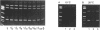Abstract
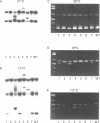
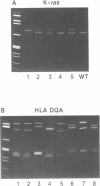
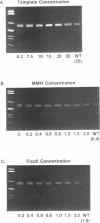
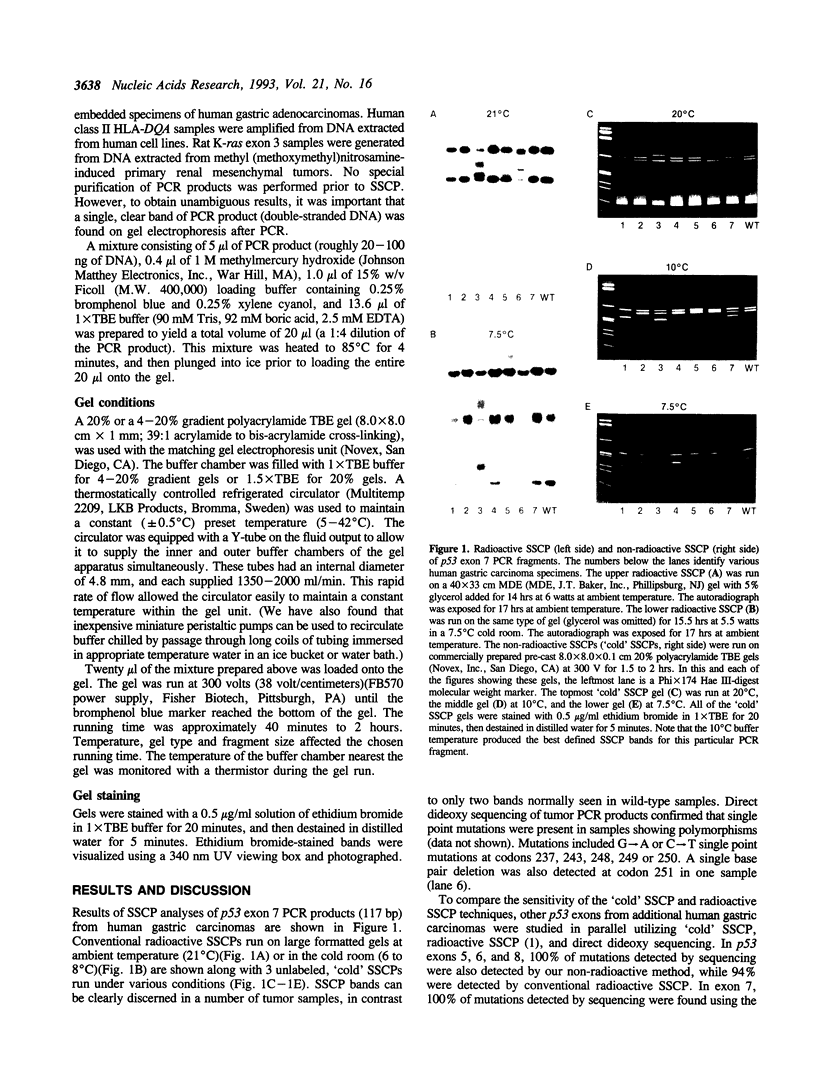
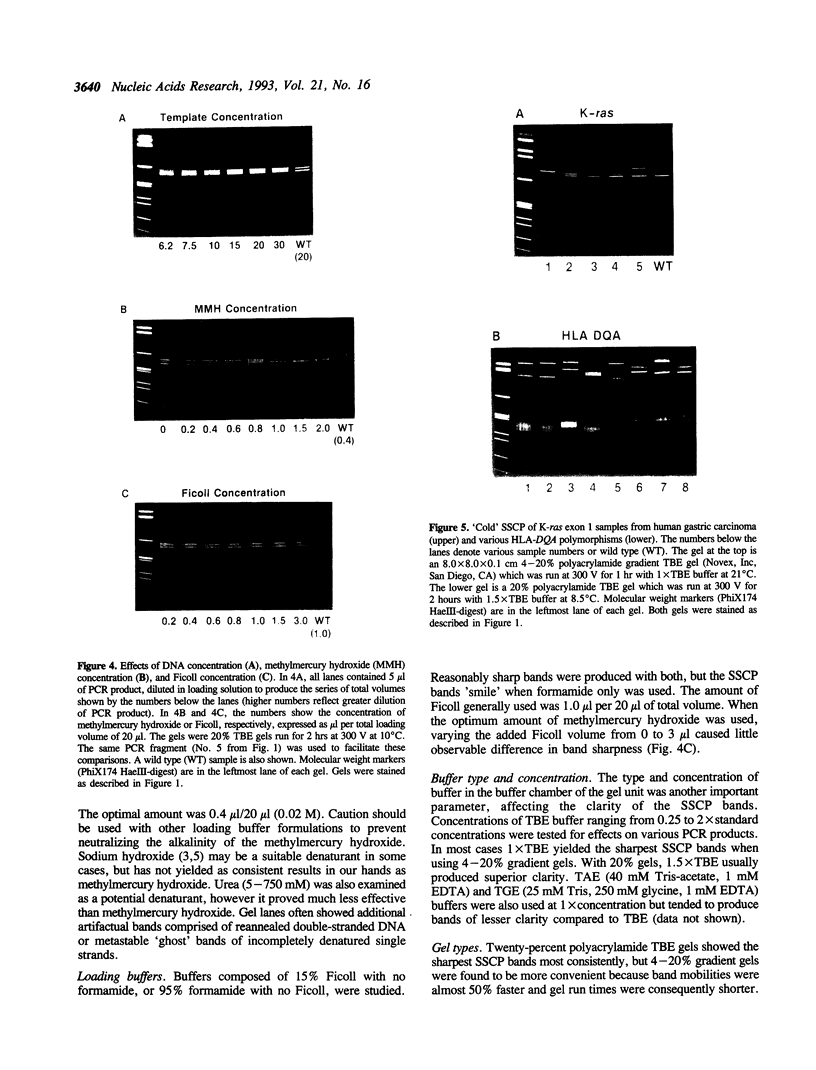
A rapid (< 2.5 hrs) method for single-strand conformation polymorphism (SSCP) analysis of PCR products that allows the use of ethidium bromide staining is described. PCR products ranging in size from 117 to 256 bp were evaluated for point mutations and polymorphisms by 'cold SSCP' in commercially available pre-cast polyacrylamide mini-gels. Several electrophoretic parameters (running temperature, buffers, denaturants, DNA concentration, and gel polyacrylamide concentration) were found to influence the degree of strand separation and appeared to be PCR fragment specific. Use of the 'cold' SSCP technique and the mini-gel format allowed us to readily optimize the electrophoretic conditions for each PCR fragment. This greatly increased our ability to detect polymorphisms compared to conventional, radioisotope-labeled 'hot' SSCP, typically run under two standard temperature conditions. Excellent results have been obtained in resolving mutant PCR fragments from human p53 exons 5 through 8, human HLA-DQA, human K-ras exons 1 and 2, and rat K-ras exon 3. Polymorphisms could be detected when mutant DNA comprised as little as 3% of the total gene copies in a PCR mixture. Compared to standard 'hot' SSCP, this novel non-isotopic method has additional advantages of dramatically increased speed, precise temperature control, reproducibility, and easily and inexpensively obtainable reagents and equipment. This new method also lacks the safety and hazardous waste management concerns associated with radioactive methods.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993 Mar 26;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Ainsworth P. J., Rodenhiser D. I., Costa M. T. Identification and characterization of sporadic and inherited mutations in exon 31 of the neurofibromatosis (NF1) gene. Hum Genet. 1993 Mar;91(2):151–156. doi: 10.1007/BF00222716. [DOI] [PubMed] [Google Scholar]
