Abstract
The obligate intracellular parasite pathogen Plasmodium falciparum is the causative agent of malaria, a disease that results in nearly one million deaths per year. A key step in disease pathology in the human host is the parasite-mediated rupture of red blood cells, a process that requires extensive proteolysis of a number of host and parasite proteins. However, only a relatively small number of specific proteolytic processing events have been characterized. Here we describe the application of the Protein Topography and Migration Analysis Platform (PROTOMAP) (Dix, M. M., Simon, G. M., and Cravatt, B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691; Simon, G. M., Dix, M. M., and Cravatt, B. F. (2009) Comparative assessment of large-scale proteomic studies of apoptotic proteolysis. ACS Chem. Biol. 4, 401–408) technology to globally profile proteolytic events occurring over the last 6–8 h of the intraerythrocytic cycle of P. falciparum. Using this method, we were able to generate peptographs for a large number of proteins at 6 h prior to rupture as well as at the point of rupture and in purified merozoites after exit from the host cell. These peptographs allowed assessment of proteolytic processing as well as changes in both protein localization and overall stage-specific expression of a large number of parasite proteins. Furthermore, by using a highly selective inhibitor of the cysteine protease dipeptidyl aminopeptidase 3 (DPAP3) that has been shown to be a key regulator of host cell rupture, we were able to identify specific substrates whose processing may be of particular importance to the process of host cell rupture. These results provide the first global map of the proteolytic processing events that take place as the human malarial parasite extracts itself from the host red blood cell. These data also provide insight into the biochemical events that take place during host cell rupture and are likely to be valuable for the study of proteases that could potentially be targeted for therapeutic gain.
Malaria is a parasitic disease that results in nearly one million deaths per year worldwide. The disease is caused by infection of the human host by the obligate intracellular parasite Plasmodium falciparum. Upon initial infection, the pathogen actively invades cells of the liver where it produces large quantities of parasites for release into the blood. Most of the disease pathology is associated with this blood stage infection. While in the blood, the parasite must establish residence in host red blood cells (RBCs) where it actively replicates to amplify the infection. One of the final steps in the 48-h life cycle is the rupture of the host RBC to produce merozoite progeny that then infect naïve RBCs to continue the cycle.
A number of recent studies have determined that both serine and cysteine proteases are key regulators of host cell rupture (1–3). In particular, the serine protease subtilisin 1 (PfSUB1) (3) and the cysteine protease dipeptidyl aminopeptidase 3 (DPAP3)1 (1) have been found to be essential for parasite-mediated host cell rupture. Presumably, these enzymes regulate parasite release by cleaving downstream protein substrates to facilitate a specific biochemical program. However, only a small number of actual proteolytic processing events have been characterized. Therefore, a more global proteomics analysis of proteolytic processing would provide vital information on the presumably large number of proteolytic processing events that take place at the time of parasite exit from the host cell. A comprehensive map of these events will help to shed light on the biochemical processes that must be orchestrated to prepare parasites for release from the host cell. In addition, such a map of proteolytic processing events may help to identify families of proteases that mediate these events. Protease targets that direct the processing of key proteolytic events required for host cell rupture would therefore be potentially valuable targets for new classes of antimalarial drugs.
Although many proteases have been studied in detail, the mapping of specific protein substrates has been limited by the lack of analytical methods that facilitate identification of a cleaved peptide product. The past few years have seen a rapid growth in the development of methods to globally identify substrates of proteases. In particular, several recently reported methods have begun to make strides to improve our knowledge of protease substrate processing (4–9). These methods make use of chemical or enzymatic tagging strategies to specifically isolate peptides that are produced as the result of proteolytic cleavage. Thus, all of these methods can be used to both identify the target protein substrate and to identify the exact site of proteolytic processing. However, all of these methods require multiple processing steps before MS analysis, and targets are often identified using only a single peptide sequence.
As an alternative, PROTOMAP (6, 10) is a standard one-dimensional SDS-PAGE method in which each lane of the gel is cut into 20–26 slices for analysis by in-gel digestion and MS/MS sequencing. Much like standard multidimensional protein identification technology two-dimensional LC methods, this allows a complex proteomic sample to be analyzed in many fractions, thereby allowing extensive sequence coverage of the identified proteins. For each protein identified, it is possible to map the location of the protein (i.e. gel slice number) and therefore its overall migration in the gel. When a protein is processed by a protease, the location of the protein fragments in the gel often changes. Because the method generates data for peptides that map to various regions of the protein, it is possible to identify the general location of the cleavage event. In some cases, the exact site of processing can be determined based on the presence of a semitryptic peptide that contains the cleavage site at one end. Because PROTOMAP does not require isolation of the peptide where processing takes place, it requires only a small amount of starting material and provides broad sequence coverage of the target proteins. This is particularly important when working with synchronously grown populations of parasites that are best isolated in relatively small quantities.
In this study, we used the PROTOMAP method to identify proteolytic events that take place in the last 6 h of the blood stage life cycle of P. falciparum. By comparing profiles at 6 h prior to rupture with those at the point of rupture and after release from host cells, we were able to identify a significant number of specific proteolytic events that are associated with rupture. In addition, by fractionating the samples into soluble and membrane-bound fractions, we could identify changes in cellular localization of proteins as the parasite prepares for release from the host. By comparing overall spectral counts for each protein at the various time points, we could also identify stage-specific protein expression. Finally, by using a highly selective inhibitor for DPAP3, an enzyme that is essential for efficient host cell rupture, we could identify proteins whose processing may be of particular importance for parasite release from the host cell.
MATERIALS AND METHODS
Parasite Maintenance and Culture
P. falciparum strain D10 was maintained and synchronized as described previously (1, 11). Specifically, synchrony was maintained using 5% sorbitol (12), and enrichment of (∼42 h postinfection) schizonts prior to the proteomics studies was conducted on a 70% Percoll gradient (13). The schizonts obtained from the Percoll gradient were transferred to media without Albumax (serum-free) for the 6-h duration of the experiment. A separate sample was cultured in the presence of 50 μm SAK-1 to provide a SAK-1-treated T = 0 sample and a SAK-1-treated supernatant sample. Merozoites were purified using a SuperMACSTM II separator (Miltemyi Biotec, Auburn, CA) as described previously (14).
