Abstract
Ubiquitinated substrates can be recruited to macromolecular complexes through interactions between their covalently bound ubiquitin (Ub) signals and Ub receptor proteins. To develop a functional understanding of the Ub system in vivo, methods are needed to determine the composition of Ub signals on individual substrates and in protein mixtures. Mass spectrometry has emerged as an important tool for characterizing the various forms of Ub. In the Ubiquitin-AQUA approach, synthetic isotopically labeled internal standard peptides are used to quantify unbranched peptides and the branched -GG signature peptides generated by trypsin digestion of Ub signals. Here we have built upon existing methods and established a comprehensive platform for the characterization of Ub signals. Digested peptides and isotopically labeled standards are analyzed either by selected reaction monitoring on a QTRAP mass spectrometer or by narrow window extracted ion chromatograms on a high resolution LTQ-Orbitrap. Additional peptides are now monitored to account for the N terminus of ubiquitin, linear polyUb chains, the peptides surrounding K33 and K48, and incomplete digestion products. Using this expanded battery of peptides, the total amount of Ub in a sample can be determined from multiple loci within the protein, minimizing possible confounding effects of complex Ub signals, digestion abnormalities, or use of mutant Ub in experiments. These methods have been useful for the characterization of in vitro, multistage ubiquitination and have now been extended to reactions catalyzed by multiple E2 enzymes. One question arising from in vitro studies is whether individual protein substrates in cells may be modified by multiple forms of polyUb. Here we have taken advantage of recently developed polyubiquitin linkage-specific antibodies recognizing K48- and K63-linked polyUb chains, coupled with these mass spectrometry methods, to further evaluate the abundance of mixed linkage Ub substrates in cultured mammalian cells. By combining these two powerful tools, we show that polyubiquitinated substrates purified from cells can be modified by mixtures of K48, K63, and K11 linkages.
The ubiquitin (Ub)1 system regulates cellular processes, such as protein degradation, endocytosis, DNA repair, and signal transduction. The central player in this system is Ub, an abundant 76-residue protein that acts as a post-translational modification (1). Conjugation of Ub to protein substrates and the assembly of polyubiquitin (polyUb) chains are catalyzed by a hierarchical system involving E1 activating, E2 conjugating, and E3 ligase enzymes. Deubiquitinating (DUB) enzymes oppose the effects of ubiquitination by hydrolyzing the bond between the C terminus of a Ub molecule and the substrate or polyUb chain to which it is conjugated (2). Protein substrates can be modified by a single Ub (monoubiquitination), by multiple Ub molecules on separate residues (multiubiquitination), and by polyUb chains (polyubiquitination).
A diverse array of structurally distinct Ub signals offers the potential for finely tuned regulation of protein stability, localization, and activity (3). Monoubiquitination has been shown to regulate endocytosis and DNA repair as well as transcription. Although polyUb chains can form via the N terminus and each of the seven lysine residues within the Ub sequence, the most widely studied are chains linked through lysine 48 (K48) and lysine 63 (K63). K48-linked polyUb plays an important role in proteasomal degradation, whereas K63 chains mediate endocytic trafficking, signal transduction, and DNA repair. Recent reports have established that lysine 11 (K11)-linked chains control the degradation of proteins in the endoplasmic reticulum-associated degradation pathway (4) and the cell cycle (5–8), whereas linear head-to-tail polyUb signals downstream of the TNF receptor (9). To a lesser extent, K63-linked chains and multiubiquitination may also target protein substrates for degradation (10–13). Myriad Ub-binding proteins function within cells by recognizing and translating these various Ub signals into biological effects (14).
Complex genetic and post-translational controls exist to ensure that proper levels of Ub are available to meet cellular requirements. Encoded by four separate genes, monomeric Ub (monoUb) protein is generated from ribosomal fusion and stress-inducible Ub-Ub fusion proteins by cotranslational processing. Co-expression of Ub with ribosomal subunits links Ub levels directly to the protein synthesis activity of a cell, whereas inducible polyUb genes increase available Ub levels in response to oxidative stress, heavy metals, and heat shock (15, 16). At the protein level, DUB enzymes recycle substrate-bound Ub to minimize its destruction via the proteasomal and lysosomal degradation pathways (17–19). This sophisticated recycling system, coupled with exquisite transcriptional and translational controls, highlights the central role of this protein within eukaryotic cells. Dysregulation of the cellular Ub pool is a common feature of xenobiotic toxicity and neurodegenerative disease (20), whereas ligase and DUB enzymes are frequently disrupted during tumorigenesis (21) and bacterial/viral infection (22).
Given the complexity of Ub signals on individual protein substrates and the biological complexity of the cellular Ub pool, robust methods for decoding Ub signals are needed to address fundamental biological questions. Early efforts to determine the functional roles and relative abundances of mono- and polyUb relied upon antibodies, mutagenesis, and/or introduction of exogenous DNA constructs (23, 24). Antibody-based approaches to profiling Ub in cells and tissues have been complicated by differences in the affinity of antibodies toward different forms of Ub. In yeast and more recently in mammalian cells, sophisticated genetics approaches have been developed to eliminate endogenous Ub expression and replace it with mutant Ub (23, 25). These approaches make it possible to directly study the effects of individual mutant forms of Ub without the confounding effects of overexpression. Recently, mass spectrometry-based methods have facilitated direct analyses of ubiquitinated proteins purified from cells, tissues, and biochemical reactions. In purified Ub conjugates from yeast, Peng et al. (26) showed the K48-, K63-, and K11-linked chains were the most abundant cellular linkages and that all seven lysines in Ub were competent for forming polyUb. K48-, K63-, and K11-linked chains have consistently been the predominant forms of polyUb detected in biological samples as was shown for Ub conjugates enriched from human cells, clinical specimens, and mouse models of Huntington disease (27).
The Ub-AQUA method (12) was established as a means of quantifying the forms of Ub bound to individual protein substrates generated in vitro (28, 29) or enriched from cells (30, 31) and has been applied to yeast cell lysates (32). The method involves using isotopically labeled internal standard peptides directed toward Ub and the individual forms of polyUb. Peptides in the sample are generated by digestion of Ub-modified proteins and polyUb chains with trypsin. Both unlabeled sample peptides and isotopically labeled internal standards can be assayed by selected reaction monitoring (SRM) on a triple quadrupole mass spectrometer or by narrow window extracted ion chromatograms on a high resolution tandem mass spectrometer, such as the LTQ-Orbitrap. Here we describe advances in the methods used for characterizing polyUb linkage profiles within simple and complex biological matrices using both approaches. We have expanded the use of isotope-labeled internal standard peptides to each of the lysine-based loci within Ub and several mutant forms of Ub. These methods, coupled with recently developed linkage-specific antibodies, have permitted us to evaluate the unexpected effects of mutant Ub in vitro and the frequency of mixed linkage substrates within the cellular Ub pool.
MATERIALS AND METHODS
Antibodies, Purified Enzymes, Ub, and Ub Chains
Purified E1, UbcH5A, UbWT, UbK11R, UbK48R, UbK6R, UbK0, K48-linked polyUb (two to seven Ubs), and K63-linked polyUb (two to seven Ubs) were purchased from Boston Biochem (Cambridge, MA). Synthesis and purification of K11-linked polyUb (33), linear polyUb (two to four Ubs) (48), and full-length c-IAP1 protein (35) have each been recently described elsewhere. Ub-conjugating enzyme UbE2S was subcloned into pST239 vector with modified N-terminal Unizyme tag and purified under native conditions over a nickel-nitrilotriacetic acid column followed by purification over an S-75 gel filtration column. The enzyme was prepared in 25 mm Tris, 0.15 m NaCl, 0.25 mm tris(2-carboxyethyl)phosphine, pH 8. Antibodies against total Ub (P4D1) were obtained from Santa Cruz Biotechnologies (Santa Cruz, CA). Ub linkage-specific antibodies against K48 linkages (α48), K63 linkages (α63), and K11 linkages (α11) were generated in house by phage display as described previously (29, 33).