- Ainsworth P. J., Surh L. C., Coulter-Mackie M. B. Diagnostic single strand conformational polymorphism, (SSCP): a simplified non-radioisotopic method as applied to a Tay-Sachs B1 variant. Nucleic Acids Res. 1991 Jan 25;19(2):405–406. doi: 10.1093/nar/19.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidis C., Jansen G., Amemiya C., Shutler G., Mahadevan M., Tsilfidis C., Chen C., Alleman J., Wormskamp N. G., Vooijs M. Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature. 1992 Feb 6;355(6360):548–551. doi: 10.1038/355548a0. [DOI] [PubMed] [Google Scholar]
- Dockhorn-Dworniczak B., Dworniczak B., Brömmelkamp L., Bülles J., Horst J., Böcker W. W. Non-isotopic detection of single strand conformation polymorphism (PCR-SSCP): a rapid and sensitive technique in diagnosis of phenylketonuria. Nucleic Acids Res. 1991 May 11;19(9):2500–2500. doi: 10.1093/nar/19.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I., Perez-Magallanes M., Rossi J. M. Neurotrophic factor-like activity in Drosophila. Biochem Biophys Res Commun. 1992 Apr 15;184(1):73–79. doi: 10.1016/0006-291x(92)91159-n. [DOI] [PubMed] [Google Scholar]
- La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991 Jul 4;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Lázaro C., Estivill X. Mutation analysis of genetic diseases by asymmetric-PCR SSCP and ethidium bromide staining: application to neurofibromatosis and cystic fibrosis. Mol Cell Probes. 1992 Oct;6(5):357–359. doi: 10.1016/0890-8508(92)90027-u. [DOI] [PubMed] [Google Scholar]
- Maekawa M., Sudo K., Kitajima M., Matsuura Y., Li S. S., Kanno T. Detection and characterization of new genetic mutations in individuals heterozygous for lactate dehydrogenase-B(H) deficiency using DNA conformation polymorphism analysis and silver staining. Hum Genet. 1993 Mar;91(2):163–168. doi: 10.1007/BF00222718. [DOI] [PubMed] [Google Scholar]
- Makino R., Yazyu H., Kishimoto Y., Sekiya T., Hayashi K. F-SSCP: fluorescence-based polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP) analysis. PCR Methods Appl. 1992 Aug;2(1):10–13. doi: 10.1101/gr.2.1.10. [DOI] [PubMed] [Google Scholar]