Sample Preparation, SDS-PAGE, and In-gel Digestion
Enriched schizont samples were prepared at two time points: 42 h postinvasion (T = −6) directly from the Percoll gradient and 48 h postinvasion (T = 0) as merozoites are egressing from the infected erythrocyte. At each time point, the enriched schizonts were isolated by centrifugation (1000 × g), washed in PBS, and then snap frozen (liquid N2) in the presence of Mini Complete protease inhibitors (Roche Applied Science). All samples derived from these experiments were supplemented with protease inhibitors unless otherwise stated. The serum-free media from the cultured schizonts were cleared by centrifugation (16,000 × g, 30 min, 4 °C) to remove debris and released merozoites before snap freezing and storage. Prior to analysis, the media were thawed and concentrated on a 10-kDa-cutoff membrane (Amicon). The enriched schizont samples were lysed by five freeze-thaw cycles (liquid N2 and 37 °C water) followed by isolation of the “cytosolic” fraction by centrifugation (16,000 × g, 20 min), and the pellet was extracted into PBS + 1% SDS (2 h, 4 °C) to yield a “membrane” fraction after clearance of the insoluble debris (16,000 × g, 20 min). SDS-PAGE separation and preparation for LC-MS analysis were conducted as described previously (6) with some modifications. Briefly, 200 μg of each protein preparation was separated via 11% SDS-PAGE. The gel was subsequently cut into 26 0.5-cm bands. Gel slices were subjected to in-gel digestion with trypsin as described (6) before analysis on an LTQ ion trap mass spectrometer (ThermoFisher). The T = −6 versus T = 0 comparison is composed of four replicates (three biological and one technical). The T = −6 versus T = 0 (SAK-1-treated) is composed of three replicates (two biological and one technical). The supernatant analysis was performed in biological duplicate. The merozoite sample is composed of a single run.
LC-MS/MS
LC-MS/MS analysis was performed on an LTQ ion trap mass spectrometer (ThermoFisher) coupled to an Eksigent nanoLC-2DTM pump. Peptides were separated on a Basic Picofrit C18 capillary column (New Objective). Peptides were eluted with an acetonitrile gradient from 0 to 60% in a 0.1% solution of formic acid over 2 h with an overall flow rate of 350 nl/min. The flow rate through the column was 250 nl/min, and the spray voltage was 2.0 kV. Data-dependent scanning was used, allowing six MS2 scans of the most abundant ions of the parent full MS scan (400–1800 m/z). Dynamic exclusion was enabled for 180 s.
Data Analysis
RAW files, generated for each gel slice by Xcalibur (version 2.0) running on an LTQ ion trap mass spectrometer, were analyzed using the SEQUEST algorithm in the Bioworks (version 3.3) software package. The searches were performed against a custom concatenated target/decoy non-redundant variant of the human Internation Protein Index database version 3.33 (22,935 non-redundant entries from the human Internation Protein Index database) and a Plasmodium-specific database generated from Swiss-Prot (5543 proteins). A total of 56,956 sequences were searched including the reversed decoy library. Initial searches were conducted with differential methionine oxidation (±16 amu) and fixed cysteine carboxyamidomethylation (+57 amu) as modifications with a requirement for the peptide to be tryptic at one end only and allowing for one missed cleavage. The peptide mass tolerance for the parent ion was 2.0 amu, and that for the fragment ions was 1.0 amu. Duplicate references were reported. SEQUEST data from each gel band were filtered and sorted using DTASelect version 1.9 (15) using default settings. Peptides in 1+, 2+, and 3+ charge states were required to have minimum XCorr values of 1.8, 2.5, and 3.5, respectively. The minimum requirement for ΔCN was 0.08. The output from DTASelect was subsequently used to generate peptographs as described previously (6). Briefly, sequence coverage maps were generated from DTASelect-filter.txt files using the custom Perl script (coverage.pl). Protomap.pl was used to combine sequence coverage data and spectral count data. This step also adds a requirement for two distinct and unique peptides to be present per locus in any given replicate and as such served to remove “singlet” and duplicate peptides from the analysis. The false discovery (FD) rate was determined for each of the data sets according to the guidelines of Elias and Gygi (16) and are reported in Table I. Finally, peptographer.pl was used to assemble individual peptographs for each protein identified. This resulted in the generation of 61 peptographs of reversed proteins. None of these peptographs contained more than four spectral counts. In total, 2078 distinct peptographs were generated; 653 of these contained four or fewer total spectral counts. All of the peptographs generated for each of the data sets are available for searching at http://www.scripps.edu/chemphys/cravatt/Bowyer2010/. Although four spectral counts constituted the limit of spectral counts occurring in any decoy protein, it is impractical to attempt to deduce information regarding proteolysis from such a small number of spectral counts. A minimum of 10 spectral counts is suggested before attempting to identify proteolytic events. However, the full searchable data contain an option to select a custom spectral count cutoff, and all generated peptographs are available. The raw data associated with this study may also be downloaded from the ProteomeCommons.org Tranche network using the following hash: Tk6MwpZKT7ZZXlepbeO/PZsc0KS4npRoxSPW9li8mOKQ5F6daqsFo0asBIt8zuVFrsJ72sKb1yZZIjW3imVW2G2ycbwAAAAAAAHPwA==.
Table I. Peptide statistics.
The total number of peptide identifications for each sample is shown. Also listed are FD rates as determined by searching against a decoy database (16).
| Sample | No. peptides | FD rate | No. semitryptic peptides | FD rate |
|---|---|---|---|---|
| % | % | |||
| Cytosolic/membrane (T = −6) | 15,744 | 0.95 | 3,329 | 3.97 |
| Cytosolic/membrane (T = 0) | 13,519 | 0.78 | 3,200 | 2.94 |
| Cytosolic/membrane (T = 0 SAK) | 8,477 | 0.71 | 1,382 | 3.62 |
| Supernatant | 3,685 | 1.52 | 720 | 7.50 |
| Supernatant (SAK) | 8,447 | 0.71 | 560 | 3.21 |
| Merozoite | 7,142 | 0.17 | 493 | 2.43 |
Western Blot Analysis
The protein extracts used in the LC-MS/MS analyses were analyzed by Western blot on a nitrocellulose membrane. Anti-falcipain-3 primary antibody (1:1000) (a gift from the laboratory of Dr. Phil Rosenthal, University of California, San Francisco) and horseradish peroxidase (HRP)-conjugated anti-rat IgG secondary (1:10,000) were detected using the ECL Advance Western blot detection kit (GE Healthcare).