Preparation of Ub-AQUA Peptide Mixtures
Concentrated stocks of isotopically labeled internal standard (heavy) peptides (K11, K27, K33, K63, K48, LIF, QLE, LI-QL, TLS, EST, LIF-R, TLS-R, IQ-EG, EGI, and TL-ES) were purchased from Cell Signaling Technologies (Danvers, MA), whereas five peptides (GGMQ, MQIF, TITLE, K6, and K29) were synthesized in house by solid phase synthesis (see Table I). Each heavy peptide had a single incorporated residue enriched in 13C/15N, and peptide concentrations were determined by amino acid analysis as described previously (36). All concentrated stocks were stored at −80 °C. From these concentrated stocks, working stock solutions of each individual peptide were prepared at 40 pmol/μl in 30% ACN, 0.1% FA and used to prepare experimental mixtures consisting of all 20 peptides at either 2000 fmol/μl in 30% ACN, 0.1% FA or 1000 fmol/μl in 15% ACN, 0.1% FA. Experimental mixtures were frozen at −80 °C in single use aliquots for direct addition to samples to avoid multiple freeze-thaw cycles.
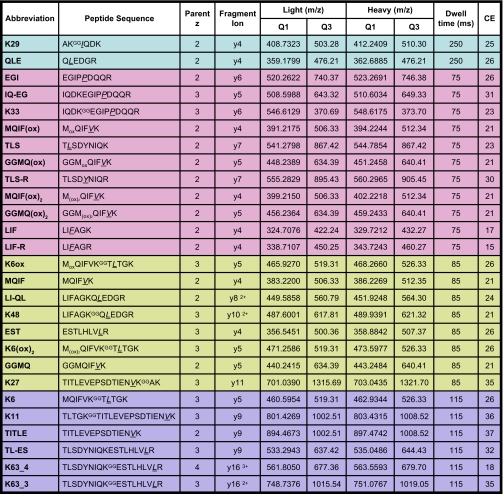
Table I. List of isotopically labeled peptides used to monitor total Ub and various polyUb linkages.
Signature peptides representing isopeptide-linked polyUb linkages are denoted by a superscript “GG” adjacent to the modified lysine. Isotopically labeled amino acids are underlined and italicized. The oxidation state of Met-containing peptides (MQIF, GGMQ, and K6) is denoted by “(ox)” for sulfoxide or “(ox)2” for sulfone. For each peptide, the optimal precursor ion, fragment ion, and collision energy (CE) used in the SRM transition are shown. Dwell times correspond to those used in a segmented SRM analyses where the four colored regions of the table represent the distribution of peptides into four segments based on chromatographic retention time. For Orbitrap data, accurate precursor ion m/z values shown in the Light and Heavy Q1 columns are used to generate narrow window extracted ion chromatograms.
Trypsin Digestion for Quantitative Analysis of Ubiquitination
In vitro ubiquitination reaction products, purified monoUb and polyUb chains, or cell lysates were separated by SDS-PAGE on 4–12% NuPAGE Bis-Tris gels (Invitrogen) and stained with SimplyBlue Coomassie (Invitrogen). Gel bands were excised, diced into 1-mm3 pieces, and destained by addition of a 10× gel volume of 50 mm AMBIC, 50% ACN, pH 8.0 with gentle agitation for 20 min. The solution was removed and replaced with a 10× gel volume of 100% ACN for 15 min. A second 100% ACN wash was performed to ensure complete gel dehydration. Trypsin digestion solution (20 ng/μl unless otherwise indicated) was prepared on ice by dilution of modified sequencing grade trypsin (Promega, Madison, WI) using prechilled 50 mm AMBIC, 5% ACN, pH 8.0. Trypsin solution was subsequently added to gel pieces at approximately equivalent volume and incubated on ice for 30 min. Another 1× gel volume of trypsin solution was added to gel samples and incubated an additional 2 h on ice for a total incubation time of 2.5 h prior to transferring samples to 37 °C for overnight digestion. Digests were quenched by addition of a 0.5× gel volume of extraction buffer (50% ACN, 5% FA) and briefly vortexed. After addition of Ub-AQUA peptides at the indicated concentration, samples were vortexed for 15 s, agitated gently for 2 min, and then centrifuged for 1 min at 13,000 × g. Digested peptides were transferred into fresh Eppendorf tubes, and two additional extraction steps were performed with shaking (a 1.5× gel volume of 50% ACN, 5% FA for 10 min and then a 1× gel volume of 100% ACN). All extracted peptides were combined, frozen, and then dried to completion in a SpeedVac. At least 30 min prior to analysis, samples were resuspended in 10% ACN, 5% FA, 0.01% H2O2 for analysis.
Preparation of Complex Mixture Lysate
Jurkat cells were grown in RPMI 1640 medium with antibiotics, 10% fetal bovine serum, and 2 mm l-glutamine to a density of 2 × 106 cells/ml. Cells were lysed in 8 m urea, 25 mm NaCl, 50 mm Tris, pH 8.0 by sonication; clarified by centrifugation (16,000 × g for 10 min); and quantified by Bradford assay. Lysate (7.3 mg) was separated on a single well 4–12% NuPAGE Bis-Tris gel and stained with SimplyBlue Coomassie. The top gel region corresponding to proteins >70 kDa was cut into 1.0-mm3 pieces and digested as described above with trypsin at a concentration of 5 ng/μl. Dried digests were resuspended in 98% H2O, 2% ACN, 0.1% FA at an approximate concentration of 1 μg/μl; aliquoted; stored at −80 °C; and thawed prior to use.
Quantitation by Selected Reaction Monitoring
SRM quantitation was performed on a 4000 QTRAP system (Applied Biosystems, Foster City, CA) with a Turboionspray source coupled to a Tempo Autosampler (Applied Biosystems) and Agilent 1200 capLC system (Santa Clara, CA). Samples were loaded via full loop injection directly onto a 2.1 × 150-mm Thermo Aquasil C18 column (Thermo Scientific, San Jose, CA) and separated by reverse phase chromatography at a flow rate of 200 μl/min where solvent A was 98% H2O, 2% ACN, 0.1% FA and solvent B was 98% ACN, 2% H2O, 0.1% FA. An 18-min two-stage linear gradient was used where in stage 1 solvent B went from 5 to 13% over 3 min, and in stage 2 solvent B went from 13 to 25% over 15 min. Total run length was 34 min per sample. The QTRAP mass spectrometer was operated in SRM mode with Q1/Q3 resolution settings of unit/unit. The most abundant charge states of heavy and light versions of each peptide were determined empirically and used for SRM transition development. For the K63 -GG signature peptide, transitions were generated for both 3+ and 4+ charge states. SRM transitions were selected to monitor the fragment ion with the highest intensity, and preference was given to fragment ions with m/z values higher than that of the parent (37). Collision energies for each transition were optimized empirically to maximize signal and improve limits of detection (see Table I). The analysis time was broken into four segments where a subset of transitions was monitored within each segment, allowing for dwell times between 75 and 250 ms. The four segment periods were 0–6.5, 6.5–15.8, 15.8–19.6, and 19.6–34 min. Peak integration was performed using MultiQuant 1.1 software (Applied Biosystems), and area under the curve (AUC) was used to determine the abundance of each light peptide in the sample relative to its corresponding heavy internal standard. For comparison, a non-segmented method was set up similarly to the segmented method with the exception that all transitions were monitored throughout the analysis with dwell times fixed at 60 ms.
Quantitation of High Mass Accuracy Precursor Ions
Analysis was performed on an LTQ-Orbitrap XL (ThermoFisher, San Jose, CA) equipped with a Michrom ADVANCE source (Auburn, CA) in combination with a Waters nanoAcquity UPLC system (Milford, MA). Samples were loaded via partial loop injection directly onto a 0.1 × 100-mm Waters 1.7-μm BEH-130 C18 column at a flow rate of 1.5 μl/min for 10 min. Peptides were separated at 1.0 μl/min across a two-stage linear gradient where solvent B ramped from 5 to 25% over 20 min and then from 25 to 50% over 2 min. The composition of solvents A and B were the same as on the QTRAP, and the total run time per injection was 45 min. The Orbitrap mass spectrometer was operated in data-dependent mode whereby the duty cycle comprised one full MS scan collected at 60,000 resolution in the Orbitrap and MS/MS scans in the ion trap for the four most intense ions observed in the full MS scan. Quantitation was performed at the MS level by integrating areas of peaks corresponding to heavy and light m/z values of the most abundant charge states of each peptide (see Table I). Peaks for integration were obtained through application of extracted ion chromatograms over 10-ppm mass intervals using QualBrowser v2.07 (ThermoFisher).
