Abstract
We describe a method for the systematic improvement of reaction coordinates in quantum mechanical / molecular mechanical (QM/MM) calculations of reaction free energy profiles. In umbrella-sampling free energy calculations, a biasing potential acting on a chosen reaction coordinate is used to sample the system in reactant, product, and transition states. Sharp, nearly discontinuous changes along the resulting reaction path are used to identify coordinates that are relevant for the reaction but not properly sampled. These degrees of freedom are then included in an extended reaction coordinate. The general formalism is illustrated for the catalytic cleavage of the RNA backbone of an RNA/DNA hybrid duplex by the RNase H enzyme of bacillus halodurans. We find that in the initial attack of the phosphate diester by water, the oxygen-phosphorus distances alone are not sufficient as reaction coordinates, resulting in substantial hysteresis in the proton degrees of freedom and a barrier that is too low (~10 kcal/mol). If the proton degrees of freedom are included in an extended reaction coordinate, we obtain a barrier of 21.6 kcal/mol consistent with the experimental rates. As the barrier is approached, the attacking water molecule transfers one of its protons to the O1P oxygen of the phosphate group. At the barrier top, the resulting hydroxide ion forms a penta-coordinated phosphate intermediate. The method used to identify important degrees of freedom, and the procedure to optimize the reaction coordinate are general and should be useful both in classical and in QM/MM free energy calculations.
Keywords: QM/MM, free energy, enzyme, catalysis, umbrella sampling, RNase H, proton transfer, reaction coordinate
INTRODUCTION
Quantum chemical calculations are often validated by comparing calculated and measured reaction rates. In the calculations, the reaction rate k is typically estimated from the height of the free energy barrier ΔG‡ by using transition state theory,
| (1) |
where kB is the Boltzmann constant, and T is the absolute temperature. A common assumption for the pre-factor is k0 = kBT/h, with h Planck's constant, which neglects re-crossings of the dividing surface between reactant and product states1 and lacks a firm theoretical foundation2. Nevertheless, in many chemical reactions involving only a small number of atoms and without strong dynamical coupling to the surrounding, a prefactor of ~1 ps−1 seems reasonable. The other, potentially more important factor concerns the height, ΔG‡, of the free energy barrier.
With reaction rates being exponentially sensitive to ΔG‡, accurate barrier heights are a major concern in rate calculations. The barrier height (or activation free energy) ΔG‡ is defined with respect to a chosen reaction coordinate, and is typically obtained either from second-order expansions of the energy surfaces, or from biased simulations using, e.g., umbrella sampling (US)3, thermodynamic integration (TI)4, lambda dynamics5, or adaptive biasing force6. The choice of coordinate is usually based on chemical intuition. However, ignoring degrees of freedom important for the reaction can produce apparent reaction free energy barriers that are too low (which would, in principle, be compensated in the rate formula Eq. (1) by a small transmission coefficient1,7 scaling down the pre-factor k0); in contrast, inefficiencies in sampling the free energy along the chosen coordinate, possibly associated with the system getting trapped in local minima, can result in apparent reaction barriers that are artificially high8.
In principle, these problems can be alleviated by using high-dimensional reaction coordinates, possibly combined with procedures such as variational transition-state theory9. However, the computational effort of US scales exponentially with the dimensionality of the reaction coordinate. Finding good one-dimensional coordinates thus represents a primary objective. Here, we propose a simple method to improve on an initially chosen one-dimensional (1D) reaction coordinate by systematically adding important degrees of freedom previously left out.
The central motivating observation is that in US simulations (and other restrained or constrained simulations), incomplete reaction coordinates are manifest in hysteresis and near-discontinuities along important degrees of freedom not included in the reaction coordinate. In a typical US simulation, the system is driven from reactant to product states by advancing the center of a harmonic biasing potential in a step-wise fashion. If the bias of the US is not properly driving the reaction, some of the missing degrees of freedom orthogonal to the chosen reaction coordinate will not be fully equilibrated10. As the umbrella window is advanced, tension builds up, metastability in the orthogonal coordinates disappears, eventually leading to sharp transitions of these coordinates from values typical of reactant states to those of product states.
