Abstract
Proteolytic processing modifies the pleiotropic functions of many large, complex, and modular proteins and can generate cleavage products with new biological activity. The identification of exact proteolytic cleavage sites in the extracellular matrix laminins, fibronectin, and other extracellular matrix proteins is not only important for understanding protein turnover but is needed for the identification of new bioactive cleavage products. Several such products have recently been recognized that are suggested to play important cellular regulatory roles in processes, including angiogenesis. However, identifying multiple cleavage sites in extracellular matrix proteins and other large proteins is challenging as N-terminal Edman sequencing of multiple and often closely spaced cleavage fragments on SDS-PAGE gels is difficult, thus limiting throughput and coverage. We developed a new liquid chromatography-mass spectrometry approach we call amino-terminal oriented mass spectrometry of substrates (ATOMS) for the N-terminal identification of protein cleavage fragments in solution. ATOMS utilizes efficient and low cost dimethylation isotopic labeling of original N-terminal and proteolytically generated N termini of protein cleavage fragments followed by quantitative tandem mass spectrometry analysis. Being a peptide-centric approach, ATOMS is not dependent on the SDS-PAGE resolution limits for protein fragments of similar mass. We demonstrate that ATOMS reliably identifies multiple proteolytic sites per reaction in complex proteins. Fifty-five neutrophil elastase cleavage sites were identified in laminin-1 and fibronectin-1 with 34 more identified by matrix metalloproteinase cleavage. Hence, our degradomics approach offers a complimentary alternative to Edman sequencing with broad applicability in identifying N termini such as cleavage sites in complex high molecular weight extracellular matrix proteins after in vitro cleavage assays. ATOMS can therefore be useful in identifying new cleavage products of extracellular matrix proteins cleaved by proteases in pathology for bioactivity screening.
Recently, considerable efforts have been deployed to develop high throughput proteomic screens to identify protease substrates in complex biological samples (1–8). Validation of substrates identified by these approaches or identification of cleavage sites by in vitro incubation of candidate substrates with the protease of interest is generally performed by SDS-PAGE analysis and Edman degradation and sequencing. However, the complexity of large modular proteins renders Edman sequencing of proteolytic fragments difficult to apply because each of the numerous proteolytic fragments should be analyzed separately, and high coverage of cleavage sites is rarely attained (9). Cleavage site identification after protein degradation is also very difficult for small peptide products less than 4 kDa. Consequently, the precise cleavage sites in complex extracellular matrix proteins such as laminin and fibronectin by important tissue and inflammatory cell proteases such as the matrix metalloproteinases (MMPs)1 and neutrophil elastase are mostly unknown.
These limitations of Edman sequencing are problematic in the study of tissue remodeling and proteolysis in pathology. Neutrophil elastase and several MMPs such as MMP2, MMP8, and MMP9 play key roles in inflammation (10, 11), tissue healing (12, 13), and carcinogenesis (14, 15) and are well known for degrading extracellular matrix proteins (16). More recently, signaling functions for MMPs are increasingly recognized as one of their most important roles by the precise processing of cytokines or their binding proteins (17). In addition, several important examples are now known of cryptic binding sites being exposed after precise protein cleavage or new proteins termed neoproteins (18) being released upon limited cleavage of extracellular matrix proteins and having completely different functions compared with their parent molecule, including several with importance in angiogenesis (19–25). Many such sites or neoproteins are generated by inflammatory proteases or proteases of the coagulation and fibrinolysis systems (24, 25), and this is a burgeoning field of discovery that is often hampered by difficulties in their N-terminal sequencing.
In light of this limitation, we developed, validated, and used a new method for targeted and simultaneous N-terminal sequencing of one or a small number of protein N termini or cleavage products we call amino-terminal oriented mass spectrometry of substrates (ATOMS). We applied ATOMS for the analysis of cleavage sites generated in laminin-1 and fibronectin-1 by neutrophil elastase and neutrophil and tissue MMPs. Laminin-1 (LM-111), a trimeric glycoprotein composed of the α1, β1, and γ1 chains, is ubiquitously expressed in epithelium and endothelium. Proteolytic processing of laminins greatly affects cellular behavior and is also implicated in cancer cell migration (20, 26–29). Another important extracellular matrix protein is plasma fibronectin (also known as fibronectin isoform 1) and its cellular isoforms, which are homodimers linked by a disulfide bridge at the C terminus (30) that are important for cell adhesion and intracellular signaling (31–34). Fibronectin is susceptible to proteolysis (35, 36), which affects its biological functions (37–39). However, the cleavage sites within these two molecules by inflammatory MMPs and neutrophil elastase are largely unknown. Here we identified a total of 55 neutrophil elastase cleavage sites in LM-111 and fibronectin-1 and 34 MMP cleavage sites, demonstrating the capacity of ATOMS to identify multiple N-terminal sequences in solution. ATOMS also outperformed N-terminal Edman sequencing with 50% more cleavage sites identified by ATOMS, representing a significant advance in N-terminal sequencing technology. The utility of the method is broadly applicable for the analysis of multiple cleavages in other very large molecules and so offers great potential to accurately identify and rapidly sequence multiple cryptic bioactive protein fragments liberated following proteolytic processing.
EXPERIMENTAL PROCEDURES
Materials
Human plasma fibronectin, LM-111 from Engelbreth-Holm-Swarm murine carcinoma and sequencing grade Staphylococcus aureus protease V8 (Glu-C) (EC 3.4.21.19) were purchased from Sigma. Isotopically labeled heavy formaldehyde (13CD2O) was purchased from Cambridge Isotope Laboratories, Inc. Sodium cyanoborohydride (NaCNBH3) was from Sterogene Bioseparations (ALD reagent). Recombinant human MMP2 (EC 3.4.24.24), MMP9 (EC 3.4.24.35), and MMP8 (EC 3.4.24.34) were expressed and purified as described previously (40, 41). Human neutrophil elastase (EC 3.4.21.37) was purchased from Elastin Products Co. Inc. Sequencing grade trypsin was purchased from Promega, and Sep-Pak light C18 cartridges were from Waters. All other reagents were the purest grade available.
Endoprotease Digestion
LM-111 and fibronectin-1 (25 μg) were each incubated with Glu-C at an enzyme to substrate ratio of 1:80 (w/w) at 25 °C for 16 h in 25 mm ammonium bicarbonate, pH 8.0. For cleavage by human neutrophil elastase (1:20), incubations were at 37 °C in 50 mm HEPES, 100 mm NaCl, 10 mm CaCl2, pH 7.8, for 16 h. MMPs were activated with 1 mm 4-aminophenylmercuric acetate for 1 h at 37 °C. MMPs 2, 8, and 9 were incubated with laminin-1 (1:20) and fibronectin-1 (1:40). LM-111 and fibronectin-1 concentrations were 0.83 mg/ml for all digestion reactions except for fibronectin digested with MMP-2 and MMP-8 (0.75 mg/ml) and denatured LM-111 and fibronectin digested with Glu-C (0.17 mg/ml).
Edman Sequencing
Protein (15 μg) treated or not with protease was separated by SDS-PAGE (10 or 15% acrylamide/bisacrylamide) and transferred to a polyvinylidene fluoride membrane. Bands were stained with Coomassie Blue R-250, excised, and submitted to five cycles of N-terminal sequencing using the Edman method (ABI 494 protein sequencer, Tufts University Core Facility).