Analysis of Peptographs for Time-dependent and SAK-1-dependent Processing Events
All the peptographs generated in this work were manually assessed for the presence of time-dependent processing events. To evaluate the peptographs, a side-by-side analysis was conducted (see http://www.scripps.edu/chemphys/cravatt/Bowyer2010). Specifically two sets of four peptographs were generated. The following peptograph series were compared.
T = 0 versus T = −6 for the cytosolic fraction, T = 0 (SAK) versus T = −6 for the cytosolic fraction, T = 0 versus T = −6 for the membrane fraction, and T = 0 (SAK) versus T = −6 for the membrane fraction.
T = 0 versus T = −6 for the cytosolic fraction and T = 0 versus T = −6 for the membrane fraction, the merozoite fraction, and the supernatant fraction.
These peptographs were analyzed for proteins that showed a peptide coverage at a lower molecular weight than the full-length/major peptide coverage region. In addition, for this to be classed as a time-dependent proteolysis event the “blue” peptides should be more abundant at the lower molecular weight (more abundant at T = 0 versus T = −6). The DPAP3 dependence of the processing was assessed by comparing the T = 0 and T = 0 (SAK) conditions. If the lower molecular weight species is not present in the in the T = 0 (SAK) condition, then the processing is classed as DPAP3-dependent. If the lower molecular weight species is present in both, then the processing is classed as DPAP3-independent. More significance can be attached to this later category as it requires the identification of processed peptides at a lower molecular weight in both conditions and therefore reduces the likelihood of a false positive annotation of processing. Lists generated as a result of this analysis are presented in the supplemental information. These can be submitted to the peptograph data sets hosted on the PROTOMAP web site.
RESULTS
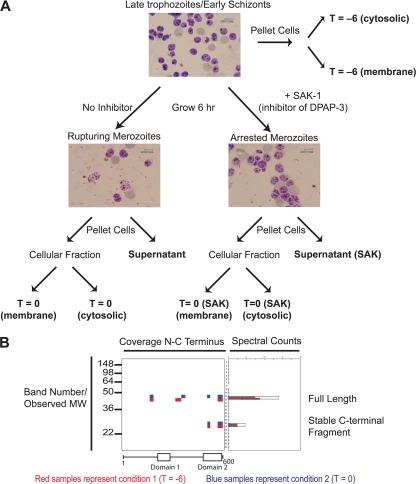
We chose to use the PROTOMAP method because it has been successfully used to determine proteolytic events occurring during apoptosis (6). In addition, the method only requires a minimal amount of protein sample so it is ideally suited for use with Plasmodium culture systems. The main work flow of sample preparation prior to SDS-PAGE analysis is shown in Fig. 1A. We started with late schizont stage parasites that were enriched using a Percoll gradient at ∼42 h postinfection. Giemsa stain analysis of the schizonts showed very few segmented merozoites at this time point. This initial sample corresponds to a time point approximately 6 h prior to rupture. We then divided the schizonts into a sample for analysis (T = −6) and a sample that was allowed to progress for an additional 6 h in serum-free media until the parasites were rupturing (T = 0). At this point we pelleted the cells and collected the cellular fraction as well as the media. The media provided a supernatant sample of proteins that were released from the parasites during rupture (“supernatant”). In addition, we performed a separate purification of the extracellular merozoites that are released after rupture from the RBC (“merozoite”). We also subjected a similar culture of late schizonts to continuous treatment with SAK-1, a recently reported highly selective inhibitor of DPAP3 (1) from the T = −6 h time point to the point of rupture (T = 0 SAK). We then performed SDS-PAGE and cut each gel lane into 26 slices for processing by in-gel digestion with trypsin followed by LC-MS/MS analysis. Table I summarizes the peptide data for each of the samples along with an FD rate generated from a decoy database (16). In these searches, we allowed for the presence of both semitryptic peptides (a requirement for lysine or arginine at one end only) and fully tryptic peptides. The FD rate increases dramatically for the semitryptic peptides, but we have included them because they can be used to map the exact site of proteolytic events. The full peptide and protein assignments for each individual gel slice and for each pooled sample are shown in supplemental Tables 1–3.
Fig. 1.
PROTOMAP analysis of parasites rupturing from host red blood cells. A, experimental setup used to generate samples for analysis by PROTOMAP. Synchronously grown parasites were isolated in the late trophozoite/early schizont stage (approximately 42 h postinfection). Samples for analysis (bold labels) were prepared as indicated. Representative stained blood smears are shown to indicate parasite morphology at each sample point. Final parasite or supernatant samples were subjected to SDS-PAGE, and each lane was excised into 26 slices followed by analysis by LC-MS/MS. B, representative peptograph generated from the data set collected from the samples in A. Peptides (colored bars) are mapped to the location on the protein sequence (top of gel image). Approximate molecular weight (MW) positions are shown at left, and total spectral counts for each peptide are shown at right. The colors are used to compare data obtained for this protein in each of two samples. In this case, peptides identified in the T = −6 are shown in red, and peptides identified in the T = 0 sample are shown in blue. A conserved domain map based on published predictions is shown at the bottom of the peptograph (21, 22). Spectral count data error bars are standard error of the mean.
Table II lists the protein identifications for each of the fractions along with the number of decoy (reversed) proteins identified. The overall number of Plasmodium proteins identified in our study is slightly higher than the number of identifications reported in PlasmoDB for the schizont stage (17–19). This confirms that the SDS-PAGE-MS approach achieves proteome coverage comparable with the pre-existing shotgun LC-MS methods (20). In addition, searching of the peptide sequences against a reverse database identified only a small number of hypothetical reversed proteins, and these all had spectral counts of less than 10, which is below a sufficient number to generate a peptograph that can be interpreted confidently.