Figure 1 illustrates the hysteresis problem for reaction paths on a schematic 2D free energy surface. The horizontal coordinate x is the chosen reaction coordinate, and the vertical coordinate y is an important coordinate left out from the US (or TI) simulations. Three relevant paths are compared: SD: the steepest-descent path from the saddle of the free energy surface to the minima as a reference; G: a path obtained by globally minimizing the energy along y for fixed values of the chosen reaction coordinate x; and L: a path obtained by minimizing the energy along y locally for fixed values of x, starting from either the reactant well or (path L') the product well. Path G mimics the situation of good statistical sampling, but a poor choice of coordinate; in path L, the sampling is poor as well. In both types of minimum-energy paths, the barrier is crossed in discontinuous transitions.
Figure 1.
Reaction paths on a schematic 2D free energy surface. SD (yellow): steepest-descent path. G (red): path obtained by minimizing energy globally along y direction (ordinate) for fixed x (abscissa). L and L' (white): paths obtained by minimizing energy locally along y for fixed x, starting from reactant (left) and product wells (right), respectively. The dashed segments of the paths indicate discontinuous changes in y. Numbers next to the paths indicate the apparent height of the free energy barrier.
The apparent barriers along the three paths differ dramatically: if the energy surface is searched globally along y for fixed x, the barrier is 11 kBT, far below the actual free energy barrier of 28 kBT, but close to the 1D free energy barrier obtained by integrating out y (see Eq. (3) below). During the transition on path G, the y coordinate appears to “tunnel” through the high-energy regions of the energy surface. In contrast, if the energy is minimized only locally along y, the system is driven up steep valleys on the free energy surface before the y-coordinate finally crosses from the reactant well to the product well (path L) or back (path L'). As a consequence, the barriers along path L and L' are vastly overestimated at 60 kBT.
Metastability and the resulting hysteresis along y are responsible for the deviations from the correct barrier for both minimum-energy paths in Figure 1, even though the globally-minimized path predicts a barrier that is too low, and the locally-minimized one a barrier that is too high. In both cases, the path is characterized by a discontinuous coordinate y. This observation is the basis for a simple method to find relevant coordinates previously left out. As the biasing function (here acting on x) is advanced to drive the system from reactant to product states, one seeks to find near-discontinuities in other coordinates (here: y), in particular as the system crosses the apparent barrier. Such discontinuous coordinates can then be included in an improved reaction coordinate, either by increasing the dimensionality of the reaction coordinate, or by creating a new, extended 1D coordinate that takes into account the extra degrees of freedom.
Continuity can also be ensured for a predefined set of variables by using optimization methods that operate on paths rather than individual structures. A number of developments (the nudged elastic band11,12, replica path13,14 or string methods15) offer such a solution for potential energy (or free energy) minimizations by simultaneously choosing a large number of atomic coordinates that are included in the reaction coordinate. Other methods use a predefined set of collective variables to create a history-dependent biasing potential to “fill-up” local reactant and product wells16–19. The present approach can complement these methods and identify additional important missing variables that were not included in the predefined set of variables.
Our primary goal here is to establish a procedure to calculate accurate free energy barriers corresponding to a proposed enzyme reaction mechanism. We illustrate the general procedure by applying it to the RNA-cleavage reaction catalyzed by RNase H of bacillus halodurans. RNase H binds an RNA/DNA hybrid duplex substrate and catalyzes the hydrolysis of the phosphate-ester bond between two nucleotides of the RNA chain. This reaction is prototypical of two-metal ion assisted catalysis, which occurs in many important phosphoryl-transfer reactions20–22. The mechanistic details of this reaction are still not fully understood. There is a long-standing debate about whether the reaction follows a one-step (concerted) or a two-step mechanism, with either a dissociative or an associative pathway23,24. Several studies proposed a dissociative mechanism with a metaphosphate-like intermediate25–27. However, recent studies found a two-step associative mechanism28–30. Another major question concerns the protonation states of the active-site groups along the reaction path. In particular, it is not clear whether the nucleophile attacking the RNA phosphate backbone is a water molecule or a hydroxide ion.