Optimization of Methionine Oxidation
Ub-AQUA peptides were mixed with complex mixture lysate in the presence of 5% FA, 10% ACN without H2O2 or with a concentration of 0.01, 1, 3, 5, or 10% H2O2 and incubated for 30 min at 25 °C. For each sample, a 5-μl injection consisting of 375 fmol of AQUA peptides and 0.125 μg of complex mixture lysate was introduced to the QTRAP for analysis using the segmented method (n = 3). At the 11- and 21.5-h time points following extended incubation at 4 °C, the samples were reinjected, mimicking sample conditions during analysis of a long sample queue. Analysis buffers comprising H2O2 and FA were freshly prepared for each experiment approximately 1 h prior to use.
Determining Total Ub from Loci Using Conservation of Mass
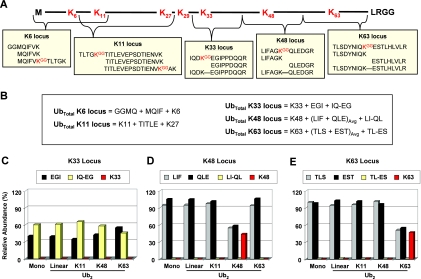
Total Ub is determined for a locus as the sum of the modified and unmodified forms based on peptides generated during trypsin digestion (12). Sequence considerations require that conservation of mass equations be tailored for each locus as the unmodified form may be represented by one or more unbranched peptides, and the modified form may yield one or more branched -GG signature peptides. At the K6 locus, the unbranched portion is represented by the MQIF peptide, whereas the branched forms are represented as the sum of K6 and GGMQ (linear chain) peptides. The C-terminal unbranched peptide at the K6 locus is only five residues (TLTGK) and has not been considered in this method. At the K11 locus, the unbranched portion is represented by the TITLE peptide from the C-terminal flank, whereas the N-terminal flank (again TLTGK) has not been considered. The sum of K11 and K27 makes up the branched portion at the K11 locus as the presence of a -GG signature on either K11 or K27 ablates signal from the TITLE peptide. Peptides derived from forked polyUb chains, stemming from concurrent modification at adjacent lysines (e.g. K6-K11 and K11-K27), were assumed in these calculations to be of negligible abundance and were not actively measured. Given sequence considerations, calculations at the K27 locus would be identical to the K11 locus. The K29 locus was not used for determining total Ub as the two unbranched peptides flanking K29 (AK and IQDK) are only two and four residues, respectively. At the K33 locus, the unbranched portion has been represented by the sum of the fully tryptic EGI peptide and the missed trypsin cleavage product IQ-EG (as described under “Results” and in Fig. 1) with the K33 -GG signature peptide comprising the branched population. At the K33 locus, it was assumed that the forked K29-K33 double -GG peptide was of negligible abundance. The K48 and K63 loci are flanked on both N- and C-terminal sides by Arg residues, and as such, digestion of their branched and unbranched peptides is unaffected by polyUb chain formation at adjacent loci. The unbranched portion of the K48 locus was accounted for as the average of the LIF and QLE peptides, whereas the LI-QL peptide was included to monitor incomplete digestion of the unbranched sequence. The unbranched portion of the K63 locus was accounted for as the average of the TLS and EST peptides, whereas the TL-ES peptide was included to monitor the unbranched sequence in case of incomplete digestion. In each case, total Ub was determined as the sum of the -GG signature peptide (K48 and K63, respectively), the incomplete digestion product (typically negligible as described under “Results” and in Fig. 1), and the average of the unbranched, fully tryptic peptides. At all loci, we have also assumed that non-Ub modifications (e.g. phosphorylation) comprise a negligible proportion of the total Ub.
Fig. 1.
Loci used to determine total Ub. A, schematic diagram of Ub amino acid sequence demonstrating that five different lysine-based loci can be used to determine the total amount of Ub in a sample. Unbranched tryptic peptides, -GG signature peptides, and/or peptides covering observed missed cleavages may be required to effectively cover a locus. The peptide used to quantify linear polyUb (GGMQ) is part of the K6 locus. B, conservation of mass equations used to determine total Ub at the K6, K11, K33, K48, and K63 loci. Equations assume complete digestion efficiency except where missed cleavage peptides are in place to make direct measurements. C, analysis of the K33, K48, and K63 loci from digested monoUb or Ub2 linked through the N terminus (linear), K11, K48, and K63. The abundance of each peptide is displayed relative to the total Ub determined from its corresponding locus. Detection of the IQ-EG missed cleavage peptide indicated that >50% missed cleavage is observed at the K33 locus, denoted by the presence of the IQ-EG missed cleavage peptide (yellow). D, the LI-QL missed cleavage peptide accounted for <1% of the total peptides from the K48 locus, whereas the K48 -GG signature peptide (red) was observed at 44% for K48 Ub2. E, the TL-ES missed cleavage peptide accounted for <1% of the total peptides from the K63 locus despite having a glutamate at the adjacent position. The K63 -GG signature peptide (red) was observed at 47% for K63 Ub2. All samples were injected in triplicate. Avg, average.
Characterization of Linear Range and Reproducibility of QTRAP Analyses
Experimental stocks of Ub-AQUA peptides at 2000 fmol/μl were serially diluted to prepare samples containing Ub-AQUA peptide mixtures at 1000, 200, 100, 20, 10, 2, 1, and 0.2 fmol/μl in a constant background of 0.03 μg/μl complex mixture lysate in 10% ACN, 5% FA, 0.01% H2O2. Individual injections of 5 μl per sample, corresponding to on-column loading of 5000, 1000, 500, 100, 50, 10, 5, and 1 fmol of Ub-AQUA peptides, each in 0.15 μg of complex mixture lysate, were analyzed using the segmented method (n = 4). Samples containing 50 fmol of Ub-AQUA peptides in 0.15 μg of lysate were also analyzed using the non-segmented method (n = 3).
Characterization of Linear Range and Reproducibility of Orbitrap Analyses
Ub-AQUA peptide mixtures at concentrations of 10, 5, 2, 1, 0.2, and 0.1 fmol/μl in a constant background of 0.03 μg/μl complex mixture in 10% ACN, 5% FA, 0.01% H2O2 were prepared by serial dilution of 2000 fmol/μl experimental stocks. On-column injections of 5 μl (n = 3) corresponding to 50, 25, 10, 5, 1, and 0.5 fmol of Ub-AQUA peptides in 0.15 μg of complex mixture were performed in order of increasing concentration.
Analysis of Ub Linkages in Serial Dilution Series of MG132-treated 293T Cell Lysate
293T cells were grown to 90% confluence in DMEM containing antibiotics, 10% fetal bovine serum, and 2 mm l-glutamine. Cells were treated for 4 h with freshly prepared 50 μm MG132 in DMSO. Cells were lifted with 1× trypsin in PBS; washed once with ice-cold PBS; and lysed in 20 mm Tris, pH 7.5, 135 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1% Triton X-100, 10% glycerol, 6 m urea, 2 mm N-ethylmaleimide, and phosphatase and protease inhibitors (Pierce) by sonication. Lysate was clarified by centrifugation at 13,000 × g for 15 min, and protein concentration determined by BCA assay. Lysate (880 μg) was partially separated on a 1.5-mm single well 4–12% NuPAGE Bis-Tris gel and stained with SimplyBlue Coomassie. The gel region containing protein was excised, diced into 1.5-mm cubes, and subjected to in-gel digestion using trypsin (20 ng/μl) as described above except that AQUA peptides were not added. Dried, digested peptides were resuspended in 10% ACN, 5% FA at ∼20 μg/μl and serially diluted to concentrations ranging from 4 to 0.02 μg/μl. At least 30 min prior to injection, these were used to prepare final lysate samples containing Ub-AQUA peptide mixtures in 10% ACN, 5% FA, 0.01% H2O2. For QTRAP segmented method analysis (n = 3), 5-μl injections containing 0.04, 0.08, 0.4, 0.8, 4, 8, 20, or 40 μg of lysate, each in the presence of 75 fmol of Ub-AQUA peptides, were loaded on the column. Likewise, 5-μl injections corresponding to 0.04, 0.08, 0.4, or 0.8 μg lysate with 75 fmol of Ub-AQUA peptides were analyzed (n = 2) on the Orbitrap.