Table II. Total number of protein identities determined for each sample.
| No. Plasmodium | No. human | No. reverse | |
|---|---|---|---|
| Cytosolic/membrane (T = −6) | 1,137 | 371 | 37 |
| Cytosolic/membrane (T = 0) | 1,024 | 334 | 27 |
| Cytosolic/membrane (T = 0 SAK) | 829 | 245 | 15 |
| Supernatant | 377 | 117 | 13 |
| Supernatant (SAK) | 254 | 108 | 5 |
| Merozoite | 841 | 115 | 3 |
To identify proteolytic cleavage events, we applied the PROTOMAP software to the resulting peptide data (http://www.scripps.edu/chemphys/cravatt/protomap/) to generate peptographs for each protein. Peptographs are, in effect, a reconstruction of the SDS-PAGE gel with peptide coverage data for each gel slice. A representative peptograph is shown (Fig. 1B). Each individual protein identified by LC-MS/MS has a corresponding peptograph that shows where specific peptides were identified (i.e. which gel slice) and how the peptides map to the full-length protein sequence. Using two different colors (i.e. red and blue) for two different conditions (i.e. T = 0 and T = −6), it is possible to directly compare migration of a protein or protein fragments in each sample. In the example provided, the full-length protein is identified in gel bands 11 and 12, and a stable C-terminal fragment is found in bands 19 and 20. The presence of a stable C-terminal fragment is indicative of an event that alters mobility on SDS-PAGE. The restriction in peptide coverage to the C terminus is consistent with a proteolytic event. Comparison of the two different conditions indicates an equal degree of proteolysis and overall abundance in the two conditions. Abundance can be determined from the average number of total spectral counts for each gel band, which is plotted on the right. Putative domains are shown at the bottom of the peptograph. For the experimental peptographs, these domain maps are derived from predictions using the conserved domain predictor (21, 22).
Peptographs for all of the identified proteins in this study can be viewed at the following link (http://www.scripps.edu/chemphys/cravatt/Bowyer2010/). All peptographs are available on this site regardless of the number of spectral counts generated. The peptographs are also linked to the malaria database, PlasmoDB, to allow direct association with specific database queries. In considering the peptographs, it is important to note that slight differences in migration rates were observed between replicate gels. Typically, a specific protein could migrate ±1 band across the replicates as suggested by molecular weight standards (data not shown). This variability is more prevalent for lower molecular weight proteins (higher band number). Although such variations in gel migration may result in broader images than would be expected from a Western blot, the resolution of the overall method is sufficient to identify many proteolytic events. In addition, nonspecific protein degradation may cause confusion in analyses of this style. However, we used protease inhibitors to limit the proteolysis after isolations of the individual samples. Regardless, several abundant proteins such as elongation factor 1-α (PF13_0304) show peptide coverage across the entire protein at many molecular weights. Thus, aspecific smearing of the peptides can be a feature for abundant proteins, but spectral counting usually differentiates the more abundant fragments, and in most cases, it is still possible to identify proteolytic processing events using peptographs.
Spectral Counting Reflects Expected Abundance Changes
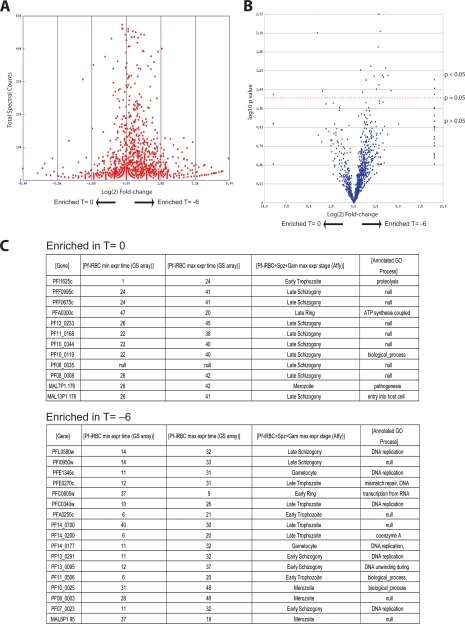
Because the PROTOMAP method provides coverage of multiple peptides for each identified protein, we could use the information from spectral counts to estimate abundance at each of the time points (23). Therefore, we could validate our approach and assess the quality of our data by analyzing predicted protein function with expression levels at the two main time points. For this analysis, we used the Student's t test to generate p values based on total spectral counts to identify proteins that changed in abundance between time points (T = −6 and T = 0). The total number of spectral counts identified in the T = −6 and T = 0 time point was 89,790 and 75,015, respectively. Thus, for any given protein, a slightly increased abundance is expected when the total spectral counts for T = −6 versus T = 0 are compared. This can be observed graphically with the slightly off center nature of the plot of the log (base 2) -fold change in abundance versus spectral counts (Fig. 2A). By plotting the -fold change in expression between the two conditions against the p value (a “volcano plot”), we could establish the degree of significance for the abundance changes for each protein (Fig. 2B). We then used the Stat-sort tool on the PROTOMAP web site to list proteins with the most significant change in abundance (p < 0.1) at each time point (Fig. 2C). This list of enriched proteins at the T = 0 time point has a peak in transcription at late schizogony, whereas proteins with the highest counts in T = −6 are mainly expressed at late trophozoite to early schizogony (11, 19, 24). Interestingly, many of these proteins from the early time point are involved in DNA replication as would be expected for parasites that are starting to produce new merozoites. Overall, these data confirm that our method is working as expected and that we have two distinct populations of parasites for comparison by the PROTOMAP method.
Fig. 2.
Analysis of PROTOMAP data set confirms stage-specific expression of proteins. A, plot of the total number of spectra counts observed for each protein versus the log2 -fold change between the early (T = −6) and late (T = 0) time points. This allows selection of proteins with the greatest life cycle stage-specific expression. B, plot of the log10 p values for each protein identified relative to the log2 -fold change in spectral counts from the early to late time points. A line of p = 0.05 is shown. C, list of the proteins most significantly enriched in the late time point (top) and early time point (bottom). Accession numbers, number of hours postinfection of maximum and minimum gene expression, and corresponding gene ontology functional annotations as reported in PlasmoDB (www.plasmodb.org) are shown. Pf-iRBC, P. falciparum-infected RBC; min expr, minimum expression; max expr, maximum expression; spz, sporozoite; Gam, gametocyte.