Isotope Labeling and Trypsin Digestion
Proteins (10 μg) were denatured with 4 m guanidine hydrochloride at 65 °C for 10 min with cysteine reduction (5 mm DTT at 65 °C for 1 h) and alkylation (15 mm iodoacetamide at room temperature in the dark for 2 h). The proteins were isotope-labeled: Endoprotease-treated samples were incubated with 100 mm 13CD2O (heavy formaldehyde) in the presence of 50 mm sodium cyanoborohydride and control samples were incubated with 12CH2O (light formaldehyde) under the same conditions. Heavy and light labeled proteins were combined; precipitated with 8 volumes of acetone and 1 volume of methanol; resuspended in 300 μl of 0.8 m guanidine hydrochloride, 100 mm HEPES, pH 8.0; and digested overnight with trypsin. Peptides were desalted by reverse-phase chromatography using Sep-Pak light C18 cartridges and resuspended in 0.1% formic acid.
Liquid Chromatography-Tandem Mass Spectrometry
All samples (1 μg) were submitted to reverse-phase liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using an Ultimate LC system coupled to either a QStar Pulsar Hybrid or a QStar XL Hybrid ESI quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems/MDS-Sciex). The mass spectrometer was operated in positive mode with a data-dependent acquisition method and with a selected precursor mass/charge range of 400–1200 m/z. Peptides with 2+ to 4+ charge state were analyzed, and the two most abundant peptides above a threshold of 30 counts were selected for MS/MS and dynamically excluded for 60 s with a 100-millimass unit mass tolerance.
Mass Spectrometry Data Analysis
Peak lists (.mgf files) were generated with Analyst 1.1 software using the default parameters and used for database searches with Mascot 2.2.2. Mass spectrometry data were searched against the mouse International Protein Index database (version 3.69, 113,982 entries, including reversed decoy sequences with scrambled Lys and Arg) for LM-111 samples or the human International Protein Index database (version 3.69, 174,499 entries, including reversed decoy sequences with scrambled Lys and Arg) for fibronectin samples. Two searches per sample were conducted; the first with lysine and N-terminal dimethylation with heavy formaldehyde (+34.0631 Da) and the second with light formaldehyde (+28.0311 Da) set as fixed modifications. To quantify all tryptic peptides with labeled lysines for isotope ratio cutoff determination, N-terminal dimethylation modification in the search parameters was omitted. Fixed modification for carboxymethylation of cysteines and variable modification of methionine oxidation were applied to all searches. Enzyme specificity was semi-Arg-C, precursor and fragment ion mass tolerance was 0.4 Da, and a maximum of three miscleavages were allowed. Results were analyzed with the Trans-Proteomic Pipeline version 4.3 (42). Using .wiff files, mzXML files were generated with mzWiff software using the default parameters. Mascot result files (.dat) were converted to pepXML, and the two pepXML files (heavy and light searches) for each sample were combined using the XInteract tool, and quantification was achieved using the automated statistical analysis on protein ratio (ASAPRatio) tool of the Trans-Proteomic Pipeline (43). All peptides originating from LM-111 and fibronectin-1 were extracted from the peptide list, and their quantification values were manually verified. The Mascot peptide ion score was used to filter the peptides identified with 95% or higher confidence (≥46 and ≥48 for LM-111 and fibronectin searches, respectively). (Note that the ion score cutoff differs between LM-111 and fibronectin due to species differences resulting in different database sizes used for Mascot searches that thereby affect the ion score parameter.) MS/MS spectra of protease-generated peptides are presented in the supplemental spectrum file.
Accession Information
All LC-MS/MS data (.wiff files) associated with this study may be downloaded from Tranche (proteomecommons.org) using the following hash: vkT00SX97cSok0ZbwTvmbFZiiQivF07UH1me8n+4lsCGcycT3xtf2Ng8j1EGqhsh/AA3Mid/DEDKZfgB0w8uQyCwPq0AAAAAAAAhAg==. The hash may be used to prove exactly what files were published as part of this study's data set, and the hash may also be used to check that the data have not changed since publication.
RESULTS
ATOMS Work Flow
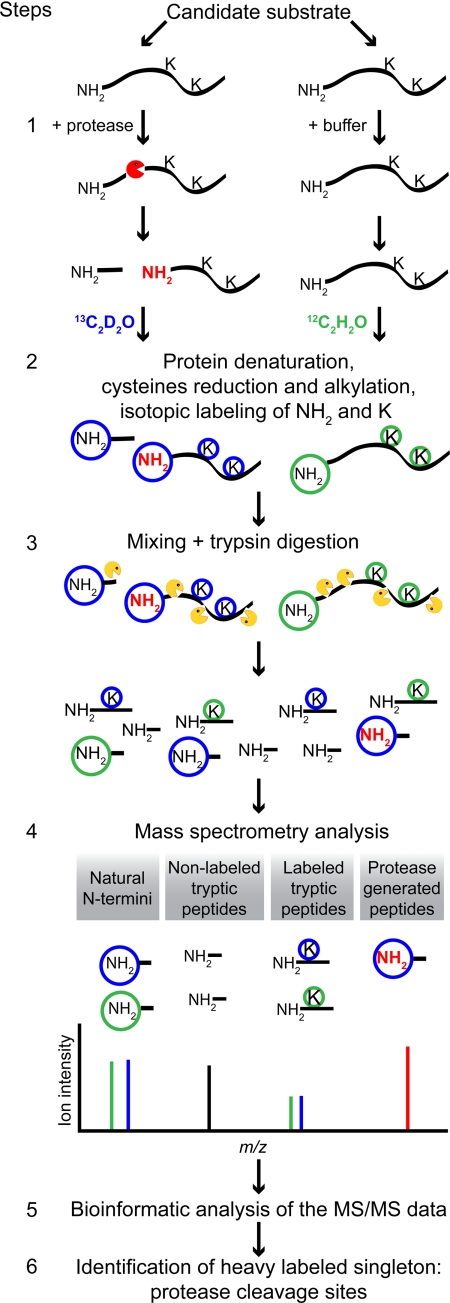
The first step in ATOMS is to incubate the substrate protein with the protease of interest or with buffer alone (control sample) (Fig. 1, Step 1). Here we used two complex extracellular matrix proteins, fibronectin-1 and LM-111, that were digested by different inflammatory and tissue MMPs and neutrophil elastase. Protein fragments bearing protease-generated neo-N termini are generated and so are only found in the protease-treated sample but not in the control sample. In ATOMS, the neo-N termini are the signature used to identify the sequence of the carboxyl side of the protease cleavage sites, also known as the prime side (P′) of proteolytic sites (see later in Step 6). In Step 2, the two samples are denatured, the cysteines are alkylated, and the protein primary amines (N termini and lysines) are isotope-labeled by reductive dimethylation using 13CD2O (heavy formaldehyde; blue circles) or 12CH2O (light formaldehyde; green circles) for the protease-treated and control samples, respectively. The mass of the peptides increases by 34.06 or 28.03 Da for each residue labeled with heavy or light formaldehyde, respectively.
Fig. 1.