In our calculations, we use an ab initio quantum mechanical / molecular mechanical (QM/MM) methodology combined with US to construct the free energy surface (or potential of mean force, PMF) along the chosen reaction coordinates. Ab initio calculations with traditional geometry minimization approaches have been demonstrated to give accurate results for phosphate hydrolysis reactions in solution using continuum solvation models31–33. In enzymatic systems, however, combined QM/MM free energy simulations, with adequate sampling, are necessary to properly account for the electrostatic effects of the protein media34.
Our study shows that the reaction follows a mechanism in which the attacking water molecule loses its proton before reaching a transition state. After careful examination of a large number of possibly relevant coordinates, we find that the proton transfer from the water to the phosphate group plays an important role, and should be included in the reaction coordinate. With the extended coordinate the free energy barrier is consistent with the experimentally reported rate measurements for the human RNase H135, which has an active site that is nearly identical to the one for bacillus halodurans36.
Overall, we find the enzymatic reaction to follow a two-step associative mechanism (AN+DN, where AN stands for a nucleophilic association, and DN for a nucleofugic dissociation occurring in a separate step between nucleophile approach and leaving-group departure). However, here we only probe the first step. To account for a complete picture of the enzymatic reaction, investigation of different proton-transfer pathways occurring within the protein environment will be necessary. Thus further studies in this direction are underway.
METHODS
Structural Model
The initial structure was based on the crystal structure of RNase H complexed with a hybrid RNA/DNA duplex substrate (Protein Data Bank code 1ZBI37, 1.85 Å resolution).
Figure 2a shows a representation of the entire system. Chain A was taken from the structure of the protein dimer together with its substrate, which was trimmed to a six base-pair long RNA/DNA complex. The experimentally mutated Asn132 residue was replaced by Asp132 to reproduce the wild-type protein. Crystallographic water molecules in the vicinity of the protein or the substrate were preserved in the initial structure.
Figure 2.
Structure and reaction mechanism. (a) Structure of RNase H (blue) and bound RNA/DNA duplex (red/gray) as ribbon diagrams, together with Mg2+ ions (green) and solvent water. The phosphate group involved in bond breaking is shown atomistically. (b) QM region and the Mg2+ ions of the RNase H active site. r1, r2, r3 and r4 are distances relevant to the reaction mechanism. (c) Schematic reaction scheme of the probed mechanism. The hydrogen involved in the proton transfer steps is shown in red.
Molecular Dynamics (MD) Simulations
After generating the coordinates of the hydrogen atoms with CHARMM38 using standard protonation states of all ionizable residues, the system was placed in a cube of TIP3P water, 69.6 Å along each axis. Water molecules were then replaced randomly by ions to account for a 0.10 M NaCl solution, adding 14 additional Na+ ions for neutralization. We used the CHARMM27 extended-atom force field for the MM interaction, with periodic boundary conditions, Ewald summation and a 12 Å cutoff for the evaluation of the nonbonded interactions. After an initial equilibration, the final structure was taken as a starting point for subsequent QM/MM simulations.
QM/MM Simulations
The equilibrated classical system was trimmed to a sphere of 32 Å radius centered at the Mg2+ ions. Atoms further than 20 Å from the active-site Mg2+ ions were fixed in space. We used the Q-Chem program package39 to perform the ab initio calculations, at the B3LYP40 6-31G(d) density functional theory (DFT) level of theory. The QM system was coupled with the CHARMM program using full electrostatic embedding41. Standard link atom treatment was used to cut bonds between the QM and MM region, adding hydrogen atoms for the missing ligands. Full QM/MM minimizations were carried out to determine starting structures for the free energy simulations.
Free Energy Simulations
In the US simulations, harmonic biasing potentials were placed along the reaction coordinate at steps of 0.1 Å (−2.1≤Qe≤0.2 Å, with Qe=r1−r2; see Figure 2b) with spring constants of 1000 kcal/(molÅ2). The QM/MM dynamics in each window was run for at least 5 ps after equilibration, again using a Langevin thermostat and a 1 fs time step.