Multienzyme in Vitro Ubiquitination
Ubiquitination assays were performed in a 50-μl reaction volume with 2 μg of wild-type or indicated mutant Ub proteins; 0.4 μg of E1; 2 μg of E2 (UbcH5A or UbE2S); 2 μg of recombinant c-IAP1 in a buffer containing 30 mm HEPES, 2 mm DTT, 20 m ZnCl2, and 5 mm MgCl2-ATP. Reactions were preincubated at 21 °C for 5 min with 0.1 μg of UbcH5A followed by the addition of indicated E2 enzymes and subsequent incubation for another 35 min at 37 °C (both time periods with shaking at 750 rpm). Reactions were stopped by addition of 4× lithium dodecyl sulfate sample buffer, boiled at 90 °C for 10 min, and resolved by SDS-PAGE. Selected samples were analyzed on the Orbitrap to identify Ub substrates. All ubiquitination reactions were analyzed on the QTRAP using the segmented method described above (n = 2) with 420 fmol of Ub-AQUA peptides loaded on the column. Total Ub was determined based on data from all five loci (K6, K11, K33, K48, and K63).
Transfection and Linkage Antibody Immunoprecipitation
Immunoprecipitation experiments with linkage-specific antibodies were performed as described previously using 293T cells (33). Briefly, cells were either mock-transfected or transfected with mutant UbK0 lacking all seven lysines. After 48 h, transfected cells were lysed in 8 m urea, 50 mm Tris, pH 7.5, 25 mm NaCl, 5 mm EDTA, 2 mm N-ethylmaleimide, 10 μl/ml 100× Halt protease and phosphatase inhibitors (Pierce) and sonicated briefly to reduce viscosity. Lysates were diluted to 4 m urea with IP buffer (20 mm Tris, pH 7.5, 135 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm EDTA, 1.5 mm MgCl2). Lysate (5 mg) was precleared with Protein A Dynabeads and incubated overnight at 25 °C with 40 μg of IgG recognizing either K48-linked or K63-linked polyUb (29). Bound proteins were washed five times with 4 m urea IP buffer and five times in PBS followed by elution in SDS sample buffer. Proteins were resolved by SDS-PAGE, stained with SimplyBlue, and digested with trypsin as described above for quantitative characterization of ubiquitination. Analysis was performed on the QTRAP using the segmented method with 333 fmol of Ub-AQUA peptides loaded on the column.
RESULTS
Peptides and Loci Used in Profiling Ub Signals
The Ub-AQUA method originally described quantification of seven -GG signature peptides from Ub as well as the unbranched peptides flanking the K63 and K11 loci (12). The total amount of Ub (total Ub) in a sample was determined as the sum of branched -GG signature (K63) and unbranched (TLS and EST) peptides surrounding the K63 locus and confirmed by measurements made at the K11 locus (K11 and TITLE). Following on these efforts, we evaluated the benefits of determining total Ub from three additional lysine-based loci of Ub (Table I and Fig. 1, A and B). The battery of isotopically labeled peptides was expanded to include internal standards toward the K33, K48, and K6 loci. At the K33 locus, the unbranched EGI peptide can be used along with the K33 -GG signature peptide to calculate total Ub, whereas at the K48 locus, LIF and QLE peptides can be used in combination with the K48 -GG signature peptide to determine total Ub. Internal standards toward the K6 locus have also been generated, including unbranched MQIF peptide directed toward the unmodified terminus and GGMQ peptide to assay linear polyUb linkages (38). In a linear polyUb chain, the -GG signature of one Ub molecule is attached to the N terminus of a second through a peptide bond. The abundance values of MQIF and GGMQ can be added to the K6 -GG signature peptide to determine total Ub from the K6 locus. Depending on experimental conditions, it can be beneficial to use a subset of lysine-based loci to uniformly quantify total Ub between samples of differing Ub compositions.
Within a Ub-AQUA experiment, the reliability of total Ub measurements depends on accurately accounting for all of the major peptides at a given locus produced during trypsin digestion. A key component of this conservation of mass approach is the assumption that complete digestion efficiency is achieved at each quantified locus such that partial digestion products impact the measurement in a negligible way. During initial characterization of the EGI peptide at the K33 locus, directed quantitation and database search results indicated that missed cleavage was common, resulting in a peptide with the sequence IQDK33EGIPPDQQR (referred to as IQ-EG). To assess the frequency of missed cleavage at this locus as well as the K48 and K63 loci, additional internal standard peptides mimicking missed cleavage at K33 (IQ-EG), K48 (LI-QL), and K63 (TL-ES) were developed. Digestion optimization experiments were carried out across a range of trypsin concentrations using monoUb and polyUb of various linkages. As previously described for K48 and K63 loci (12), we found that digestion with 20 ng/μl trypsin diluted in 50 mm AMBIC, 5% ACN, pH 8 yielded nearly complete proteolysis. Following digestion of monoUb and Ub2 species with 20 ng/μl trypsin, we examined the abundances of IQ-EG, LI-QL, and TL-ES relative to fully digested peptides from each of their respective loci. In the case of the K33 locus, the abundance of IQ-EG relative to the EGI peptide indicated that 50–60% of Ub molecules observed within each sample were not fully digested regardless of linkage type (Fig. 1C). This may be attributable to the presence of acidic residues flanking K33 on both sides that impair trypsin activity. In contrast, based upon the level of the LI-QL peptide, near complete digestion efficiency was observed at K48 (Fig. 1D). Interestingly, although K63 resides next to a glutamate residue, >99% of Ub molecules were also fully digested at this locus to generate TLS and EST peptides (Fig. 1E).
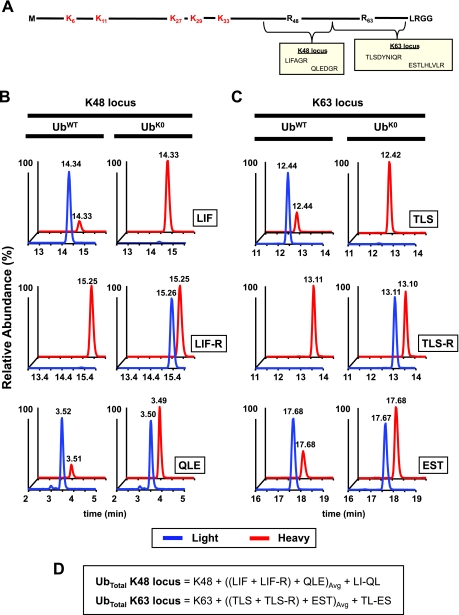
Mutant forms of Ub are frequently used in vitro to prevent polyUb chain formation through defined lysines. Likewise, forms of Ub missing between one and seven lysines can be overexpressed in cells to act as dominant negatives of linkage-dependent processes. Because it can be important within these studies to measure the total amount of Ub in a mixed population containing both UbWT and Ubmutant, two additional isotopically labeled peptides have been developed to permit direct measurement of Ubmutant (e.g. point mutant UbK48R and lysineless UbK0) and determine its abundance relative to UbWT (Fig. 2; also see Fig. 7). The two peptides come from the K48 and K63 loci (LIF-R and TLS-R, respectively) (Fig. 2A) and can be used interchangeably or in combination with their wild-type counterparts (Fig. 2, B and C). Whereas in UbWT complete digestion should yield equivalent amounts of light TLS and EST peptides, complete digestion of UbK63R or UbK0 yields equivalent amounts of light TLS-R and EST. Likewise when UbK48R or UbK0 are digested, the abundances of LIF-R and QLE peptides should be equivalent. When determining total Ub in a mixed population, the sum of TLS and TLS-R can be used to replace TLS alone in the conservation of mass equation for the K63 locus. Likewise, LIF and LIF-R can be used in place of LIF when determining total Ub at the K48 locus (Fig. 2D).
Fig. 2.