ATOMS work flow. Step 1, one aliquot of the candidate substrate is incubated with the protease (red), and the second aliquot is incubated with buffer only (control). The protease treatment generates proteolytic fragments bearing neo-N termini not found in the control aliquot. Step 2, the proteins are denatured, their cysteines are reduced and alkylated, the amine groups (N termini and lysine side chains) of the peptides in the protease-treated aliquot are labeled with heavy formaldehyde (blue circles), and the amino groups of the control aliquot are labeled with light formaldehyde (green circles). This results in a mass difference of 6 Da/label for similar peptides labeled with heavy versus light formaldehyde, which is easily detectable by MS. Step 3, the two aliquots are mixed together and digested with trypsin (yellow), which cuts only after arginine residues due to the lysine dimethylation. This is advantageous as it generates longer peptides, facilitating their identification. Step 4, this peptide mixture is submitted to LC-MS/MS where four different types of peptides are observed. (i) Protein natural N termini (green/blue) as well as peptides with an N terminus resulting from proteolysis already having taken place in the sample before assay yield doublets of equal intensity separated by the mass difference induced by the isotope labeling. (ii) Tryptic peptides without lysine in their sequence are found as unlabeled singletons (black). (iii) Internal tryptic peptides containing one or more lysines in their sequence are found in equal amounts in the protease- and buffer-treated sample, resulting in doublets of the same intensity (green/blue) in the MS1 spectra. (iv) Neo-N-terminal peptides, found only in the protease-treated samples, are identified as heavy labeled singletons in the MS1 spectra, which is the signature indicating that the peptide originates from proteolysis (red). Step 5, the LC-MS/MS results are searched using the Mascot database search engine, and the peptides are quantified using the ASAPRatio software tool. Step 6, the heavy labeled singletons are identified, and their position in the full-length protein is determined, allowing the identification of the cleavage site.
In Step 3 of the ATOMS work flow, the two samples are combined and digested with trypsin. In this protocol, trypsin cleaves only after arginine residues because the lysines are blocked by dimethylation. This is advantageous as it generates longer peptides, facilitating their identification. The tryptic digestion generates two types of peptides. 1) Semitryptic peptides are those where the peptides have a neo-N terminus generated by the protease of interest and a C terminus generated by trypsin. Such peptides have a labeled N terminus and so can be identified later in the work flow as the cleavage site. Protein natural N and C termini are also semitryptic peptides, and if the original N terminus of the protein is not naturally blocked, such as by acetylation or cyclization, it too will be labeled. 2) Fully tryptic peptides are those cleaved by trypsin at both termini and are the most abundant in the sample after tryptic digestion. Being generated in a separate step after labeling, they are therefore not labeled at the N terminus.
The peptides are then subjected to LC-MS/MS (Step 4). The ASAPRatio quantification value of each peptide is its relative abundance in the protease-treated/control samples. Protein natural N termini and any degradation products present in the sample before protease digestion as well as tryptic peptides containing one (or more) lysines are found as doublets of equal intensity in the MS1 spectra as they are equally present in both the protease-treated and control samples. Tryptic peptides without lysine (i.e. not labeled) generate non-quantifiable peptide ions. Neo-N-terminal peptides are found in the MS1 spectra as signature heavy labeled single, non-paired peptide ions called singletons. In the case where the protein N terminus is processed by the protease under study, these will be present at lower abundance in the protease-treated sample and will also generate a neo-N terminus represented by a singleton in the MS1 spectra. The LC-MS/MS data are analyzed using the database search engine Mascot (44), specifying dimethylation of the N termini and lysines as fixed (obligatory) modifications (Step 5). This key feature means that fully tryptic peptides, which canonically lack a dimethylated N terminus, are ignored by the search engine, leading to data enriched for protease-generated peptides labeled both at their N terminus and lysines if present. Identification of the peptides giving rise to heavy labeled singletons gives the P′ sequence of the protease cleavage site (Step 6). The P side of the cleavage site is then filled in bioinformatically using the sequence of the protein.
Quantification Validation
To be effective in distinguishing neopeptides with increased abundance generated by proteolysis from those present at equal abundance, that is tryptic peptides or protein N-terminal peptides, whether present as the original mature protein terminus or degradation products present in the protein sample before assay, isotopic labeling was used for relative quantification of the peptides. We took two equal aliquots of LM-111 and labeled one with light formaldehyde and the other with heavy formaldehyde and processed the samples by ATOMS. This experiment was performed in duplicate and repeated using fibronectin-1. As determined by database searches, labeling efficiency was 97.9 and 99.0% for the light and heavy labels, respectively. We predicted that in these controls the heavy and light versions of all peptide pairs should be found in equal amounts as none of the samples were exposed to proteolysis and so could be used as a statistical classifier. The database searches were modified to identify and quantify all the peptides labeled on lysine residues (see supplemental Tables 1–4). A mean quantification value of 1.14 ± 0.47 was obtained from 600 independent peptide quantifications (331 unique peptides) from four separate experiments having individual values of 1.14 ± 0.26, 1.23 ± 0.65, 1.15 ± 0.25, and 0.85 ± 0.1 (45). According to this result, when a sample is treated with a protease, a peptide with a protease/control ratio >2.5 (mean + 3 standard deviations) is considered significantly elevated in the protease-treated sample and thus was generated by proteolysis. Conversely, peptides with a protease/control ratio <0.4 are highly enriched in the control sample and so were depleted by proteolysis.
We also performed database searches of these control LC-MS/MS data sets with N-terminal dimethylation as a fixed modification (the parameters used to identify protease substrates). Several semitryptic internal peptides with dimethylated N termini and an isotope ratio centered on 1.0 were identified (supplemental Tables 5 and 6) and interpreted as natural degradation products present in the protein samples before the assay. The semitryptic peptide of the natural N terminus of the LM-111 γ1 chain (AMDECADEGGRPQR) was identified in the two replicates; it was labeled with both the heavy and light formaldehyde and displayed a protease/control ratio of 1.0. Several peptides with ratios greatly deviating from 1 (supplemental Tables 7 and 8) were outliers caused by low peptide ion intensities close to the noise level (below 300 with some in the range of 25–55) or by misidentification of the peptide by the database search engine, which leads to inaccurate peptide quantification. These outliers were identified and removed from other data sets.
Method Validation: Identification of Protease Cleavage Sites Using Glu-C
Before analyzing the digestion patterns and cleavage sites of the broad cleavage site specificity MMPs and elastase, it was important to validate ATOMS using a protease of known specificity. We used Glu-C, which specifically cleaves after glutamate residues and rarely after aspartate (46), thus allowing for manual validation of the identified cleavage sites.
The database search engine Mascot was used to identify the peptides from the experimental MS/MS spectra. Of note, Mascot was developed to identify proteins, and thus, all peptides attributed to the same protein are regrouped and listed irrespective of their individual level of confidence. However, for analysis of cleavage sites, we are interested in individual peptides to precisely determine the proteolytic cleavage sites. For this reason, we consider only the peptides that either 1) have a Mascot ion score associated with ≥95% confidence at the peptide identification level (ion scores 46 and 48 for LM-111 and fibronectin-1 database searches, respectively) or 2) were identified in two or more forms by multiple MS/MS spectra. The latter assumes that it is unlikely that two different spectra are matched to the same peptide by pure chance, and thus, this increases the confidence in the peptide identification.
Denatured LM-111 and fibronectin-1 were incubated with Glu-C, and 31 peptides were identified having a dimethylated N terminus and a protease/control ratio >2.5 (Table I and supplemental Tables 9 and 10). The location of the cleavage sites in the protein amino acid sequence is presented in the supplemental material. All conformed to the known Glu-C cleavage specificity, thus validating the cutoff ratio we experimentally determined. These numbers are conservative as other peptides with a Glu at P1 and a high ratio were identified, indicating that they were products of Glu-C cleavage but did not reach the 95% confidence in peptide identification.
Table I. Cleavage sites generated by Glu-C in LM-111 and fibronectin-1.