In addition to the standard US/PMF simulations we also carried out simulations in conjunction with Hamiltonian replica exchange (RE)42 between the neighboring windows. RE/US allowed us to enhance the sampling, while only marginally increasing the simulation time. To increase the acceptance ratio of RE attempts, we used softer harmonic biasing potentials with spring constants of 50 kcal/(molÅ2) that were spaced more closely at steps of 0.05 Å (−1.25≤Qe≤−0.7 Å). The RE/US simulations were initialized from structures of the US/PMF simulations. RE between neighboring windows was attempted every 20 MD steps. Free energy profiles were evaluated globally by using the multidimensional weighted histogram analysis method (WHAM)43.
RESULTS AND DISCUSSION
In the QM/MM calculations, the QM system was defined as the phosphate group (Figure 2b) and the attacking water molecule. Figure 2b shows four distances relevant to the phosphate hydrolysis mechanism. The initial reaction coordinate Qe= r1− r2 was chosen as the difference in length of the breaking and forming bonds, i.e., the phosphorus atom and the leaving O3' oxygen atom (r1), and the oxygen of the attacking water molecule and the phosphorus atom (r2). Qe probes primarily the electron-transfer part of the hydrolysis reaction. This choice of reaction coordinate allows us to distinguish between dissociative and associative paths. As the reaction is forced to progress by advancing the windows in the US simulations, the two extreme responses of the system are either an increase in the distance between the phosphorus and the O3' oxygen of the leaving group (which would lead to a dissociative mechanism), or a decrease in the distance between the attacking water oxygen and the phosphorus.
To initialize the US/PMF simulations, we prepared starting structures of the system through a series of energy minimizations along the reaction coordinate Qe. Our preliminary QM/MM minimization studies showed hysteresis when the Qe reaction coordinate was used (data not shown). We started minimizations from the reactant state and moved towards the product state and vice versa. We found that the forward and backward minimum energy pathways differed at the transition state region, although the endpoints (RS and PS) were identical.
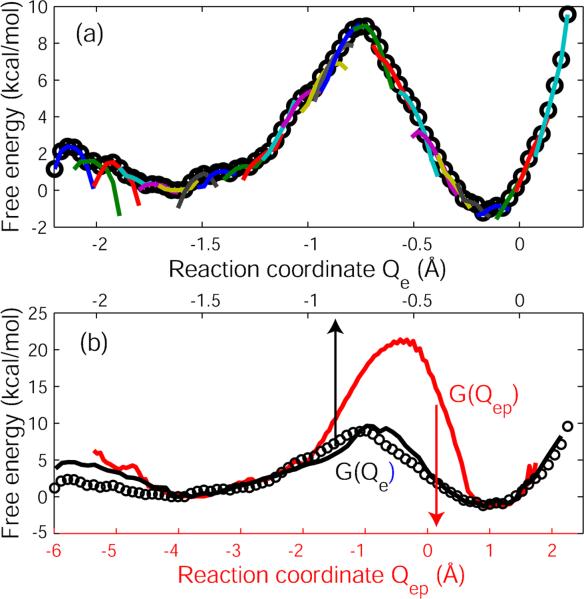
The PMF profile obtained from the standard US simulations along Qe appears well converged, as shown in Figure 3 (a). The individual free energy curves for different US windows overlap, have similar local curvatures, and the overall free energy profile appears well connected. The PMF suggests an associative mechanism with a low barrier for the first step of the hydrolysis reaction, resulting in a stable penta-coordinated intermediate. The barrier for this step is about 9.5 kcal/mol, significantly lower than the experimental estimate of about 21 kcal/mol35.
Figure 3.
One-dimensional free energy profiles of RNase H reaction. (a) Free energy profile along the reaction coordinate Qe obtained from PMF/US simulations. The overall free energy profile (circles) was obtained by using WHAM43. The lines show the unbiased free energy curves of the individual umbrella sampling windows. (b) Free energy profiles along the reaction coordinates Qe and Qep. The solid lines show the results obtained by projecting the 2D free energy profile onto Qe (black, top axis) and Qep (red, bottom axis), respectively. The black circles show the free energy profile obtained in (a) using only Qe as the reaction coordinate.