Use of Semitryptic Peptides to Identify Signal Peptide Removal and Proteolytic Processing Events
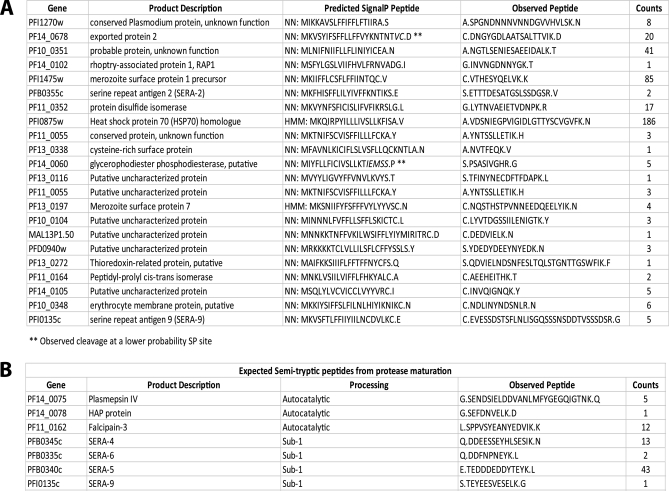
As a further validation of the complete peptide data set, we compiled a list of semitryptic peptides to determine whether we could identify known proteolytic processing events. Because these peptides result from either a natural N-terminal peptide or are produced by the action of a protease at one end of the peptide, they can be used to identify the exact site of protease processing. In a first pass analysis, we searched for semitryptic peptides that were produced as the result of removal of the signal peptide (Fig. 3A). By searching PlasmoDB, we determined that 1043 proteins are expected to have a signal peptide (25, 26). Of that total, 118 proteins were identified in our data set and had semitryptic peptides assigned. Of these 118 proteins, 14 have predicted signal peptides that are cleaved such that they result in an apparent tryptic peptide (i.e. the residue N-terminal to the cut site is naturally a Lys or Arg residue). These would therefore not yield a semitryptic terminus. We then searched the remaining 104 putative signal peptides against the full list of semitryptic peptides identified. In all, we identified 22 of the 104 predicted peptides (Fig. 3B). Most of these peptides exactly match the neural network predictions of the SignalP prediction software. We only found four discrepancies, and in two of these cases (PFI075w and PF13_0197), the hidden Markov model exactly predicts the peptides we observed. In the remaining two differences (PF14_0060 and PF14_0678), the observed peptides agree with a lower scoring potential cleavage site (25, 26). Thus, we have identified at least two signal peptide cleavage sites that would not be predicted to be the most optimal. Single identifications of the unique semitryptic peptide were tolerated in the analysis of these potential signal peptides. In all cases, other peptides matching to the protein were identified.
Fig. 3.
Identification of signal peptide removal and proteolytic maturation of proteins using PROTOMAP data. A, list of predicted signal peptides for which corresponding semitryptic peptides corresponding to the resultant mature N terminus were found. Predicted signal peptides were determined from neural network (NN) and hidden Markov model (HMM) predictions obtained from PlasmoDB. Protein accession numbers, gene ontology annotations, and total spectral counts for each peptide are also listed. Note that two peptides did not match the top predicted signal peptide cleavage site but matched a second, more weakly predicted site (indicated with **). B, list of identified proteolytic maturation sites of seven proteins known to undergo proteolytic maturation. The gene accession numbers and proteins names are shown along with the type of predicted processing expected and total number of spectral counts for identified semitryptic peptides. Periods in sequences represent cleavage sites. HAP, is plasmepsin III or histo-aspartic protease.
In a second analysis, we searched for peptides that would be generated as the result of known proteolytic maturation events of a protein target. We focused on the plasmepsins, falcipain-3, and SERA family proteases as each of these enzymes is processed in multiple locations during maturation to produce the active protease (3, 27, 28). Fig. 3B lists the semitryptic peptides that we observed in our data set that matched the predicted peptides for each of the target proteases. We identified one of the sites of autocatalytic processing of falcipain-3 and one expected semitryptic peptide for each of two plasmepsin proteases (PF14_0075 and PF14_0078). In addition, we identified four of the predicted subtilisin-1 cleavage sites for several of the SERA family proteins (3). Taken together, these results confirm that the PROTOMAP method can be used to directly identify sites of proteolytic processing. Furthermore, we can identify a significant number of cleavage events that are consistent with known processing events, thus validating the approach.
Peptographs Identify Time-dependent and Time-independent Proteolytic Events
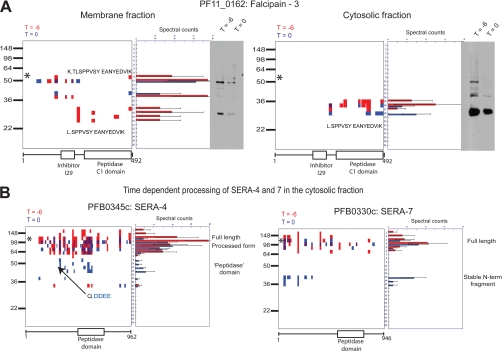
After analysis of the raw peptide data, we next focused our attention on the peptographs to identify specific processing events. To identify time-dependent proteolytic events, we manually examined all of the peptographs generated (see “Materials and Methods”). Within the data, there are examples of stable fragments occurring mostly in the early time point, mostly in the late time point, and in both time points. As an example, Fig. 4A shows peptographs comparing the T = −6 with T = 0 samples for the cysteine protease falcipain-3 (PF11_0162) for both the membrane and cytosolic fractions. These plots show multiple processing events that produce three primary fragments. Comparison with Western blots using an anti-falcipain-3 antibody raised against the C-terminal catalytic domain confirms that the fragments identified in the peptograph match the predicted size and abundance of the fragments in the blot. The inability to detect the precursor forms of falcipain-3 observed in the Western blot of the cytosolic fraction is most likely due to the fact that most of the protease in the cytosol is in the active form, and the larger precursor fragments are not sufficiently abundant to detect by in-gel digest. Falcipain-3 contains a transmembrane domain near the N terminus and is transported to the food vacuole as a result of a bifunctional motif on either side of the transmembrane domain (29). The protein undergoes autocatalysis to give the mature protease domain at acidic pH (28). Consistent with these findings, peptographs from the membrane fraction show two precursor proforms that are only weakly detected by overexposure of the Western blot of the cytosolic fraction. The largest of these fragments (∼50 kDa) likely corresponds to the full-length protein, whereas the next smaller fragment (40 kDa) likely corresponds to a loss of some of the N terminus. The catalytic domain (∼28 kDa) is found in both the cytosolic and membrane fractions. This mature form of the protease is the only form found in the cytosolic fraction most likely because loss of the N-terminal prodomain releases it from the membrane. Furthermore, the mature form of the protease is only found in the membrane fraction at the earlier T = −6 time point, suggesting that it may be cleaved. It also associates with the membrane, but this association is lost as the parasite matures at the end of the blood stage life cycle.