Top, neo-N-terminal peptides from denatured LM-111 and fibronectin-1 partially digested with Glu-C. Bottom, neo-N-terminal peptides from native LM-111 and fibronectin-1 partially digested with Glu-C. Cleavage sites were identified with ATOMS. Neo-N-terminal peptides from LM-111 are presented on the left side of the table, and peptides from fibronectin are on the right. Positions of the neo-N-terminal peptides in the precursor proteins are identified by the numbers in superscript at the beginning of the peptides.
| P1 | Identified LM-111 peptides | P1 | Identified fibronectin-1 peptides |
|---|---|---|---|
| Proteolysis of denatured LM-111 and fibronectin-1 with Glu-C | |||
| α1 | |||
| Glu | 1368LCVCPPGTAGHSCQDCAPGYYR | Glu | 67RTYLGNALVCTCYGGSR |
| Glu | 1730LKAAKDLLSR | Glu | 89SKPEAEETCFDKYTGNTYR |
| Glu | 2029SAVKTLEDVLALSLR | Glu | 93AEETCFDKYTGNTYR |
| Glu | 2036DVLALSLR | Glu | 272RHTSVQTTSSGSGPFTDVR |
| Glu | 2084MQANLLLDR | Glu | 393QDQKYSFCTDHTVLVQTR |
| Glu | 2873IMVNGQQLDKDRPLSASAVDR | Glu | 566TGTFYQIGDSWEKYVHGVR |
| Glu | 2901GTFFEGSGYAALVKEGYKVR | Glu | 756LSEEGDEPQYLDLPSTATSVNIPDLLPGR |
| β1 | |||
| Glu | 33GSCYPATGDLLIGR | Glu | 1256SVPISDTIIPAVPPPTDLR |
| Glu | 172SSFPGISTGPMKKVDDIICDSR | Glu | 2227SGFKLLCQCLGFGSGHFR |
| Glu | 1006GDHCQLCQYGYYGDALR | Glu | 2311YLGAICSCTCFGGQR |
| Glu | 1099FTGQCQCMPGFGGR | Glu | 2344GTTGQSYNQYSQR |
| Glu | 1120LFWGDPDVECR | ||
| Glu | 1559VILQQSAADIAR | ||
| γ1 | |||
| Glu | 392ACSPCHCSPVGSLSTQCDSYGR | ||
| Glu | 538WSSDRQDIAVISDSYFPR | ||
| Glu | 804LCDDGYFGDPLGSNGPVR | ||
| Glu | 847CLKCIYNTAGFYCDR | ||
| Glu | 1050LESLIANLGTGDDMVTDQAFEDR | ||
| Glu | 1087AQEVKDVDQNLMDR | ||
| Glu | 1361ANDILNNLKDFDR | ||
| Proteolysis of native LM-111 and fibronectin-1 with Glu-C | |||
| β1 | |||
| Glu | 26FSYGCAEGSCYPATGDLLIGR | Glu | 619TPSQPNSHPIQWNAPQPSHISKYILR |
| Glu | 573RQYIQDRIPSWTGPGFVR | Pro | 1178TNLHLEANPDTGVLTVSWER |
| γ1 | |||
| Val | 1037KDKAAEHRVKLQELESLIANLGTGDDMVTDQAFEDR | Glu | 2197GLNQPTDDSCFDPYTVSHYAVGDEWER |
| Glu | 1589DIKKTLPTGCFNTPSIEKP | Glu | 2227SGFKLLCQCLGFGSGHFR |
| Glu | 2344GTTGQSYNQYSQR |
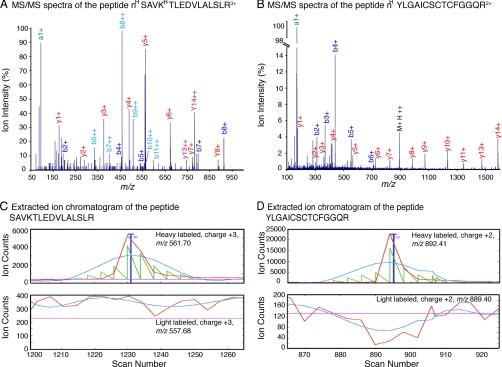
Next, the digestion with Glu-C was conducted on native LM-111 and fibronectin-1 in triplicate, and nine Glu-C-generated peptides were identified (Table I and supplemental Tables 11 and 12), four in LM-111 and five in fibronectin-1. Seven of the nine cleavage sites are exclusive to the proteins digested in the native state and are not found in the proteins if denatured prior to proteolysis (Table I and supplemental material). A strong a1+ ion, a high intensity y ion series, and a very low background are the signature of N-terminal dimethylated peptides (47). This is demonstrated by the MS/MS spectra of two Glu-C-generated peptides, nHSAVKHTLEDVLALSLR and nHYLGAICSCTCFGGQR (Fig. 2, A and B), and these characteristics offer an extra layer of confidence at the peptide identification level. The two peptides are heavy labeled singletons as demonstrated by their respective extracted ion chromatograms in Fig. 2, C and D, where their theoretical light labeled counterparts were not found in the same elution time frame.
Fig. 2.
Tandem mass spectra and quantification of two peptides generated by Glu-C. Examples of two tandem mass spectra from heavy labeled singletons generated by the proteolysis of LM-111 and fibronectin-1 by Glu-C are shown: SAVKTLEDVLALSLR from LM-111 (A) and YLGAICSCTCFGGQR from fibronectin (B), where superscript H indicate the residues labeled with heavy formaldehyde. The b and y ion series are indicated in red and blue, respectively. The strong a1+ ion peak indicative of N-terminal dimethylation is identified in green. Elution profiles of the heavy labeled peptides (top panel) are shown by their extracted ion chromatograms (C and D). The light labeled version of this peptide was not found (bottom panel). The raw chromatograms are the red traces, the smoothed chromatograms are in blue, the areas used to calculate the isotopic ratios are in green, and the background cutoff is in cyan.
Identification of Neutrophil Elastase Cleavage Sites in LM-111 and Fibronectin-1
LM-111 and fibronectin-1 were digested with neutrophil elastase in duplicate and analyzed by ATOMS. Eleven proteolytic sites distributed along the three chains were identified in LM-111 (supplemental Table 13 and supplemental material). Fibronectin-1 was very susceptible to elastase proteolysis with 44 cleavage sites identified (supplemental Table 14 and supplemental material). Analysis of the cleaved peptides indicates that elastase cleavage specificity is dictated by the amino acid at the P1 position with the amino acids Val, Ile, Thr, and Ala preferred as depicted in the WebLogo (supplemental Fig. 1). This result is in very good agreement with the published elastase cleavage specificity (48) and the specificity obtained from the total of 107 neutrophil elastase cleavage sites reported in MEROPS, the protease database (49). That ATOMS identified 55 new neutrophil elastase cleavage sites reveals the power of the approach in rapidly generating valuable new cleavage site data compared with previous methods.
Identification of MMP2 and MMP9 Cleavage Sites in LM-111 and Fibronectin-1
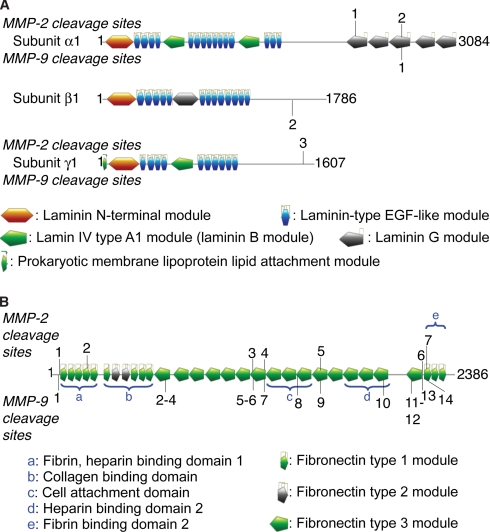
Native LM-111 and fibronectin-1 were subjected to limited proteolysis by MMP2 and MMP9, two closely related gelatinases exhibiting broad substrate specificity (48, 50), that cleave a variety of extracellular matrix proteins and signaling proteins (8, 51, 52). MMP9 is also a prominent protease in inflammatory cells, and so its cleavage products are biologically important in inflammation and healing processes. In total, five cleavage sites were identified in LM-111, including one site common to MMP2 and MMP9 (Fig. 3 and supplemental Tables 15 and 16 and supplemental material). Three sites are found in the C-terminal laminin G module of the α1 subunit.