Peptides used to directly monitor lysine-to-arginine mutant forms of Ub. A, experiments frequently utilize mutant forms of Ub where K48, K63, or all lysines (K0) are mutated to arginine. The schematic diagram shows the mutant UbK48R,K63R sequence and the derived tryptic peptides from K48R and K63R mutant loci. These mutations prevent formation of polyUb linkages through these lysines. B, extracted ion chromatograms of unbranched peptides from the K48 locus in samples containing either UbWT or UbK0 (for each, data are from Fig. 7, gel region 1). Pairs of co-eluting light (blue) and heavy (red) peaks are shown on the same relative scale (defined by the more abundant of the two peaks) with retention times shown for each. In UbWT-containing sample, equivalent amounts of the light forms (blue) of the LIF and QLE peptides are observed, whereas in the UbK0 sample, the light LIF-R to QLE ratio is near equivalence. Each is compared with a constant amount of heavy (red) internal standard. C, analysis of the K63 locus in the same two samples reveals a similar pattern where equivalent amounts of the light TLS and EST peptides are observed for the UbWT sample, whereas the TLS-R and EST light peptides are equivalent for the UbK0 sample. D, in a mixed population of UbWT and Ubmutant (e.g. UbK0), the total Ub can be computed at each locus using a combined equation. Furthermore, the LIF-R/(LIF + LIF-R + K48) and TLS-R/(TLS + TLS-R + K63) ratios directly define the fraction of Ubmutant in a population. Avg, average.
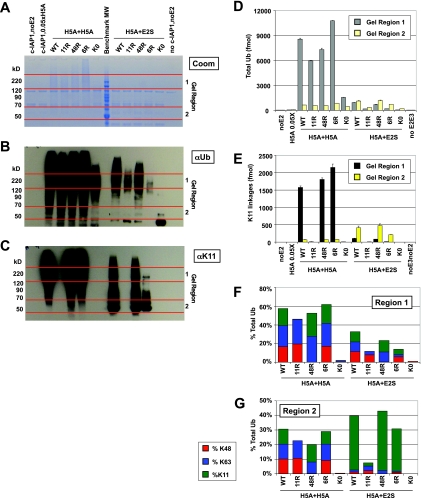
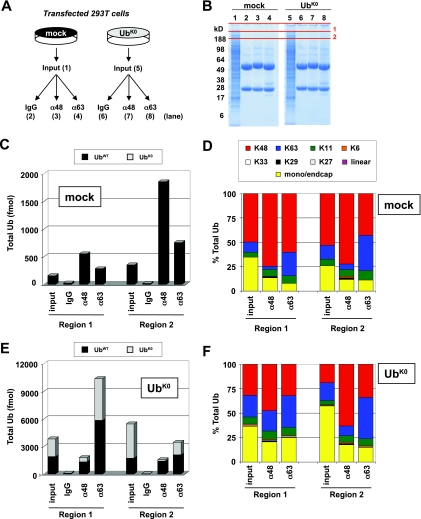
Fig. 7.
Advanced MS method reveals that UbE2S can modulate function of UbcH5A in K11-dependent manner. In vitro ubiquitination reactions containing Uba1 (E1) and c-IAP1 (E3) were performed in two steps with either UbcH5A or UbE2S, both following preincubation with a 0.05× amount of UbcH5A, which alone was insufficient to generate chains. Samples were separated by SDS-PAGE for Coomassie staining (A), and Western blots against total Ub (αUb) (B) and K11 linkages (α11) (C) were run in parallel. Regions 1 and 2 on the Coomassie gel (denoted by red lines) were selected for digestion and MS analysis based upon αUb and α11 Western blot signals. A benchmark molecular weight marker was loaded in the middle lane (every other band is annotated). D, total Ub reported as the average of K48 and K63 loci for representative regions 1 and 2 with error bars representing S.D. of duplicate injections. E, K11 linkages reported for regions 1 and 2. F, abundances of K48, K63, and K11 relative to the total Ub reported for region 1. The mixed linkage profile suggests that UbE2S activates UbcH5A rather than directly ubiquitinating c-IAP1 or extending chains generated by UbcH5A. G, abundances of K48, K63, and K11 relative to the total Ub reported for region 2. For UbE2S-stimulated reactions, Western blots and total Ub measurements indicate that these chains are almost exclusively K11 linkages. UbK11R and UbK0 reactions with UbE2S do not generate these chains, indicating synthesis directly by UbE2S.
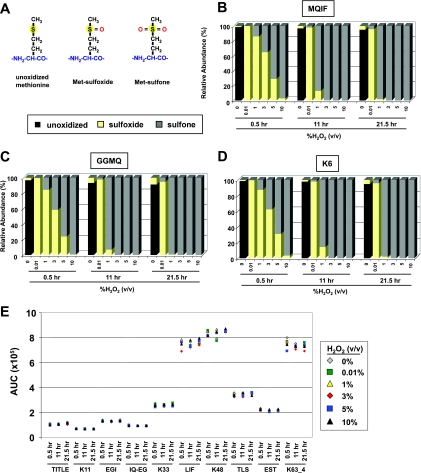
One challenge that comes with quantifying peptides covering the N terminus of Ub (MQIF, K6, and GGMQ) is the presence of a methionine (Met). It is possible for Met residues to become modified within a biological sample or during sample handling by one or two oxygen atoms, yielding Met sulfoxide (ox) or Met sulfone ((ox)2) (Fig. 3A). Met oxidation poses a problem for peptide-based quantification when the analyte is distributed between oxidation states or when the analyte is in a different form than the internal standard. A series of conditions were evaluated to find a condition that would quantitatively convert peptides into either Met(ox) or Met(ox)2 forms. As described previously (39), the combination of FA and H2O2 can be a potent oxidizer of Met residues. In the presence of 5% FA, we confirmed that 30-min incubation of the MQIF, K6, and GGMQ peptides with 0.01% H2O2 converted >95% of the unoxidized peptide population to the Met(ox) form (Fig. 3, B–D). In the case of MQIF, the AUC signal for the unoxidized peptide decreased from 3.1 × 105 to undetectable levels (<3 × 102), whereas the signal for MQIF(ox) increased from 1.5 × 103 to 1.1 × 105. Once Met was converted to Met(ox) under these conditions, the Met(ox) form remained stable out to 21.5 h. This allows for the analysis of large sample sets without concern over shifting oxidation states midseries. For conversion into the Met(ox)2 form, H2O2 concentrations above 5% H2O2 in 5% FA or incubations longer than 30 min were required (Fig. 3, B–D) to quantitatively oxidize Met-containing peptides. Even under the harshest oxidizing conditions tested (10% H2O2, 5% FA for 21.5 h), collateral oxidation was not observed for other peptides from Ub (Fig. 3E). These conditions have been shown to oxidize cysteine and tryptophan residues (40), but neither occurs in the Ub sequence.
Fig. 3.
Quantitative oxidation of methionine-containing peptides. A, peptides containing Met residues can be observed by LC-MS in the unoxidized, Met sulfoxide, or Met sulfone forms when modified by zero, one, or two oxygens, respectively. B, controlled oxidation of the N-terminal MQIF peptide from Ub. Quantitative conversion of unoxidized Met to Met sulfoxide is observed in 0.01% H2O2, 5% FA after 30 min and remains stable out to 21.5 h. The maximal signals for unoxidized and Met sulfone-containing peptides were normalized based upon Met sulfoxide signal (0.01% H2O2; 30 min) based on the observation that Met sulfoxide conversion was essentially complete under these conditions. Further oxidation to Met sulfone is observed beginning at concentrations of ∼1% H2O2, 5% FA for 30 min, whereas complete conversion was only seen at higher concentrations and with longer incubation times. C and D, controlled oxidation of the GGMQ linear polyUb signature peptide and K6 isopeptide-linked -GG signature peptide reveals similar kinetics to MQIF with Met sulfoxide observed with 0.01% H2O2, 5% FA after 30 min and increasing amounts of Met sulfone observed at higher concentrations and longer times. E, signal intensities for a selection of Ub peptides (unbranched and -GG signature) show consistent signal intensity across the range of oxidation conditions used. Values represent the mean area under the curve for n = 3 at each data point.