Selected average atomic distances, r1, r2, r3 and r4 (Figure 2) observed along Qe are shown in Figure 4 (panels a and c for standard US and RE/US, respectively). Consistent with an associative mechanism, the distance between the water molecule and the phosphate (r2) decreases, while the leaving-group distance (r1) remains nearly constant as the reaction progresses. However, other relevant distances, such as r3 and r4 (involving the hydrogen atom of the attacking water molecule) present discontinuous jumps along Qe.
Figure 4.
Average atomic distances r1, r2, r3 and r4 (Figure 2) observed along the reaction coordinates Qe (panels a and c) and Qep (panels b and d). Results from (a,b) US and (c,d) RE/US.
Such discontinuities in distances not included in the reaction coordinate Qe can artificially lower the apparent free energy barrier. Thus, we reexamined the free energy simulations at the transition state region for discontinuities, following the arguments given in the discussion of Figure 1. To systematically search for additional important coordinates, we developed the following simple analysis. We calculated the averages and the standard deviations of the atom-atom distances that involved each of the QM atoms and all atoms within 4.0 Å distance of the QM subsystem (a total of about 1000 atom pair distances, dij). For each of the neighboring US windows, we analyzed the difference between the average dij values and considered the path to be continuous if the difference did not exceed twice the sum of the standard deviations of dij in the two neighboring windows. By using this procedure, we discovered that several distances at the transition state were not continuous. The majority of these distances involved the hydrogen atom of the attacking water molecule that was transferred to the O1P oxygen of the phosphate group in a sharp, jump-like transition (curves for r3 and r4 in Figure 4a and c, with r3 being the distance between the hydrogen and the oxygen of the attacking water, and r4 the distance between the same hydrogen and the O1P oxygen atom of the phosphate).
To include these missing degrees of freedom, we extended the reaction coordinate Qe by combining it with Qp = r3 − r4 in a new coordinate Qep = Qe + Qp. The combined coordinate Qep accounts for the forming and breaking bonds by adding the distance of the hydrogen and the oxygen of the water molecule (r3) and by subtracting the distance between the hydrogen of the water and the oxygen of the phosphate (r4) (Figure 2b).
We performed a new set of US/PMF simulations along Qep with 33 windows along this reaction coordinate. The centers of the harmonic biasing potentials were equally spaced from Qep = −2.6 Å to 0.6 Å, with a force constant of 2000 kcal/(molÅ2). In addition to the umbrella potentials on the new coordinate Qep, we also applied a harmonic potential at a constant value of Qe=−0.70 Å, with a 100 kcal/(molÅ2) force constant to keep the geometries near the transition state along Qe. RE/US simulations were also carried out. We used a force constant of 100 kcal/(molÅ2) for the biasing potential centered at Qep between −1.75 Å and 0.15 Å in 0.05 Å increments. In the US simulations biasing Qep, we found that all distances varied continuously, including r3 and r4, as shown in Figure 4b and d.
Data from all four simulations were combined to determine the two-dimensional free energy surface in the Qe−Qep plane. As shown in Figure 5, our best estimate of the barrier of the first step of the hydrolysis reaction is about 22 kcal/mol, in excellent agreement with the experimental value of 20.5 kcal/mol35, assuming a pre-factor of k0=1/ps. Importantly, we found that extending the 1D reaction coordinate from Qe to Qep increased the barrier by more than 10 kcal/mol. We note that an earlier study30 also had used Qe, and found a barrier that was too low compared to experiment.
Figure 5.
Two-dimensional free energy surface for the first step of the hydrolysis reaction of RNase H. All simulation data (both standard US and RE/US, with either Qe or Qep as the reaction coordinate) were included when determining the free energy surface (kcal/mol) using 2D-WHAM43. Three representative structures of the QM region are shown for the reactant and transition state, and the intermediate penta-coordinated state.
The projected one-dimensional free energy profiles along Qe (red, bottom axis) and Qep (black, top axis) are shown in Figure 3(b). Note that the barrier shown in the one-dimensional profile for Qe is consistently lower than that for Qep, and it is in quantitative agreement with the profile in Figure 3(a) obtained by using US along Qe alone. We also consistently find that the intermediate state formed as the product of the first step of the reaction has a free energy about 1 kcal/mol below that of the reactant state.