Fig. 3.
Identification of MMP2 and MMP9 cleavage sites in LM-111 and fibronectin-1. LM-111 (A) and fibronectin-1 (B) were submitted to limited proteolysis by MMP2 (indicated on the top of the schematics (modified from the PROSITE domain view, ExPASy proteomics server)) or MMP9 (indicated at the bottom of the schematics), and the cleavage sites were identified by ATOMS. The neo-N-terminal peptides and their positions in the protein are presented in supplemental Tables 15–18. The domains and modules of LM-111 and fibronectin-1 precursors are indicated with the amino acid numbers at the ends of each chain.
Fibronectin-1 was cleaved at seven sites by MMP2 and at 14 sites by MMP9 (Fig. 3 and supplemental Tables 17 and 18 and supplemental material). The two MMPs have four common cleavage sites in fibronectin-1, and several other sites are in close proximity, many of which are located between the N-terminal collagen binding domain and the cell binding domain. MMP9 cleaves at several sites in the C-terminal portion of fibronectin-1 between positions 2148 and 2224, which is downstream of the cysteine residues (positions 2367 and 2371) important in fibronectin dimerization, thus leading to dissociation of the fibronectin-1 dimer to a monomer. Six MMP9 autoproteolytic fragments were also identified in the samples, and all have ratios higher than 4 (supplemental Table 18).
Comparison of Cleavage Sites Identified by ATOMS and Edman Degradation in Fibronectin-1 Digested with MMP2 and MMP8
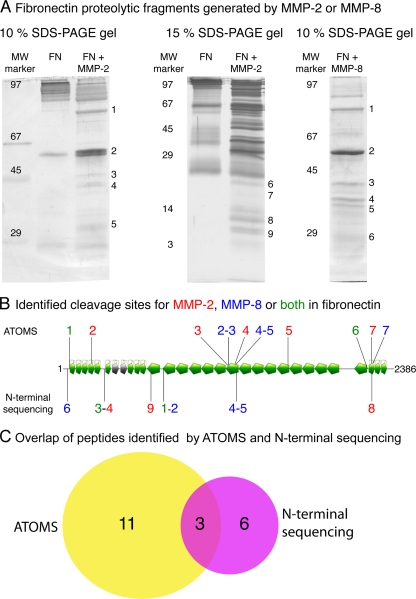
We compared the results obtained with ATOMS and automated N-terminal sequencing of cleavage fragments by Edman chemistry (9). After incubation of fibronectin-1 (25 μg) with MMP2 or MMP8 or with buffer alone, samples were divided as follows. 1) 10 μg were processed by ATOMS. 2) 15 μg were prepared for Edman sequencing. Eight MMP2-generated neo-N-terminal peptides were identified by ATOMS (Table II and supplemental Table 17 and supplemental material) with one peptide unambiguously assigned to isoform 8 of fibronectin. MMP2-digested fibronectin-1 was separated by SDS-PAGE, and proteolytic fragments from 3 to over 100 kDa were observed (Fig. 4A). The N-terminal sequencing of nine MMP2 proteolytic products (Table II) resulted in the identification of four cleavage sites (Fig. 4B) from fragments 1, 3, 4, 8, and 9. Band 8 had the N-terminal sequence YAVGD, and the neo-N-terminal peptide YAVGDEWER was also identified with high confidence by ATOMS. Bands 5 and 7 were identified as autoproteolytic fragments of MMP2. The proteolytic fragments of bands 3 and 4 have different molecular weights (Fig. 4A) but the same N-terminal sequence (AVYQP), indicating different C-terminal cleavages. This N-terminal sequence was not found by ATOMS. Band 2 could not be interpreted possibly because it was a poorly resolved doublet.
Table II. Cleavage sites generated by MMP2 and MMP8 in fibronectin-1 identified by ATOMS or Edman chemistry.
Fibronectin-1 was digested with MMP2 (top portion) and MMP8 (bottom portion), and the cleavage sites were identified with ATOMS (left) and N-terminal sequencing using Edman degradation (right). Positions of the neo-N-terminal peptides in the precursor proteins are identified by the numbers in superscript at the beginning of the peptides.
| ATOMS |
Edman degradation |
||
|---|---|---|---|
| Sequenced peptide | Cleavage site no. | Sequenced peptide | Fragment and cleavage sites no. |
| Cleavage by MMP2 | |||
| 48SKPGCYDNGKHYQINQQWER | 1 | 721LVATS | 1 |
| 209MMVDCTCLGEGSGR | 2 | NDa | 2 |
| 1180LHLEANPDTGVLTVSWER | 3 | 291AVYQP | 3 |
| 1248YTVKDDKESVPISDTIIPAVPPPTDLR | 4 | 291AVYQP | 4 |
| 1548MQVTDVQDNSISVKWLPSSSPVTGYR | 5 | LYGAS (MMP2) | 5 |
| 2198LNQPTDDSCFDPYTVSHYAVGDEWER | 6 | TDQXD | 6 |
| 2215YAVGDEWER | 7 | AFAPX (MMP2) | 7 |
| 1715QTAVTTIPAPTDLKFTQVTPTSLSAQWTPPNVQLTGYRb | 2214YAVGD | 8 | |
| 631XNAPQ | 9 | ||
| Cleavage by MMP8 | |||
| 48SKPGCYDNGKHYQINQQWER | 1 | 721LVATS | 1 |
| 1182LEANPDTGVLTVSWER | 2 | 721LVATS | 2 |
| 1187DTGVLTVSWER | 3 | 291AVYQP | 3 |
| 1250VKDDKESVPISDTIIPAVPPPTDLR | 4 | 1250VKDDK | 4 |
| 1259ISDTIIPAVPPPTDLR | 5 | 1259ISDTI | 5 |
| 2198LNQPTDDSCFDPYTVSHYAVGDEWER | 6 | 44VSQSK | 6 |
| 2231LLCQCLGFGSGHFR | 7 | ||
a Not determined.
b Isoform 8.
Fig. 4.
Comparative analysis of cleavage sites identified by ATOMS and N-terminal sequencing. Fibronectin-1 was cleaved by MMP2 or MMP8 after which the reaction mixtures were separated in two aliquots and subjected to ATOMS or SDS-PAGE and N-terminal sequencing by Edman degradation. A, SDS-PAGE gels of fibronectin-1 (FN) digested with MMP2 and MMP8. The band numbers are associated with the peptides sequenced and presented in Table II. Molecular weight markers are indicated on the left. B, schematic of the cleavage sites identified by ATOMS and N-terminal sequencing. Cleavage sites identified by ATOMS and N-terminal sequencing are labeled on top and bottom of the schematic, respectively. MMP2 cleavage sites are blue, MMP8 sites are red, and cleavage sites common to MMP2 and MMP8 are green. A Venn diagram of the number of cleavage sites identified by ATOMS (yellow) and N-terminal sequencing (magenta) is shown.
For MMP8, ATOMS identified seven peptides and thus seven cleavage sites of fibronectin-1 (supplemental Table 19 and supplemental material), two of which were also identified by N-terminal sequencing of bands 4 and 5 from the SDS-PAGE gel (Table II and Fig. 4A). In comparison, five individual MMP8 cleavage sites were identified from Edman sequencing of six proteolytic fragments (Fig. 4). Bands 1 and 2 have different molecular weights but the same N-terminal sequence (LVATS), indicating different C-terminal cleavages (Fig. 4A). In total, ATOMS identified 15 proteolytic sites and N-terminal sequencing identified nine cleavage sites for fibronectin-1 treated with MMP2 and MMP8, demonstrating the usefulness of ATOMS. When both methods were combined, 17 unique cleavage sites were identified (Fig. 4C), showing the complementarity of the two methods.