Fig. 4.
Peptographs identify time-dependent proteolytic processing events. A, peptographs for falcipain-3 (PF11_0162) at time T = −6 (red) and T = 0 (blue) for membrane (left) and cytosolic (right) samples. The sequences of the semitryptic peptide identified as the N-terminal most peptide in the mature 25-kDa fragment are shown in both membrane and cytosolic samples. The tryptic peptide corresponding to the mature N terminus plus an additional two N-terminal residues was identified in the full-length proform as shown. A Western blot of the same sample used for PROTMAP analysis using an anti-falcipain-3 antibody is shown for comparison with location of peptide identified in the peptographs. B, peptographs showing time-dependent processing of SERA-4 and SERA-7. The predicted Sub-1 cleavage site on SERA-4 that was identified as a semitryptic peptide is shown. * identifies the predicted size of full-length proteins (PF11_0162, 56.6 kDa; PFB0345c, 108.6 kDa; PFB0330c, 109.6 kDa). Periods in sequences represent cleavage sites.
In addition, we observed that the most N-terminal peptide identified in the 28-kDa protein is a semitryptic peptide (L↓SPPVSY…K↓) that likely constitutes the physiological N terminus of the catalytic domain. Interestingly, an in vitro study with the recombinant protease suggested that the product of autocatalysis contains an additional two residues (28). Although it is possible that the peptide has undergone processing after or during preparation of the sample, it should be noted that we identified the tryptic peptide that corresponds to the expected autocatalytic product in the 50-kDa sample (K↓TLSPPVSY….K↓). These data suggest that the catalytic domain undergoes specific trimming in the parasite as it matures. A potential candidate for this event would be dipeptidyl aminopeptidase 1 (DPAP1), which is also present in the food vacuole and whose processing activity would likely be blocked after one cleavage event by the proline residues found in the N-terminal sequence. These peptographs demonstrate the benefits of studying the membrane fractions and cytosolic fraction separately and also confirm that native proteolytic events can be observed.
Another example of proteins that undergo processing events that could be observed in the peptographs is the SERA family proteins. The SERA family consists of nine members, eight of which are expressed in the schizont stages (3). Peptographs of SERA-4 and SERA-7 from the cytosolic fraction show the time-dependent formation of a stable fragment (Fig. 4B). This 50-kDa fragment of SERA-4 corresponds to the putative protease domain of this protein and is likely a product of the time-dependent maturation of this protein because it is more abundant at T = 0. The semitryptic peptide in the most N-terminal peptide corresponds to the predicted cleavage site of subtilisin-1, a serine protease that was recently shown to be a key regulator of host cell rupture (3). Similarly, we found a time-dependent production of a protein fragment of SERA-7 corresponding to a stable N-terminal fragment of ∼38 kDa. Surprisingly, the putative peptidase domain is only detected in the full-length protein and not in the fragment. This raises the possibility that this N-terminal fragment may have a biological role that does not require the potential enzyme activity of the protein. We were also able to identify additional members of the SERA family with the exception of SERA-1 and -8 (supplemental Fig. 1). To our knowledge, SERA-1 and -2 have not been identified in existing proteomics data sets, and SERA-8 plays a vital role during the insect stages and therefore may not be expressed during the blood stages (30). Interestingly, in merozoites, we only detected SERA-5 and -7, and we only observed the smaller N-terminal fragment of SERA-7. The absence of any full-length SERA-7 in this sample suggests that this fragment may be specifically retained by the merozoite. Furthermore, because there are very few other SERA peptides in the merozoite sample, we believe that the SERA-7 fragment is indeed merozoite-associated.
Peptographs Can Detect DPAP3-dependent Processing Events
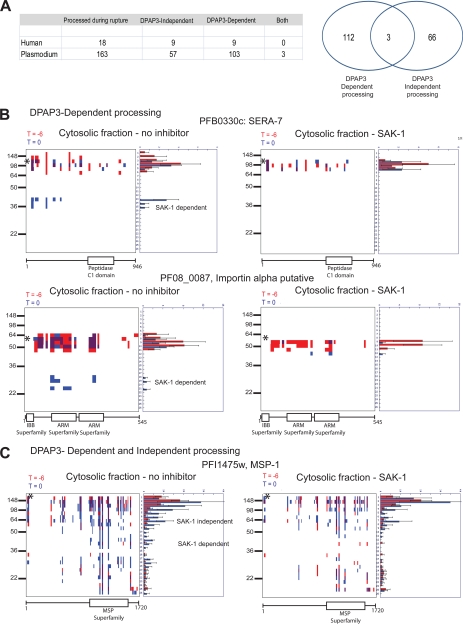
It is clear that many proteins are proteolytically cleaved during the process of host cell rupture. Therefore, we wanted to focus on processing events that were likely to have a specific role in the process of parasite egress. Analysis of the peptographs revealed 181 time-dependent processing events in which the fragment is more abundant at T = 0 (for peptographs see http://www.scripps.edu/chemphys/cravatt/Bowyer2010/). To further characterize these, we chose to perform PROTOMAP analysis on samples of parasites that had been treated with a specific inhibitor of DPAP3, a cysteine protease that we previously showed is a key regulator of host cell rupture. After manual comparison of peptographs, we determined that, of the 181 proteins that showed some form of proteolytic processing, 112 proteins showed processing events that were blocked by inhibition of DPAP1, and a further three proteins had multiple processing events of which at least one was DPAP3-dependent (Fig. 5A). These data suggest that inhibition of DPAP3 blocks the majority of the proteolytic events that occur in the course of egress from the erythrocyte, suggesting that it is a key regulator of proteolysis that may act by regulating proteins at the top of the proteolytic cascade. Interestingly, one of the most dramatic examples of DPAP3-dependent processing was observed for SERA-7 (Fig. 5B). We observed a complete loss of the N-terminal 38-kDa fragment of this protein seen in the untreated control samples. This DPAP3-dependent processing was observed in both membrane and cytosolic fractions as well as in the supernatant (data not shown). In addition to SERA-7, we also identified a number of proteins such as PF08_0087 that showed clear DPAP3-dependent processing (Fig. 5B). We also observed proteins such as MSP-1 that showed a DPAP3-independent processing event to produce an 80-kDa protein fragment as well as a 50-kDa fragment that is produced by a DPAP3-dependent mechanism (Fig. 5C). Interestingly, this fragment is an abundant fragment found in purified merozoites, suggesting that it may be one of the “mature” forms of MSP-1 (supplemental Fig. 2A). The banding patterns seen in the merozoite fraction are consistent with the expected subtilisin-1 (Sub-1)-derived fragments (31). Because we previously have shown that DPAP3 inhibition leads to loss of mature Sub-1 (1), the production of any Sub-1-dependent fragments would be expected to be lost when DPAP3 is inhibited. In agreement with this finding, we also observed loss of the mature MSP-1 fragment in the culture supernatant upon DPAP3 inhibition. An example protein in the DPAP3-independent processing category is calpastatin. This is a red blood cell-derived protein that is detected mostly in the cytosolic fraction. We observed a processing event at T = 0 to yield a slightly smaller fragment irrespective of the presence of SAK-1 (supplemental Fig. 2B).