Improvements over standard US often come from using RE/US, which provides enhanced sampling by accelerating equilibration without adding extra cost to the calculation. Compared to standard US, the average values of r1 to r4 in the RE/US simulations follow the reaction coordinate more continuously as the bias changes (Figure 4). Nevertheless, replica exchange alone could not account for the missing degrees of freedom when only Qe was used, as the discontinuity remained between the US windows at the transition state region for the r3 and r4 distances. By sampling the transition state region using the improved reaction coordinate Qep, we can see in Fig. 4d that proton transfer (Qep = −0.8 Å) precedes the electron transfer between the hydroxide and the phosphate (Qep = −0.2 Å, at the barrier top).
The results obtained by searching along Qep show a clear improvement in calculating the free energy profile. However, as shown in Figure 5, the orientation of the saddle is not parallel to Qep in the Qe−Qep plane, and Qep is not the optimal one-dimensional choice. Ignoring anisotropy in the dynamics, an ideal 1D reaction coordinate for a single saddle point is given by the eigenvector of the second-derivative matrix of the free energy, for which the corresponding eigenvalue is negative. In practice, however, it is often not feasible to evaluate multidimensional Hessian matrices. Here, we note that the r1−r2 (Qe) and r3−r4 (Qp) coordinates can be considered as independent, and we search for a new, optimized reaction coordinate, Qopt, as their best linear combination by varying the angle θ in
| (2) |
We integrate the 2D free energy surface to obtain the projected one-dimensional free energy profile along Qopt
| (3) |
and determine the corresponding barrier height as a function of the angle θ. In Figure 6, the Qe coordinate is represented by θ = 0° and the corresponding barrier is 9.5 kcal/mol, whereas Qep corresponds to θ = 45°, with a barrier height of 20.6 kcal/mol. The highest barrier (Qopt) is at an angle of about θ = 20° with a height of 21.6 kcal/mol, our best estimate of a 1D free energy barrier. We note, however, that the inclusion of additional degrees of freedom beyond Qe and Qp can further increase this barrier, even within the assumptions of our QM/MM model. These results show that at the transition state of the catalytic reaction the optimal coordinate consists of about 30% “proton transfer” and 70% “electron transfer”, which allows us to define a quantitative measure of the coupling between proton transfer and electron transfer processes.
Figure 6.
Optimal 1D reaction coordinate Qopt (θ) = Qe cosθ+Qp sinθ. Integrated 1D free energy barriers are calculated as a function of θ by numerical integration using the 2D free energy surface of Figure 5.
One of the limitations of our method to improve the reaction coordinate is that not in all cases will atomic pair distances be sufficient to capture the important degrees of freedom; instead, collective coordinates may need to be considered44. Although our method can be useful in diagnosing whether a proposed collective coordinate could be important or not, we have no means of a priori list and test all possible collective coordinates. Another limitation is concerned with the sensitivity of our analysis. If the US windows are very closely spaced, one may observe quasi-continuous trajectories for the average values of the coordinates even if the underlying distribution is bistable. In such cases it will be useful to obtain more detailed information about the actual distribution of the proposed reaction coordinate. In particular, a spike in the variance of a proposed additional coordinate along the reaction could indicate bi-stability.
With regards to the RNase H reaction studied here, we caution against over-interpreting the simulation results despite the apparent agreement with experiment. Besides the choice of the QM system and the reaction coordinate, the size of the basis set and the level of theory also affect the barrier height. To extend the QM/MM model, and to give further details of the complete enzymatic reaction, the dissociation of the leaving sugar group has to be simulated as well. With the current setup, we found that a very high barrier is obtained for the second step, unless additional water molecules or protein atoms are included in the quantum region. Thus for accurate mechanistic details more simulations are underway with an increased size of the QM system.