DISCUSSION
We describe the development, validation, and use of a new proteomics approach we call ATOMS that circumvents the limitations of SDS-PAGE and Edman sequencing analysis of multiple proteolytic fragments present in solution and that increases coverage of cleavage sites in large complex proteins. The extracellular proteins fibronectins and laminins are long known to be susceptible to proteolysis (27, 35–39, 53), generating multiple proteolytic fragments and affecting their functions. Surprisingly, their cleavage sites have proven difficult to sequence, and indeed, only a few studies have made this attempt with only two to five cleavage sites being identified per study (20, 37, 54–56). In comparison, ATOMS identified 34 cleavage sites generated by MMPs and 55 from elastase in LM-111 and fibronectin-1. To put this result in context, MEROPS, the protease database, lists a total of 10 cleavage sites generated by MMPs 2, 8, and 9 and elastase in fibronectins and laminins (49). ATOMS also easily identified the numerous neutrophil elastase cleavage products typical of protein degradation, which is not possible by microsequencing due to the general inability of SDS-PAGE to resolve protein fragments <4 kDa. We also identified a naturally blocked (cyclized N-terminal glutamine residue) N-terminal peptide (data not shown) that cannot be directly sequenced by Edman chemistry. Whereas current N-terminomics (8, 52) and C-terminomics (57) approaches are best suited to identify one or a few cleavage sites in multiple proteins in complex samples, ATOMS is designed to identify multiple cleavage sites in a few proteins that are complex.
Being a peptide-centric method, ATOMS can easily identify N-terminal peptides from fragments of similar mass. For instance, fragments with cleavage sites at positions 1167, 1171, and 1172 were identified in fibronectin-1 digested with neutrophil elastase (supplemental Table 14). These fragments differ by only a few amino acids and would not be resolved by SDS-PAGE and so would be uninterpretable by Edman sequencing. Other examples of cleavage sites spaced by just a few amino acids are found in supplemental Table 13. Limited proteolysis of fibronectin-1 by MMP2 or MMP8 generated multiple proteolytic fragments, not all of which were submitted to Edman sequencing for comparison due to the difficulty in cutting out so many tightly spaced bands, another limitation of SDS-PAGE-based sequencing. MMP2, MMP8, and MMP9 have a broad cleavage specificity, but they all show a preference for Pro, Gly, and Leu at positions P3, P1, and P1′ in the cleavage site, respectively (49). Accordingly, these three enzymes generated some common proteolytic fragments.
Quantification is mandatory to distinguish peptides of elevated abundance in the protease-treated sample from peptides found in similar amounts before protease assay or generated by trypsin in the preparation of the sample for MS analysis. Quantification accuracy is demonstrated by four ATOMS analyses of paired samples not treated with protease that resulted in a mean peptide quantification value of 1.14 ± 0.47. Most bona fide protease-generated peptides have ion intensities higher than 3000 with some as high as 25,000, enabling accurate quantification, and all were manually verified. Compared with large scale proteomics experiments, this postanalysis manual validation can be readily performed for such a relatively small number of peptides and thus greatly reduces the possibility of misquantification and hence false positives.
The cutoff ratio of 2.5 to describe a peptide enriched in the protease-treated sample is supported by the data from the Glu-C digestion where 38 of 40 peptides (95%) demonstrated Glu-C cleavage specificity when this cutoff was applied. The presence of a proline and valine in P1 of two cleavage sites can be explained by the presence of a limited number of cleavable sequences in the folded protein (50, 58), an enzyme cleavage specificity <100%, or the 5% chance of misidentification of the peptide sequence at the database search level. Analysis of the elastase cleavage sites using the 2.5 cutoff for neo-N termini results in a cleavage specificity that is identical to that of other studies (48, 49), reinforcing the use of this cutoff and demonstrating that ATOMS can identify proteolytic sites generated by proteases with broad cleavage specificity.
A limitation of the peptide-based analysis by ATOMS is its inability to determine the molecular weight of the fragment generated by the proteolytic cleavages. ATOMS is also affected by the limitations inherent to mass spectrometry. The majority of the peptides possess two or three positive charges, which means that peptides smaller than about 6 amino acids and longer than 25 amino acids are not readily amenable to LC-MS/MS. Hence, if the cleavage site is too close or too far from an arginine (the site of trypsin cleavage in ATOMS used to prepare the samples for mass spectrometry) then too short or long a semitryptic peptide, respectively, would result and so might not be identified. This is shown by the identification of the cleavage site AVYQP in fibronectin-1 by Edman sequencing but not by ATOMS. The first arginine is 77 amino acids C-terminal of AVYQP in the protein sequence, thus resulting in a semitryptic peptide of ∼8.5 kDa, which is too big for MS/MS identification. This situation also applies to the fragment with the N-terminal sequence LVATS (fragment 1 for both MMP2 and MMP8), which would result in a semitryptic peptide of 30 amino acids. Hence, ATOMS and N-terminal sequencing are complementary methods as neither of the techniques can identify all the cleavage fragments from large, modular proteins, and the use of both methods increases the number of cleavage sites identified. However, if only one technique is to be used, a high throughput technique such as ATOMS offers many advantages over N-terminal sequencing.
The proteolytic cleavage sites identified here were generated in vitro, and this model system does not replicate the in vivo conditions where some of the cleavage sites might be masked and others exposed. However, they are in agreement with proteolytic fragments identified in vivo and reported in the literature. Proteolysis of the C-terminal domain of the LM-111 γ1 chain generates two peptides spanning positions 2121–2139 and 2747–2757 in the precursor protein (59–61). Here we demonstrate that both MMP2 and MMP9 cleave between the two peptides (positions 2223 and 2584) in the C-terminal domain of the LM-111 chain α1. The absence of fibronectin fibrils at the surface of cancer cells was the first molecular difference observed between normal and tumor cells, and fragmentation of pericellular fibronectin is an early sign of cell transformation toward a malignant phenotype (62, 63). The N-terminal 70-kDa fragment of fibronectin is known to prevent the formation of fibronectin fibrils (64–67). Our study shows that MMP2, MMP8, and MMP9 all cleave between the N-terminal module and the cell attachment domain of fibronectin-1 (Figs. 3 and 4), generating N-terminal fragments. Many MMPs are overexpressed in cancer or reactive stroma, and this suggests that the mechanism by which MMPs decrease cell surface fibronectin involves both degradation and prevention of the assembly of new fibronectin fibrils. Also, a 120-kDa fibronectin proteolytic fragment with the cell binding domain increases the expression of MMP1, MMP3, and MMP9 in fibroblast cells (68). The cleavage sites identified by ATOMS show that MMPs can cleave fibronectin-1 and release different proteolytic fragments spanning the cell binding domain. Hence, in cancer and inflammation, processing of LM-111 and fibronectin-1 by MMP2 and MMP9 may act in a feed forward mechanism where the ECM protein digestion by MMPs affects cell attachment and intracellular signaling and increases the expression of these proteases, leading to more disruption of the ECM; both favor angiogenesis and metastasis.