Instrument Platforms for Profiling Ub Signals
For quantitative characterization of Ub linkage profiles using the battery of internal standard peptides described above, we took advantage of two complementary instrument platforms with chromatographic systems tailored either for low end detection from small sample amounts or robust, higher throughput analyses of abundant samples. To evaluate the linear range of each peptide on both instrument setups, a series of experiments were performed where mixtures of Ub-AQUA peptides were serially diluted in a constant amount of complex mixture lysate (0.15 μg) for replicate injection. Ub-AQUA peptide mixtures were analyzed in this low level complex mixture background to minimize the effects of peptide loss during and after sample preparation.
For routine analyses, particularly from samples where >1 μg of digested peptides are analyzed, peptides are separated at 200 μl/min on a 2.1-mm column using an Agilent 1200 HPLC and then delivered to a 4000 QTRAP mass spectrometer operating in SRM mode. The higher flow rates afforded by the 2.1-mm column permit chromatographic resolution of early eluting peptides, such as K29 and QLE, that can escape detection on nanoflow systems. Likewise, sample carryover on this system is <0.01% for the majority of transitions even when multiple picomoles of analyte are injected on the column. For K48, LI-QL, K6(ox), and K63 peptides, carryover of 0.2–0.3% can be seen following a 5-pmol load. Peak areas for SRM transitions, quantified automatically using MultiQuant 1.1 software, demonstrate that the most sensitive SRM transition (K48) is detectable down below 1 fmol, and all peptides display linearity of R2 > 0.997 across at least 3 orders of linear range (Fig. 4). Additional linear range above the 5-pmol level has been observed but has not been fully elaborated.
Fig. 4.
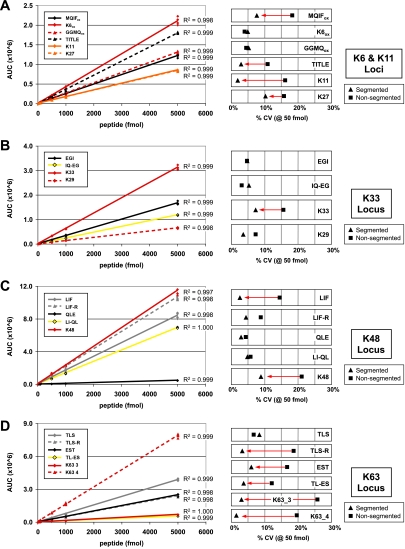
Linear range of Ub peptides on QTRAP mass spectrometer at 200 μl/min chromatographic separation. A mixture of isotopically labeled Ub peptides was prepared in a constant background of 150 ng of complex mixture digest for repeat injection (n = 4). AUC is plotted for each SRM transition monitored across a concentration range from 5 to 5000 fmol using a segmented method (four segments). A, standard curves for peptides from the K6 and K11 loci are shown on the left. On the right is a comparison of the intermeasurement variability (CV in percent) for the segmented method (triangle) versus a non-segmented method (square). An R2 value is shown for each linear fit. Four of six peptides showed a decrease of >5% in CV with the segmented method (red arrow). B and C, standard curves for peptides from the K33 and K48 loci where one of four and two of five peptides showed a decrease of >5% in CV with segmenting, respectively. For the sake of presentation, K29 has been grouped with the K33 locus. D, standard curve for peptides from the K63 locus where five of six peptides displayed a decreased CV of >5% upon segmenting.
Because elution times for each peptide are well characterized and highly reproducible from run to run (±0.1 min; n > 50), we generated a method wherein SRM transitions were divided among four segments (Table I; 0–6.5 min, blue; 6.5–15.8 min, pink; 15.8–19.6 min, yellow; 19.6–34.0 min, purple) such that only a subset of SRM transitions are monitored during any point in the run. Compared with the non-segmented method where CV ranged from 5 to 20% per peptide, the coefficient of variation decreased by more than 5% for 12 of 21 transitions in segmented runs (Fig. 4, A–D). Most greatly affected were hydrophobic peptides at the end of the chromatographic gradient where additional dwell time per transition and data points across the chromatographic peaks accounted for the improvement. Segmented methods were used for all subsequent experiments because of performance advantages.
For low level samples and samples where data-dependent MS/MS are desired in combination with quantitative data, peptides are analyzed on an LTQ-Orbitrap XL. Here, peptides are separated by a nanoAcquity UPLC system across a 0.1-mm column at pressures of ∼5500 p.s.i. and then delivered through an ADVANCE nanospray ionization source for detection. On this system, we detected multiple -GG signature peptides at levels below 1 fmol (Fig. 5) with the greatest response for the K48 -GG signature peptide. For K48, we could detect an AUC signal of 1.7 × 106 for a 0.5-fmol injection, suggesting a limit of detection between 50 and 100 amol. Standard curves on the Orbitrap detector were obtained with R2 > 0.98 for all Ub peptides except LI-QL. One limitation observed with this setup was that injections containing >1 pmol of a peptide analyte resulted in carryover that influenced quantitation of lower abundance samples in subsequent runs. However, for samples where <500 fmol are injected on the column, we find that carryover is consistently below 0.2%.
Fig. 5.
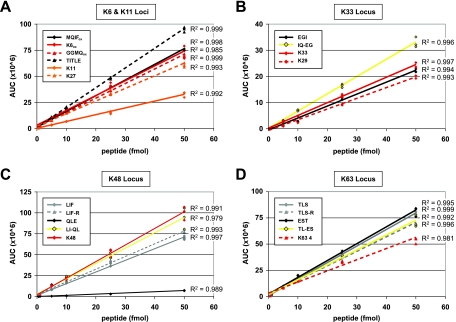
Linear range of Ub peptides on Orbitrap mass spectrometer with UPLC separation at 1 μl/min. A mixture of isotopically labeled Ub peptides was prepared at various concentrations in a constant background of 150 ng of complex mixture digest for repeat injection (n = 3). A–D, AUC is plotted for each peptide from the K6 and K11, K33, K48, and K63 loci across a range from 0.5 to 50 fmol. An R2 value is shown for each linear fit. For presentation, K29 peptide has been included in K33 locus, although it is not included in total Ub calculations at this locus. Linear ranges generated for K29 and QLE peptides include only values obtained for ≥5-fmol injections because at lower concentrations chromatographic peaks for these early eluting peptides were not well resolved. Similar to the QTRAP, the K48 peptide is the most sensitive with an AUC of 1.7 × 106 for a 0.5-fmol injection.
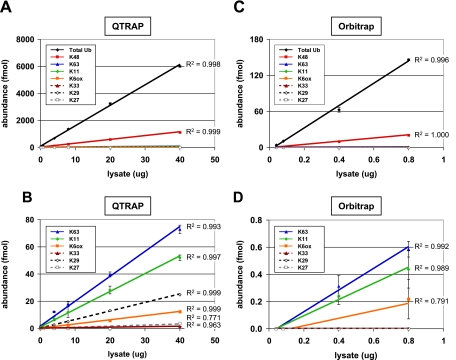
To test the ability of both analytical platforms to detect total Ub and cellular linkages from cell lysate, an experiment was designed where the amount of heavy peptides injected on the column was kept constant at 75 fmol, whereas a peptide mixture from proteasome inhibitor-treated 293T cell lysate was titrated across a range from 40 ng to 40 μg. All samples were injected on the QTRAP, whereas samples containing <1 μg were injected on the Orbitrap (Fig. 6). In the lowest level sample (40 ng of lysate), both instruments were able to detect total Ub (5.7 and 4.0 fmol) and K48 (0.9 and 0.5 fmol), whereas other linkages were below the limits of detection. On the QTRAP setup, K63 and K11 linkages were each detectable from 0.8 μg of digested lysate, whereas on the Orbitrap system, it was possible to detect K63 and K11 linkages from 0.4 μg. K6 linkages were first observed following a 4-μg injection in the QTRAP but could be detected from 0.8 μg of digested lysate on the Orbitrap. Interestingly, K29 linkages could be detected on the QTRAP when injecting a 4-μg eq of lysate but were not detected using the Orbitrap setup. This is due to the improved peak resolution and signal to noise afforded to this hydrophilic peptide by the high flow chromatography of the QTRAP platform. K33 was likewise detected on the QTRAP but only when 40 μg of digested peptides were injected on the column. The abundance of each peptide increased linearly in QTRAP analyses up to 40 μg of injected peptides. By stitching together the observed linear ranges for the Orbitrap and QTRAP instruments, quantitation can be performed across at least 4 orders of magnitude with the majority of peptides being quantifiable from 0.5 to >5000 fmol.