Fig. 5.
Identification of proteolytic processing events that depend on DPAP3 activity. A, list and Venn diagram of the total number of proteins identified that were either dependent or independent of DPAP3 activity. Proteins listed as both showed multiple processing events with at least one event dependent on DPAP3 activity. B, examples of peptographs that show DPAP3-dependent processing events. C, example of a peptograph that shows multiple processing events with only one showing DPAP3 dependence. * identifies the predicted size of full-length proteins (PFB0330c, 109.6 kDa; PF08_0087, 61.1 kDa; PFI1475w, 195.7 kDa).
Peptographs Also Reveal Localization Changes and Time-independent Processing
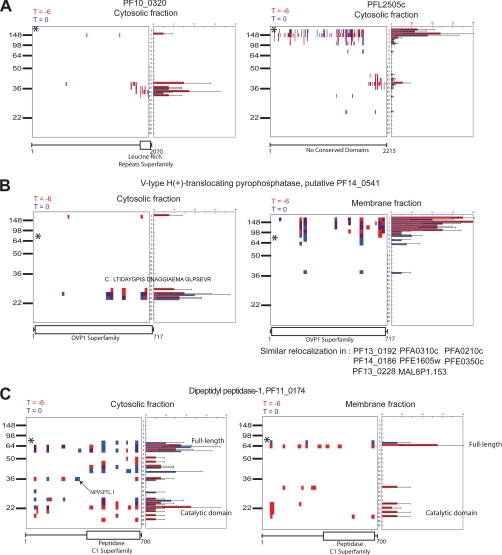
Although we were primarily interested in proteolytic processing events that specifically took place during the final hour the parasite spends in the host RBC, we also found that other biologically important data could be extracted from our data set. Specifically, we found a number of apparently constitutive processing events (Fig. 6A). This included proteins that show precursor and product at both time points as well as proteins that show only the resultant fragments. In addition, we identified a number of proteins that are constitutively processed from a membrane-associated full-length form to a C-terminal fragment that is found only in the cytosol (Fig. 6B). In one example, PF14_0541 undergoes a release of a smaller fragment into the cytosolic fraction, and a semitryptic peptide is produced at the N terminus of this fragment. This cut site may be the site of processing to release this fraction to the cytosol. A further interesting finding from this protein is the disagreement between the predicted molecular mass (76.4 kDa) and the peak of the peptide detection, which is nearer to 150 kDa. In general, the observed molecular weight for observed proteins correlates well with the predicted molecular weight. These data can be searched for each of the individual data sets on the accompanying web site (http://www.scripps.edu/chemphys/cravatt/Bowyer2010/) using the MW-plot tool.
Fig. 6.
Mining PROTOMAP data set for additional biologically relevant data. A, peptographs of two proteins comparing T = 0 (blue) with T = −6 (red) samples to demonstrate constitutive processing to produce two stable fragments. B, peptographs comparing T = 0 (blue) with T = −6 (red) for both membrane and cytosolic samples, demonstrating a time-independent processing event that results in conversion of a full-length membrane-bound protein to a cytosolic peptide fragment. C, peptographs from the cytosolic fraction showing greater processing of DPAP1 at T = −6 compared with T = 0. * identifies the predicted size of full-length proteins (PF10_0320, 246.7 kDa; PFL2505c, 263.1 kDa; PF14_0541, 76.4 kDa; PF11_0174, 80.4 kDa).
We also identified proteins, such as PF11_0174, that appear to be more processed at the early time point (Fig. 6C). The presence of increased levels of the C-terminal fragment (∼20 kDa) in the earlier time points (red peptides) is perhaps indicative of greater processing occurring earlier in the life cycle. This could be expected for a protease such as DPAP1 that is located in the food vacuole with a role in hemoglobin degradation (32). An alternative explanation could be that the mature forms of the protease, localized in the food vacuole, are lost during rupture, but the immature forms may be stored within the developing merozoite. Overall, these data demonstrate the value of the PROTOMAP method, which provides extensive coverage of each protein, thus making it possible to extract many different types of data that are likely to have biological significance.
DISCUSSION
Parasite release from the host cell is an essential part of the blood stage life cycle. Furthermore, this process leads to the majority of the pathology associated with malaria. Therefore, a better understanding of the biochemical events that take place as the parasite prepares to leave the host RBC will likely be very valuable for the identification of therapeutic strategies that can be used to treat malaria. In this study, we chose to use the newly developed PROTOMAP proteomics method to begin to map out the specific proteolytic events that take place during the final 6 h of the intraerythrocytic stage of the parasite life cycle. By using a method that requires relatively small amounts of sample, we were able to directly compare membrane and cytosolic fractions as well as samples derived from parasites treated with a specific inhibitor of a protease previously shown to be a regulator of host cell rupture. The resulting data set allowed us to extract multiple layers of biologically relevant information.