Overall, ATOMS is a simple method harnessing the power of quantitative mass spectrometry to unambiguously identify proteolytic cleavage sites generated in vitro. Because LC-MS/MS can identify hundreds of peptides in a single analysis, it is well suited for the identification of multiple cleavage sites of single proteins. Core facilities have access to open source free software such as X!Tandem (a database search engine comparable with Mascot used here) and the Trans-Proteomics Pipeline (data analysis software used for this study), thus rendering ATOMS readily accessible to many research groups. The development of ATOMS to rapidly identify protease cleavage sites in large proteins or the N terminus of proteins in liquid samples has the potential to significantly contribute to the identification of new bioactive cleavage fragments in different ECM molecules and therefore is a valuable new tool in the mechanistic analysis of the effects of proteolysis on cell function in health and disease.
Acknowledgments
We thank Dr. Wei Chen from the University of British Columbia Centre for Blood Research Mass Spectrometry Suite for excellent mass spectrometry analyses. We acknowledge Dr. Georgina Butler for critical reading of the manuscript.
Footnotes
* This work was supported in part by a grant from the Canadian Institutes of Health Research, a program project grant in breast cancer metastases from the Canadian Breast Cancer Research Alliance with funds from the Canadian Breast Cancer Foundation and the Cancer Research Society, and an infrastructure grant from the Michael Smith Foundation for Health Research.
 This article contains supplemental Fig. 1, Tables 1–19, spectra, and locations of the cleavage sites.
This article contains supplemental Fig. 1, Tables 1–19, spectra, and locations of the cleavage sites.
1 The abbreviations used are:
- MMP
- matrix metalloproteinase
- ATOMS
- amino-terminal oriented mass spectrometry of substrates
- Glu-C
- S. aureus protease V8
- LM-111
- laminin isoform 1
- ASAPRatio
- automated statistical analysis on protein ratio
- ECM
- extracellular matrix.
REFERENCES
- 1. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 2. Tam E. M., Morrison C. J., Wu Y. I., Stack M. S., Overall C. M. (2004) Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. U.S.A. 101, 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Timmer J. C., Enoksson M., Wildfang E., Zhu W., Igarashi Y., Denault J. B., Ma Y., Dummitt B., Chang Y. H., Mast A. E., Eroshkin A., Smith J. W., Tao W. A., Salvesen G. S. (2007) Profiling constitutive proteolytic events in vivo. Biochem. J. 407, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vande Walle L., Van Damme P., Lamkanfi M., Saelens X., Vandekerckhove J., Gevaert K., Vandenabeele P. (2007) Proteome-wide Identification of HtrA2/Omi Substrates. J. Proteome Res. 6, 1006–1015 [DOI] [PubMed] [Google Scholar]
- 5. Dean R. A., Overall C. M. (2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQTM labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 6. Dix M. M., Simon G. M., Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 9. Edman P., Begg G. (1967) A protein sequenator. Eur. J. Biochem. 1, 80–91 [DOI] [PubMed] [Google Scholar]
- 10. Tester A. M., Cox J. H., Connor A. R., Starr A. E., Dean R. A., Puente X. S., López-Otín C., Overall C. M. (2007) LPS responsiveness and neutrophil chemotaxis in vivo require PMN MMP-8 activity. PLoS One 2, e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cox J. H., Dean R. A., Roberts C. R., Overall C. M. (2008) Matrix metalloproteinase processing of CXCL11/I-TAC results in loss of chemoattractant activity and altered glycosaminoglycan binding. J. Biol. Chem. 283, 19389–19399 [DOI] [PubMed] [Google Scholar]
- 12. Overall C. M., Wrana J. L., Sodek J. (1989) Independent regulation of collagenase, 72-kDa progelatinase, and metalloendoproteinase inhibitor expression in human fibroblasts by transforming growth factor-beta. J. Biol. Chem. 264, 1860–1869 [PubMed] [Google Scholar]
- 13. Overall C. M., Wrana J. L., Sodek J. (1991) Transcriptional and post-transcriptional regulation of 72-kDa gelatinase/type IV collagenase by transforming growth factor-beta 1 in human fibroblasts. Comparisons with collagenase and tissue inhibitor of matrix metalloproteinase gene expression. J. Biol. Chem. 266, 14064–14071 [PubMed] [Google Scholar]
- 14. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Overall C. M., Kleifeld O. (2006) Tumour microenvironment—opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 6, 227–239 [DOI] [PubMed] [Google Scholar]
- 16. Nagase H., Woessner J. F., Jr. (1999) Matrix metalloproteinases. J. Biol. Chem. 274, 21491–21494 [DOI] [PubMed] [Google Scholar]
- 17. Morrison C. J., Butler G. S., Rodríguez D., Overall C. M. (2009) Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr. Opin. Cell Biol. 21, 645–653 [DOI] [PubMed] [Google Scholar]
- 18. Overall C. M., Tam E. M., Kappelhoff R., Connor A., Ewart T., Morrison C. J., Puente X., López-Otín C., Seth A. (2004) Protease degradomics: mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol. Chem. 385, 493–504 [DOI] [PubMed] [Google Scholar]
- 19. Morla A., Zhang Z., Ruoslahti E. (1994) Superfibronectin is a functionally distinct form of fibronectin. Nature 367, 193–196 [DOI] [PubMed] [Google Scholar]
- 20. Giannelli G., Falk-Marzillier J., Schiraldi O., Stetler-Stevenson W. G., Quaranta V. (1997) Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science 277, 225–228 [DOI] [PubMed] [Google Scholar]
- 21. Yi M., Ruoslahti E. (2001) A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc. Natl. Acad. Sci. U.S.A. 98, 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Faisal Khan K. M., Laurie G. W., McCaffrey T. A., Falcone D. J. (2002) Exposure of cryptic domains in the alpha 1-chain of laminin-1 by elastase stimulates macrophages urokinase and matrix metalloproteinase-9 expression. J. Biol. Chem. 277, 13778–13786 [DOI] [PubMed] [Google Scholar]
- 23. Pozzi A., LeVine W. F., Gardner H. A. (2002) Low plasma levels of matrix metalloproteinase 9 permit increased tumor angiogenesis. Oncogene 21, 272–281 [DOI] [PubMed] [Google Scholar]
- 24. Scapini P., Nesi L., Morini M., Tanghetti E., Belleri M., Noonan D., Presta M., Albini A., Cassatella M. A. (2002) Generation of biologically active angiostatin kringle 1–3 by activated human neutrophils. J. Immunol. 168, 5798–5804 [DOI] [PubMed] [Google Scholar]
- 25. Rege T. A., Fears C. Y., Gladson C. L. (2005) Endogenous inhibitors of angiogenesis in malignant gliomas: nature's antiangiogenic therapy. Neuro Oncol. 7, 106–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Z. L., Strickland S. (1997) Neuronal death in the hippocampus is promoted by plasmin-catalyzed degradation of laminin. Cell 91, 917–925 [DOI] [PubMed] [Google Scholar]
- 27. Goldfinger L. E., Stack M. S., Jones J. C. (1998) Processing of laminin-5 and its functional consequences: role of plasmin and tissue-type plasminogen activator. J. Cell Biol. 141, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakagami Y., Abe K., Nishiyama N., Matsuki N. (2000) Laminin degradation by plasmin regulates long-term potentiation. J. Neurosci. 20, 2003–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato N., Motoyama T. (2009) Relation between laminin-5 gamma 2 chain and cell surface metalloproteinase MT1-MMP in clear cell carcinoma of the ovary. Int. J. Gynecol. Pathol. 28, 49–54 [DOI] [PubMed] [Google Scholar]