- Mohabeer A. J., Hiti A. L., Martin W. J. Non-radioactive single strand conformation polymorphism (SSCP) using the Pharmacia 'PhastSystem'. Nucleic Acids Res. 1991 Jun 11;19(11):3154–3154. doi: 10.1093/nar/19.11.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Suzuki Y., Sekiya T., Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989 Nov;5(4):874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Reiss J., Lenz U., Rininsland F., Ballhausen P., Drews D., Posselt H. G. A novel CFTR mutation, 4035delA, detected by non-radioactive SSCP analysis. Hum Genet. 1992 Nov;90(3):303–304. doi: 10.1007/BF00220085. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Heitz D., Mandel J. L. The unstable and methylatable mutations causing the fragile X syndrome. Hum Mutat. 1992;1(2):91–96. doi: 10.1002/humu.1380010202. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Ueno S., Yorifuji S., Sugiyama Y., Ide Y., Matsuzawa Y. New mutant gene (transthyretin Arg 58) in cases with hereditary polyneuropathy detected by non-isotope method of single-strand conformation polymorphism analysis. Biochem Biophys Res Commun. 1991 Oct 15;180(1):380–385. doi: 10.1016/s0006-291x(05)81304-x. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Yoon H. S., Sommer S. S. Screening for mutations by RNA single-strand conformation polymorphism (rSSCP): comparison with DNA-SSCP. Nucleic Acids Res. 1992 Feb 25;20(4):871–878. doi: 10.1093/nar/20.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano K., Kyogoku A., Fukayama N., Ohkura H., Shimosato Y., Sekiya T., Hayashi K. Methods in laboratory investigation. Rapid and simple detection of c-Ki-ras2 gene codon 12 mutations by nonradioisotopic single-strand conformation polymorphism analysis. Lab Invest. 1993 Mar;68(3):361–366. [PubMed] [Google Scholar]
- Yap E. P., McGee J. O. Nonisotopic SSCP and competitive PCR for DNA quantification: p53 in breast cancer cells. Nucleic Acids Res. 1992 Jan 11;20(1):145–145. doi: 10.1093/nar/20.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap E. P., McGee J. O. Nonisotopic SSCP detection in PCR products by ethidium bromide staining. Trends Genet. 1992 Feb;8(2):49–49. doi: 10.1016/0168-9525(92)90339-6. [DOI] [PubMed] [Google Scholar]