Fig. 6.
Dilution series of proteasome inhibitor-treated 293T cell lysate on multiple instrument platforms. A, a series of lysate samples ranging from 0.04 to 40 μg was prepared and analyzed on a QTRAP system in the presence of 75 fmol of Ub-AQUA peptide mixture. The amount of lysate loaded on the column is plotted against the measured abundance for total Ub and each polyUb linkage. Total Ub was calculated as the average of all loci (K6, K11, K33, K48, and K63). Error bars represent S.D. (n = 3), and R2 values are shown for each linear fit. Total Ub was detectable in all injections, increasing linearly (R2 = 0.998) from 5.7 fmol in 0.04 μg of lysate to 6004 fmol in 40 μg of lysate. Similarly, 0.9 fmol of K48 was quantified in 0.04 μg of lysate up to 1126 fmol in 40 μg of lysate. B, 100× magnification of data from A showing detection of lower abundance polyUb linkages. R2 values greater than 0.99 can be obtained from 40 μg down to a 0.4-μg injection for K63, a 0.8-μg injection for K11, and a 4.0-μg injection for K6(ox) and K29. K27 and K33 signal was only detectable from 20- or 40-μg injection levels, whereas linear polyUb chains were not detected in any of the samples. C, subset of lysate samples (with 75 fmol of Ub-AQUA peptide mixture) used in A (0.04–0.8 μg) was analyzed on the Orbitrap (n = 2). A total Ub level of 4.0 fmol was measured in 0.04 μg of lysate, increasing linearly (R2 = 0.996) up to 146 fmol in 0.8 μg of lysate. D, 225× magnification of C. K63 and K11 were detected only at 0.4- and 0.8-μg lysate eq. K6(ox) was detectable only at a 0.8-μg lysate load, accounting for an R2 value of 0.79. K33, K29, K27, and linear chains were not detectable even at the highest injection levels.
Profiling Multienzyme in Vitro Ubiquitination Reactions
Initial work with the Ub-AQUA method examined cyclin B1 ubiquitination by the anaphase-promoting complex (APC). In those studies, mass spectrometry data revealed two stages of ubiquitination whereby a foundation of monoUb was applied to the substrate in the first stage, and then polyUb chains assembled in the second stage (12). Work on many substrates now suggests that multistage ubiquitination involves substrate priming by an E2 enzyme, such as those in the UbcH5 family, followed by chain extension by dedicated enzymes from other E2 families (41, 42) or by E4 ligase enzymes (43, 44). To demonstrate the utility of quantitative mass spectrometry methods for dissecting multistep ubiquitination by sequentially acting enzymes, we decided to investigate the E2 enzyme UbE2S. UbE2S was recently shown to function in concert with UbcH10 to generate K11-linked polyUb chains on substrates of the APC (5, 6, 8). It is unclear whether UbE2S functions as a chain-extending enzyme in the context of other E2s, such as those in the UbcH5 family, or with E3 ligases besides the APC.
To examine whether UbcH5A and UbE2S can act sequentially, two-step in vitro ubiquitination reactions were performed using the autoubiquitinating E3 c-IAP1 as both ligase and model substrate (Fig. 7). Previous characterization of these reaction conditions indicated that UbcH5A alone, but not UbE2S alone, was able to catalyze substrate ubiquitination.2 Conditions were identified here whereby a low level of UbcH5A (0.05×) was insufficient to catalyze appreciable ubiquitination. Two-step reactions were then set up by first performing 0.05× UbcH5A preincubation followed by addition of UbcH5A or UbE2S. Two-step reactions were performed in the presence of either UbWT, UbK11R, UbK48R, UbK6R, or UbK0 (lysineless Ub). Reaction products were subjected to SDS-PAGE to separate subpopulations of Ub substrates and either Coomassie-stained (Fig. 7A) or subjected to Western blot using antibodies recognizing total Ub (Fig. 7B) and K11 linkages (Fig. 7C) (29, 33). The Coomassie-stained gel was cut into regions based on the signals observed in corresponding Western blots and digested with trypsin for MS analysis. Given the series of Ubmutants used, total Ub was calculated from the K48 and K63 loci using the wild-type and mutant peptides described above (Fig. 7D). Specifically, (LIF + LIF-R + QLE)/2 and (TLS + TLS-R + EST)/2 were used to account for the unbranched forms of Ub (monoUb and end cap Ub) at these two loci.
Western blot and MS analysis both showed that 0.05× UbcH5A alone was insufficient to catalyze appreciable ubiquitination despite incubation for 35 min at 37 °C. In contrast, two-step ubiquitination reactions using 1× UbcH5A in the second step (H5A + H5A) demonstrated robust activity with UbWT and all Ubmutants by Western blot and MS (Fig. 7, B and D). In H5A + H5A reactions with UbK11R and UbK0, α11 Western blot (Fig. 7C) and MS measurements (Fig. 7E) demonstrated the absence of K11 linkages. Reactions with UbK48R failed to generate K48 linkages (Fig. 7, F and G), whereas the linkage profile of H5A + H5A reactions with UbK6R mirrored that of UbWT with nearly equivalent levels of K48, K63, and K11 (Fig. 7, F and G). K6 and K33 linkages were detected for UbWT, but each represented <1% of the total Ub and are not shown. The GGMQ peptide corresponding to linear chains was monitored during these runs and was not detected. Reactions with UbK0 generated Western blot and MS signal indicative of extensive multiubiquitination with a faster migrating Ub smear than UbWT (Fig. 7B) reactions and an absence of polyUb linkages (Fig. 7, F and G).
As with H5A + H5A reactions, when UbE2S was added following 0.05× UbcH5A preincubation (H5A + E2S), substrate ubiquitination was observed by Western blotting and MS (Fig. 7, B and D). This indicated that UbE2S had the ability to promote conjugation of UbWT. Because UbE2S is known to specifically generate K11 chains (33, 45), we expected that H5A + E2S reactions performed with UbK11R and UbK0 might be impaired. Indeed, H5A + E2S reactions performed using UbK11R and UbK0 yielded less total Ub signal than similar H5A + E2S reactions performed with UbWT (Fig. 7, B and D). H5A + E2S reactions utilizing UbK6R displayed Western blot (Fig. 7, B and C) and MS signal (Fig. 7, D and E) comparable with UbWT in gel region 2 but were unexpectedly impaired in high molecular weight Ub substrates in gel region 1.
The ability to dissect two populations of Ub substrates from within a single reaction for MS analysis is a key advantage of the polyUb linkage profiling methods described here. Ub substrates present in gel region 1 of H5A + E2S samples were expected to consist of primarily K11-linked chains because the reactions were dependent on addition of UbE2S. Instead, a mixture of K48, K63, and K11 linkages was observed (Fig. 7F) similar to what might be expected for UbcH5A-generated chains. One possible explanation for the mixed linkage profile observed in gel region 1 is that, in this reconstituted system, UbE2S may be modulating the Ub-conjugating activity of UbcH5A rather than assembling polyUb chains itself. This is in contrast to H5A + E2S reaction products migrating in gel region 2 where MS analysis indicated that Ub signals comprised almost exclusively K11-polyUb (Fig. 7G). Alternative explanations have not been formally tested, including whether UbcH5A modulates UbE2S activity. Ub linkage profiles in gel region 2 did differ dramatically between H5A + H5A and H5A + E2S reactions for the series of Ubmutants and suggest that UbE2S was catalytically active in these in vitro reactions. Further experiments would be required to formally establish whether UbE2S catalytic activity is required to generate the reaction products observed in gel regions 1 and 2 of the H5A + E2S reactions. Data-dependent MS/MS on the Orbitrap, performed in a follow-up analysis, did identify UbE2S peptides in gel region 2, suggesting that UbE2S is the major Ub substrate observed in this gel region (data not shown).