- 30. Garcia-Pardo A., Pearlstein E., Frangione B. (1984) Primary structure of human plasma fibronectin—characterization of the 6,000 dalton C-terminal fragment containing the interchain disulfide bridges. Biochem. Biophys. Res. Commun. 120, 1015–1021 [DOI] [PubMed] [Google Scholar]
- 31. Pearlstein E., Gold L. I. (1978) High-molecular-weight glycoprotein as a mediator of cellular adhesion. Ann. N.Y. Acad. Sci. 312, 278–292 [DOI] [PubMed] [Google Scholar]
- 32. Ruoslahti E., Engvall E., Hayman E. G. (1981) Fibronectin: current concepts of its structure and functions. Coll. Relat. Res. 1, 95–128 [DOI] [PubMed] [Google Scholar]
- 33. Hynes R. O. (1986) Fibronectins. Sci. Am. 254, 42–51 [DOI] [PubMed] [Google Scholar]
- 34. Ruoslahti E. (1988) Fibronectin and its receptors. Annu. Rev. Biochem. 57, 375–413 [DOI] [PubMed] [Google Scholar]
- 35. Furie M. B., Rifkin D. B. (1980) Proteolytically derived fragments of human plasma fibronectin and their localization within the intact molecule. J. Biol. Chem. 255, 3134–3140 [PubMed] [Google Scholar]
- 36. Gold L. I., Schwimmer R., Quigley J. P. (1989) Human plasma fibronectin as a substrate for human urokinase. Biochem. J. 262, 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horowitz J. C., Rogers D. S., Simon R. H., Sisson T. H., Thannickal V. J. (2008) Plasminogen activation induced pericellular fibronectin proteolysis promotes fibroblast apoptosis. Am. J. Respir. Cell Mol. Biol. 38, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Daudi I., Gudewicz P. W., Saba T. M., Cho E., Vincent P. (1991) Proteolysis of gelatin-bound fibronectin by activated leukocytes: a role for leukocyte elastase. J. Leukoc. Biol. 50, 331–340 [DOI] [PubMed] [Google Scholar]
- 39. Kulkarni M. M., Jones E. A., McMaster W. R., McGwire B. S. (2008) Fibronectin binding and proteolytic degradation by Leishmania and effects on macrophage activation. Infect. Immun. 76, 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Butler G. S., Tam E. M., Overall C. M. (2004) The canonical methionine 392 of matrix metalloproteinase 2 (gelatinase A) is not required for catalytic efficiency or structural integrity: probing the role of the methionine-turn in the metzincin metalloprotease superfamily. J. Biol. Chem. 279, 15615–15620 [DOI] [PubMed] [Google Scholar]
- 41. McQuibban G. A., Gong J. H., Wong J. P., Wallace J. L., Clark-Lewis I., Overall C. M. (2002) Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood 100, 1160–1167 [PubMed] [Google Scholar]
- 42. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li X. J., Zhang H., Ranish J. A., Aebersold R. (2003) Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal. Chem. 75, 6648–6657 [DOI] [PubMed] [Google Scholar]
- 44. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 45. Pflieger D., Jünger M. A., Müller M., Rinner O., Lee H., Gehrig P. M., Gstaiger M., Aebersold R. (2008) Quantitative proteomic analysis of protein complexes: concurrent identification of interactors and their state of phosphorylation. Mol. Cell. Proteomics 7, 326–346 [DOI] [PubMed] [Google Scholar]
- 46. Houmard J., Drapeau G. R. (1972) Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc. Natl. Acad. Sci. U.S.A. 69, 3506–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hsu J. L., Huang S. Y., Chow N. H., Chen S. H. (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 48. Schilling O., Overall C. M. (2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26, 685–694 [DOI] [PubMed] [Google Scholar]
- 49. Barrett A. J. (2004) Bioinformatics of proteases in the MEROPS database. Curr. Opin. Drug Discov. Devel. 7, 334–341 [PubMed] [Google Scholar]
- 50. Turk B. E., Huang L. L., Piro E. T., Cantley L. C. (2001) Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 19, 661–667 [DOI] [PubMed] [Google Scholar]
- 51. Overall C. M. (2002) Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 22, 51–86 [DOI] [PubMed] [Google Scholar]
- 52. Prudova A., auf dem Keller U., Butler G. S., Overall C. M. (2010) Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteomics 9, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Timpl R., Rohde H., Robey P. G., Rennard S. I., Foidart J. M., Martin G. R. (1979) Laminin—a glycoprotein from basement membranes. J. Biol. Chem. 254, 9933–9937 [PubMed] [Google Scholar]
- 54. Unger J., Tschesche H. (1999) The proteolytic activity and cleavage specificity of fibronectin-gelatinase and fibronectin-lamininase. J. Protein Chem. 18, 403–411 [DOI] [PubMed] [Google Scholar]
- 55. Rostagno A. A., Frangione B., Gold L. I. (1989) Biochemical characterization of the fibronectin binding sites for IgG. J. Immunol. 143, 3277–3282 [PubMed] [Google Scholar]
- 56. Lambert Vidmar S., Lottspeich F., Emod I., Planchenault T., Keil-Dlouha V. (1991) Latent fibronectin-degrading serine proteinase activity in N-terminal heparin-binding domain of human plasma fibronectin. Eur. J. Biochem. 201, 71–77 [DOI] [PubMed] [Google Scholar]
- 57. Schilling O., Barré O., Huesgen P. F., Overall C. M. (2010) Proteome-wide analysis of protein carboxy termini: C terminomics. Nat. Methods 7, 508–511 [DOI] [PubMed] [Google Scholar]
- 58. Hubbard S. J. (1998) The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta 1382, 191–206 [DOI] [PubMed] [Google Scholar]
- 59. Weeks B. S., Nomizu M., Ramchandran R. S., Yamada Y., Kleinman H. K. (1998) Laminin-1 and the RKRLQVQLSIRT laminin-1 alpha1 globular domain peptide stimulate matrix metalloproteinase secretion by PC12 cells. Exp. Cell Res. 243, 375–382 [DOI] [PubMed] [Google Scholar]
- 60. Khan K. M., Falcone D. J. (2000) Selective activation of MAPK(erk1/2) by laminin-1 peptide alpha1: Ser(2091)-Arg(2108) regulates macrophage degradative phenotype. J. Biol. Chem. 275, 4492–4498 [DOI] [PubMed] [Google Scholar]
- 61. Nakamura M., Yamaguchi K., Mie M., Nakamura M., Akita K., Kobatake E. (2009) Promotion of angiogenesis by an artificial extracellular matrix protein containing the laminin-1-derived IKVAV sequence. Bioconjug. Chem. 20, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 62. Hynes R. O. (1973) Alteration of cell-surface proteins by viral transformation and by proteolysis. Proc. Natl. Acad. Sci. U.S.A. 70, 3170–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Labat-Robert J. (2002) Fibronectin in malignancy. Semin. Cancer Biol. 12, 187–195 [DOI] [PubMed] [Google Scholar]
- 64. McDonald J. A., Quade B. J., Broekelmann T. J., LaChance R., Forsman K., Hasegawa E., Akiyama S. (1987) Fibronectin's cell-adhesive domain and an amino-terminal matrix assembly domain participate in its assembly into fibroblast pericellular matrix. J. Biol. Chem. 262, 2957–2967 [PubMed] [Google Scholar]
- 65. Schwarzbauer J. E. (1991) Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J. Cell Biol. 113, 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sechler J. L., Takada Y., Schwarzbauer J. E. (1996) Altered rate of fibronectin matrix assembly by deletion of the first type III repeats. J. Cell Biol. 134, 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sottile J., Mosher D. F. (1997) N-terminal type I modules required for fibronectin binding to fibroblasts and to fibronectin's III1 module. Biochem. J. 323, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Werb Z., Tremble P. M., Behrendtsen O., Crowley E., Damsky C. H. (1989) Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J. Cell Biol. 109, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]