10.1016/s0925-4773(98)00180-4. [DOI] [PubMed] [Google Scholar]
- McGuinness TL, Lai Y, Greengard P. Ca2+/calmodulin-dependent protein kinase II. Isozymic forms from rat forebrain and cerebellum. J. Biol. Chem. 1985;260:1696–1704. [PubMed] [Google Scholar]
- Meador-Woodruff JH. Update on dopamine receptors. Ann. Clin. Psychiatry. 1994;6:79–90. doi: 10.3109/10401239409148986. [DOI] [PubMed] [Google Scholar]
- Minthon L, Edvinsson L, Gustafson L. Somatostatin and neuropeptide Y in cerebrospinal fluid: correlations with severity of disease and clinical signs in Alzheimer's disease and frontotemporal dementia. Dement. Geriatr. Cogn. Disord. 1997;8:232–239. doi: 10.1159/000106636. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol. Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Nakajima D, Ohara O, Nakayama M. Mammalian fat3: a large protein that contains multiple cadherin and EGF-like motifs. Biochem. Biophys. Res. Commun. 2002;290:1260–1266. doi: 10.1006/bbrc.2002.6338. [DOI] [PubMed] [Google Scholar]
- Mohler H, Malherbe P, Draguhn A, Richards JG. GABAA-receptors: structural requirements and sites of gene expression in mammalian brain. Neurochem. Res. 1990;15:199–207. doi: 10.1007/BF00972210. [DOI] [PubMed] [Google Scholar]
- Murphy DB, Wiese S, Burfeind P, et al. Human brain factor 1, a new member of the fork head gene family. Genomics. 1994;21:551–557. doi: 10.1006/geno.1994.1313. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Muraoka S, Sanokawa R, Mori N. N-Shc and Sck, two neuronally expressed Shc adapter homologs: their differential regional expression in the brain and roles in neurotrophin and Src signaling. J. Biol. Chem. 1998;273:6960–6967. doi: 10.1074/jbc.273.12.6960. [DOI] [PubMed] [Google Scholar]
- O'Hara BF, Andretic R, Heller HC, Carter DB, Kilduff TS. GABAA, GABAC, and NMDA receptor subunit expression in the suprachiasmatic nucleus and other brain regions. Brain Res. Mol. Brain Res. 1995;28:239–250. doi: 10.1016/0169-328x(94)00212-w. [DOI] [PubMed] [Google Scholar]
- Pati D, Keller C, Groudine M, Plon SE. Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol. Cell Biol. 1997;17:3037–3046. doi: 10.1128/mcb.17.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Represa A, Deloulme JC, Sensenbrenner M, Ben-Ari Y, Baudier J. Neurogranin: immunocytochemical localization of a brain-specific protein kinase C substrate. J. Neurosci. 1990;10:3782–3792. doi: 10.1523/JNEUROSCI.10-12-03782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas M, Duprat F, Heurteaux C, Hugnot JP, Lazdunski M. New modulatory alpha subunits for mammalian Shab K+ channels. J. Biol. Chem. 1997;272:24371–24379. doi: 10.1074/jbc.272.39.24371. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, et al. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc. Natl. Acad. Sci. USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguela P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J. Neurosci. 1993;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Bissette G, Janicak P, Davis JM, Nemeroff CB. Cerebrospinal fluid somatostatin concentrations in schizophrenia and schizoaffective disorder: the effects of antipsychotic treatment. Schizophr. Res. 1994;13:173–177. doi: 10.1016/0920-9964(94)90099-x. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Bissette G, Janicak PG, Davis JM, Nemeroff CB. Elevation of CSF somatostatin concentrations in mania. Am. J. Psychiatry. 1995;152:1807–1809. doi: 10.1176/ajp.152.12.1807. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Misaki A, Sonobe H, Liang SB, Ohtsuki Y. Is T-cadherin (CDH13, H-cadherin) expression related to lung metastasis of osteosarcoma? Histopathology. 2000;37:193–194. doi: 10.1046/j.1365-2559.2000.00985-5.x. [DOI] [PubMed] [Google Scholar]
- Tang XX, Pleasure DE, Ikegaki N. cDNA cloning, chromosomal localization, and expression pattern of EPLG8, a new member of the EPLG gene family encoding ligands of EPH-related protein–tyrosine kinase receptors. Genomics. 1997;41:17–24. doi: 10.1006/geno.1997.4615. [DOI] [PubMed] [Google Scholar]
- Tighilet B, Hashikawa T, Jones EG. Cell- and lamina-specific expression and activity-dependent regulation of type II calcium/calmodulin-dependent protein kinase isoforms in monkey visual cortex. J. Neurosci. 1998;18:2129–2146. doi: 10.1523/JNEUROSCI.18-06-02129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Briere R, Dewar K, et al. Prediction of level of serotonin 2A receptor binding by serotonin receptor 2A genetic variation in postmortem brain samples from subjects who did or did not commit suicide. Am. J. Psychiatry. 1999;156:1456–1458. doi: 10.1176/ajp.156.9.1456. [DOI] [PubMed] [Google Scholar]
- Vallano ML, Beaman-Hall CM, Mathur A, Chen Q. Astrocytes express specific variants of CaM KII delta and gamma; but not alpha and beta, that determine their cellular localizations. Glia. 2000;30:154–164. doi: 10.1002/(sici)1098-1136(200004)30:2<154::aid-glia5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Evans SJ, Choudary PV, et al. Gender specific gene expression in postmortem human brain: localization to sex chromosomes. 2003. In Submission. [DOI] [PMC free article] [PubMed]
- Vogt BA, Finch DM, Olson CR. Functional heterogeneity in cingulate cortex: the anterior executive and posterior evaluative regions. Cereb. Cortex. 1992;2:435–443. doi: 10.1093/cercor/2.6.435-a. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Egan MF, Bertolino A, et al. Prefrontal neurons and the genetics of schizophrenia. Biol. Psychiatry. 2001;50:825–844. doi: 10.1016/s0006-3223(01)01252-5. [DOI] [PubMed] [Google Scholar]
- Zhao X, Lein ES, He A, Smith SC, Aston C, Gage FH. Transcriptional profiling reveals strict boundaries between hippocampal subregions. J. Comp. Neurol. 2001;441:187–196. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Kreiman G, Anderson DJ. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proc. Natl. Acad. Sci. USA. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]