In this study, we show examples of data that reveal constitutive, time-dependent/independent and DPAP3-dependent/independent processing events. In addition, we used our data to identify specific processing events that lead to changes in protein localization. Finally, we used spectral counting to assess overall abundance of specific proteins at two distinct time points near the end of the blood stage life cycle. Because this data set is extensive and genetic validation of specific processing events in P. falciparum parasite is difficult, we relied upon established processing events to validate our approach. Interestingly, even within these few chosen examples, we were able to identify novel cleavage sites and also validate previously uncharacterized protein fragments. In addition, by making this data set available to the malaria research community through PlasmoDB, it will be possible to link additional processing events to other proteins of interest. Hopefully, this will lead to community-based validation of the proteolysis map that increases its resolution. This long term validation process, coupled with further proteomics efforts, should further enrich our understanding of specific processing events that are required for parasites to prepare themselves for release from host RBCs. This information may lead to the identification of proteases that, like DPAP3 and Sub-1, either directly or indirectly mediate proteolytic processing events that are required for host cell rupture. These proteases could serve as valuable targets for future drug discovery efforts.
Acknowledgments
We acknowledge Phil Rosenthal for assistance with anti-falcipain antibodies and Andrew Guzzetta and Melissa Dix for assistance with mass spectrometry and PROTOMAP analysis.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI078947 (to M. B.). This work was also supported by a Burroughs Wellcome new investigator in pathogenesis award (to M. B.).
 This article contains supplemental Tables 1–3 and Figs. 1 and 2.
This article contains supplemental Tables 1–3 and Figs. 1 and 2.
1 The abbreviations used are:
- DPAP
- dipeptidyl aminopeptidase
- PROTOMAP
- Protein Topography and Migration Analysis Platform
- FD
- false discovery
- Sub-1
- subtilisin-1
- SERA
- serine-repeat antigen.
REFERENCES
- 1. Arastu-Kapur S., Ponder E. L., Fonoviæ U. P., Yeoh S., Yuan F., Fonoviæ M., Grainger M., Phillips C. I., Powers J. C., Bogyo M. (2008) Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 4, 203–213 [DOI] [PubMed] [Google Scholar]
- 2. Chandramohanadas R., Davis P. H., Beiting D. P., Harbut M. B., Darling C., Velmourougane G., Lee M. Y., Greer P. A., Roos D. S., Greenbaum D. C. (2009) Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science 324, 794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yeoh S., O'Donnell R. A., Koussis K., Dluzewski A. R., Ansell K. H., Osborne S. A., Hackett F., Withers-Martinez C., Mitchell G. H., Bannister L. H., Bryans J. S., Kettleborough C. A., Blackman M. J. (2007) Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell 131, 1072–1083 [DOI] [PubMed] [Google Scholar]
- 4. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 5. Impens F., Colaert N., Helsens K., Ghesquière B., Timmerman E., De Bock P. J., Chain B. M., Vandekerckhove J., Gevaert K. (2010) A quantitative proteomics design for systematic identification of protease cleavage events. Mol. Cell. Proteomics 9, 2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dix M. M., Simon G. M., Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Timmer J. C., Enoksson M., Wildfang E., Zhu W., Igarashi Y., Denault J. B., Ma Y., Dummitt B., Chang Y. H., Mast A. E., Eroshkin A., Smith J. W., Tao W. A., Salvesen G. S. (2007) Profiling constitutive proteolytic events in vivo. Biochem. J. 407, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 10. Simon G. M., Dix M. M., Cravatt B. F. (2009) Comparative assessment of large-scale proteomic studies of apoptotic proteolysis. ACS Chem. Biol. 4, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 [DOI] [PubMed] [Google Scholar]
- 12. Lambros C., Vanderberg J. P. (1979) Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65, 418–420 [PubMed] [Google Scholar]
- 13. Pasvol G., Wilson R. J., Smalley M. E., Brown J. (1978) Separation of viable schizont-infected red cells of Plasmodium falciparum from human blood. Ann. Trop. Med. Parasitol. 72, 87–88 [DOI] [PubMed] [Google Scholar]
- 14. Taylor H. M., Grainger M., Holder A. A. (2002) Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 70, 5779–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 17. Florens L., Liu X., Wang Y., Yang S., Schwartz O., Peglar M., Carucci D. J., Yates J. R., 3rd, Wub Y. (2004) Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol. Biochem. Parasitol. 135, 1–11 [DOI] [PubMed] [Google Scholar]
- 18. Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 19. Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J., Jr., Treatman C., Wang H. (2009) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cottingham K. (2010) 1DE proves its worth … again. J. Proteome Res. 9, 1636. [DOI] [PubMed] [Google Scholar]
- 21. Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Lu S., Marchler G. H., Mullokandov M., Song J. S., Tasneem A., Thanki N., Yamashita R. A., Zhang D., Zhang N., Bryant S. H. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37, D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchler-Bauer A., Bryant S. H. (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 24. Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 26. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 27. Banerjee R., Francis S. E., Goldberg D. E. (2003) Food vacuole plasmepsins are processed at a conserved site by an acidic convertase activity in Plasmodium falciparum. Mol. Biochem. Parasitol. 129, 157–165 [DOI] [PubMed] [Google Scholar]
- 28. Sijwali P. S., Shenai B. R., Gut J., Singh A., Rosenthal P. J. (2001) Expression and characterization of the Plasmodium falciparum haemoglobinase falcipain-3. Biochem. J. 360, 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian S., Sijwali P. S., Rosenthal P. J. (2007) Falcipain cysteine proteases require bipartite motifs for trafficking to the Plasmodium falciparum food vacuole. J. Biol. Chem. 282, 24961–24969 [DOI] [PubMed] [Google Scholar]
- 30. Aly A. S., Matuschewski K. (2005) A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koussis K., Withers-Martinez C., Yeoh S., Child M., Hackett F., Knuepfer E., Juliano L., Woehlbier U., Bujard H., Blackman M. J. (2009) A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 28, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klemba M., Gluzman I., Goldberg D. E. (2004) A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. J. Biol. Chem. 279, 43000–43007 [DOI] [PubMed] [Google Scholar]