CONCLUSIONS
We present ab initio QM/MM simulations of the phosphodiester hydrolysis reaction catalyzed by the RNase H enzyme with a catalytic water molecule. We found a two-step AN+DN mechanism for the overall cleavage reaction, which is in agreement with recent calculations30. Our results show that a direct proton transfer from the water to the phosphate group in the first step gives a barrier of 21.6 kcal/mol, which is consistent with the experimental rate found for human RNase H135. We also observed for the first time that the electron transfer step corresponds to the barrier top and it is preceded by the proton transfer step in which the attacking water loses its proton. With the QM/MM model described here, we failed to obtain a low barrier for the second (dissociation) step of the reaction, indicating that a direct proton transfer from the phosphate to the leaving group is not a preferred path. Further simulations using a larger quantum system are thus necessary to probe for possible mechanisms of this reaction step, which can include participation of protein groups and water molecules.
It is often assumed that inadequate low-dimensional reaction coordinates result in reaction barriers that are too high. Here we report a case in which the neglect of important degrees of freedom result in an apparent free energy barrier that is too low. We developed a method to extend the 1D coordinate for US/PMF simulations. Our approach is based on a statistical search for discontinuous atomic distances in the US/PMF simulations. If such distances are found, they are added to the original reaction coordinate. Further US simulations are then performed along a new, combined coordinate Qep = Qe+Qp. By using the 2D free energy profile in the Qe−Qep plane, an “optimal” one-dimensional coordinate Qopt can be found as the linear combination of Qe and Qp that maximizes the barrier in one dimension.
We also examined the use of Hamiltonian RE42 in conjunction with US simulations. We found that average distances as a function of the reaction coordinate were much smoother using RE/US, demonstrating enhanced equilibration, without adding significant extra cost to the simulations. However, we also found that replica exchange could not correct for the missing coordinate Qp when only Qe was used as a reaction coordinate. During the relatively short time scales typical of QM/MM simulations, and with the bias acting only on the poor coordinate Qe, replicas did not cross the Qp reaction barrier. “Equilibration” between the reactant and product sides of the barrier thus occurred entirely by replica exchange, such that the fractions of reactant and product replicas remained constant and the barrier region was not sampled.
ACKNOWLEDGMENT
E.R. and G.H. thank Dr. Wei Yang (NIH), Dr. Marcin Nowotny (Warsaw), and Dr. Attila Szabo (NIH) for helpful discussions. E.R. thanks Dr. Yihan Shao, Dr. Sergio Hassan and Dr. Peter Steinbach for their valuable help and comments and the NCI, CCR Fellows Editorial Board for help in editing the manuscript. This research used the Biowulf Linux cluster at the NIH, and was supported by the Intramural Research Program of the NIDDK and NHLBI, NIH.
Footnotes
Contribution to the “Special Issue: Free Energy Simulation” of the Journal of Computational Chemistry
REFERENCES
- 1.Chandler D. J Chem Phys. 1978;68:2959–2970. [Google Scholar]
- 2.Zwanzig R. Nonequilibrium Statistical Mechanics. Oxford University Press; USA: 2001. [Google Scholar]
- 3.Torrie GM, Valleau JP. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 4.Kirkwood JG. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 5.Kong X, Brooks CL., III J Chem Phys. 1996;105:2414–2423. [Google Scholar]
- 6.Darve E, Pohorille A. J Chem Phys. 2001;115:9169–9183. [Google Scholar]
- 7.Wigner E. J Chem Phys. 1937;5:720–725. [Google Scholar]
- 8.Klahn M, Braun-Sand S, Rosta E, Warshel A. J Phys Chem B. 2005;109:15645–15650. doi: 10.1021/jp0521757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truhlar DG, Garrett BC. Annu Rev Phys Chem. 1984;35:159–189. [Google Scholar]
- 10.Ensing B, Meijer EJ, Blochl PE, Baerends EJ. J Phys Chem A. 2001;105:3300–3310. [Google Scholar]
- 11.Henkelman G, Uberuaga BP, Jónsson H. J Chem Phys. 2000;113:9901–9904. [Google Scholar]
- 12.Henkelman G, Jónsson H. J Chem Phys. 2000;113:9978–9985. [Google Scholar]
- 13.Woodcock HL, Hodoscek M, Sherwood P, Lee YS, Schaefer HF, III, Brooks BR. Theor Chem Acc. 2003;109:140–148. [Google Scholar]
- 14.Woodcock HL, Hodoscek M, Brooks BR. J Phys Chem A. 2007;111:5720–5728. doi: 10.1021/jp0714217. [DOI] [PubMed] [Google Scholar]
- 15.Huber T, Torda AE, van Gunsteren WF. J Comput-Aided Mol Des. 1994;8:695–708. doi: 10.1007/BF00124016. [DOI] [PubMed] [Google Scholar]
- 16.Ensing B, Laio A, Parrinello M, Klein ML. J Phys Chem B. 2005;109:6676–6687. doi: 10.1021/jp045571i. [DOI] [PubMed] [Google Scholar]
- 17.Laio A, Parrinello M. Proc Natl Acad Sci U S A. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.E W, Ren W, Vanden-Eijnden E. Phys Rev B. 2002;66:052301. [Google Scholar]
- 19.Zheng L, Chen M, Yang W. Proc Natl Acad Sci U S A. 2008;105:20227–20232. doi: 10.1073/pnas.0810631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steitz TA, Steitz JA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Lee JY, Nowotny M. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamerlin SCL, Wilkie J. Org Biomol Chem. 2007;5:2098–2108. doi: 10.1039/b701274h. [DOI] [PubMed] [Google Scholar]
- 24.Williams NH. Biochim Biophys Acta. 2004;1697:279–287. doi: 10.1016/j.bbapap.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 25.Zalatan JG, Herschlag D. J Am Chem Soc. 2006;128:1293–1303. doi: 10.1021/ja056528r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Zhang Z-Y. Biochemistry. 1996;35:11797–11804. doi: 10.1021/bi960471r. [DOI] [PubMed] [Google Scholar]
- 27.Admiraal SJ, Herschlag D. Chem Biol. 1995;2:729–739. doi: 10.1016/1074-5521(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Yu X, Hu P, Broyde S, Zhang Y. J Am Chem Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cisneros GA, Perera L, García-Díaz M, Bebenek K, Kunkel TA, Pedersen LG. DNA Repair. 2008;7:1824–1834. doi: 10.1016/j.dnarep.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Vivo M, Dal Peraro M, Klein ML. J Am Chem Soc. 2008;130:10955–10962. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cramer CJ, Truhlar DG. In: Solvent Effects and Chemical Reactivity. Tapia O, Bertrán J, editors. Kluwer; Dordrecht: 1996. pp. 1–80. [Google Scholar]
- 32.Klahn M, Rosta E, Warshel A. J Am Chem Soc. 2006;128:15310–15323. doi: 10.1021/ja065470t. [DOI] [PubMed] [Google Scholar]
- 33.Rosta E, Kamerlin SCL, Warshel A. Biochemistry. 2008;47:3725–3735. doi: 10.1021/bi702106m. [DOI] [PubMed] [Google Scholar]
- 34.Rosta E, Klahn M, Warshel A. J Phys Chem B. 2006;110:2934–2941. doi: 10.1021/jp057109j. [DOI] [PubMed] [Google Scholar]
- 35.Lima WF, Wu H, Nichols JG, Prakash TP, Ravikumar V, Crooke ST. J Biol Chem. 2003;278:49860–49867. doi: 10.1074/jbc.M306543200. [DOI] [PubMed] [Google Scholar]
- 36.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 38.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 39.Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O'Neill DP, R. A. D, Jr, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Voorhis TV, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu C-P, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Simmonett AC, Subotnik JE, Woodcock HL, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Schaefer HF, Kong J, Krylov AI, Gill PMW, Head-Gordon M. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 40.Becke AD. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 41.Woodcock HL, Hodoscek M, Gilbert ATB, Gill PMW, Schaefer HF, III, Brooks BR. J Comput Chem. 2007;28:1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 42.Fukunishi H, Watanabe O, Takada S. J Chem Phys. 2002;116:9058–9067. [Google Scholar]
- 43.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 44.Thorpe IF, Brooks CL., III Proteins: Struct, Funct, Bioinf. 2004;57:444–457. doi: 10.1002/prot.20219. [DOI] [PubMed] [Google Scholar]