Lysine-to-arginine and lysine-free Ubmutant proteins have been important experimental tools for understanding linkage specificity of enzymes in the Ub pathway. In reactions with lysine-to-arginine mutant forms of Ub, failure to generate Ub substrates has occasionally been interpreted to mean that a certain lysine is the residue through which linkages are generated. Given the potential roles of surface-exposed Ub residues in directing polyUb chain formation at adjacent sites, caution must be used in interpreting such indirect results. In the example above, H5A + E2S reactions with UbK6R generated abundant K11-linked chains in gel region 2 but almost no Ub substrates or K11 linkages in gel region 1 (Fig. 7, A–C). Although this result might be interpreted to mean that the Ub substrates in gel region 1 comprised K6 linkages, MS analysis of UbWT reactions demonstrated that the majority of linkages were in fact through K48, K63, and K11 with K6 linkages representing a negligible fraction (Fig. 7F). Although much remains to be elucidated about the roles that individual residues of Ub play in directing polyUb linkage formation, this finding reinforces the idea that direct, quantitative analysis of Ub linkages generated by UbWT will be essential for characterizing the mechanisms of polyUb synthesis.
Profiling Mixed Linkage Substrates from Cells
In addition to characterizing in vitro reaction products, our new methods are useful for characterizing ubiquitination profiles directly from cell lysates. In cells, one question yet to be addressed is whether mixed linkage substrates, which are commonly seen in vitro (12, 28, 29, 46), are prevalent or functionally significant. A mixed linkage substrate is a protein molecule that is modified by more than one polyUb linkage. The mixed linkages on a substrate may be in the form of a single heterogeneous chain (e.g. K48 → K63 → K11) or multiple homogeneous chains of different linkages (e.g. K48 → K48 and K63 → K63) on separate substrate lysines. To evaluate the prevalence of mixed linkage substrates, we decided to combine our mass spectrometry methods with linkage-specific antibody enrichment. Linkage-specific antibodies have been shown previously to capture Ub-modified proteins carrying the linkage epitope with a high degree of specificity (29, 33). In immunoprecipitation experiments, these antibodies are capable of capturing mixed linkage substrates so long as they carry the target linkage.
An experiment was designed to compare mock-transfected cells with cells overexpressing UbK0 (Fig. 8A). Transfected 293T cells were lysed and then subjected to linkage-specific immunoprecipitation with antibodies against K48 linkages (α48), K63 linkages (α63), or an isotype control antibody (IgG). Immunoprecipitated samples, as well as unenriched lysate (input), were separated by SDS-PAGE and Coomassie-stained. Selected regions were excised and subjected to in-gel trypsin digestion and MS analysis on the QTRAP (Fig. 8B). Peptides covering the wild-type and mutant forms of the K48 and K63 loci were used to determine the abundances of endogenous UbWT and exogenous UbK0.
Fig. 8.
Mutant Ub overexpression alters composition of cellular pool and cellular mixed linkage substrates. A, 293T cells were either mock-transfected or transfected with UbK0. Cells were lysed under denaturing conditions and subjected to immunoprecipitation with antibodies directed against K48 linkages (α48), K63 linkages (α63), or an isotype control (IgG). B, Coomassie-stained gel of input lysates and IP samples. Denoted gel regions were digested with trypsin and analyzed by SRM on the QTRAP. C, abundances of total Ub for gel regions 1 and 2 were quantified for mock-transfected cells as the average of K48 and K63 loci. D, abundances of each linkage shown relative to total Ub in the mock-transfected sample. Mono/end cap represents the difference between total Ub and the sum of all linkages. In the α48 and α63 linkage IP samples, enrichment of the target linkage is observed relative to input material. The complex linkage profile with K48, K63, and K11 indicates that mixed linkage substrates are prevalent. E, following UbK0 transfection, endogenous UbWT and exogenous mutant Ub (UbK0) were observed in gel regions 1 and 2. Increased levels of endogenous UbWT could be detected compared with mock-transfected cells. The ratio of UbK0 to UbWT was determined based on peptides from the K48 and K63 loci using the equations LIF-R/(K48 + LIF + LIF-R) and TLS-R/(K63 + TLS + TLS-R). F, UbK0-transfected cells display a linkage profile similar to mock cells. Under all conditions, IP samples display a mixed linkage profile comprising primarily K48, K63 and K11 with low but detectable levels of K6, K33, and K29. Linear and K27-linked chains were not observed.
Compared with mock-transfected cells (Fig. 8C, input), overexpression of exogenous UbK0 caused a >15-fold increase in total Ub for regions 1 and 2 (Fig. 8E, input). Besides supplementing the Ub pool with exogenous UbK0, the overexpression of this construct increased UbWT (stemming from chromosomal sources) from 159 to 1885 fmol in region 1 and 352 to 1727 fmol in region 2. This increase is most likely attributable to impairment of proteasomal degradation, although other possible contributions from increased Ub gene expression and impaired DUB activity have not been examined in detail. Although UbK0 transfection could be expected to dramatically alter the cellular Ub pool by capping polyUb chains and preventing proper chain elongation, only modest effects were observed. Analysis of the Ub linkage profiles for mock (Fig. 8D) and UbK0 (Fig. 8F) input lysates showed a mixture comprising primarily K48, K63, and K11 linkages. K48 was the most abundant form of polyUb, and lower abundance linkages, such as K6, K33, and K29, were also detectable in the input. Of note, no signal was observed for the light forms of the GGMQ peptide, indicating that linear polyUb chains were either not present or below the limit of detection in these samples. The relative abundances of each linkage type differed slightly between gel regions 1 and 2, particularly for UbK0-transfected cells (Fig. 8F). In this UbK0 input sample, lower molecular weight species in gel region 2 displayed 57% monoUb and end caps (terminal unit of a polyUb chain where all seven lysines are unmodified; indistinguishable from substrate-bound monoUb in our analysis; Ref. 12) (Fig. 8F) compared with just 26% monoUb and end caps in mock input (Fig. 8D). Interestingly, this increased percentage of monoUb and chain end caps in UbK0-expressing cells came at the expense of K48, but not K63, linkages.
In both linkage immunoprecipitation experiments using mock- or UbK0-transfected cells, we found as expected that Ub proteins captured by α48 and α63 were enriched in the target linkage relative to the input material. For α48, a 3.3-fold enrichment was observed for UbK0 cells in region 2 relative to the input with 1.3–1.5-fold enrichment in other α48 immunoprecipitation analyses relative to their respective inputs. For α63, enrichment of K63 linkages was between 1.4- and 2.6-fold relative to the corresponding input material (Fig. 8, D and F). Despite this enrichment, mixtures of K48, K63, and K11 with lower levels of K6, K33, and K29 were still observed. The prevalence of mixed linkage substrates in immunoprecipitated samples demonstrated that complex Ub signals containing multiple linkages may be common for stable, cellular Ub substrates. However, the question of whether in vivo substrates are modified by single chains containing varied linkages has been difficult to resolve by genetics, antibody-based, or mass spectrometry approaches. We found that in the UbK0-transfected samples the percentage of mutant Ub for gel region 1 is 51% (Fig. 8E, input), indicating that only 49% of the Ub is capable of forming polyUb linkages. However, the Ub linkage profile indicates that 64% of the Ub in this region is in some form of polyUb chain with only 36% in the form of end caps and monoUb. This is noteworthy because if the percentage of total Ub in the form of polyUb is greater than the fraction of UbWT it indicates the presence of forked polyUb chains (through K48, K63, and K11) among the protein aggregates residing at the top of the gel (26, 46). Additional work remains to be done, but the ability to determine the total Ub and the relative abundances of endogenous and exogenous forms of Ub within cells opens the door to a full understanding of Ub dynamics and the nature of Ub signals on substrates in vivo.
DISCUSSION
Advances in the methods used to monitor signal transduction have enabled the detailed mapping of physiological and pathological processes in eukaryotic cells. Among these, developments in mass spectrometry-based proteomics have played a central role in characterizing post-translational modifications and the effects they have on cellular proteins. This has been particularly apparent for processes driven by phosphorylation and ubiquitination. Ub signals on individual substrates can be heterogeneous in terms of size (number of Ubs) and composition (polyUb linkage profile) such that even in vitro reactions can contain multiple Ub substrates with distinct Ub signals. In optimizing a quantitative MS method for characterizing Ub signals, we have focused on an approach that utilizes SDS-PAGE and in-gel digestion. Separation of Ub-modified proteins by molecular weight permits focused analysis of defined subpopulations within each sample as guided by Ub Western blots. As shown in our multienzyme in vitro experiments, gel-based separation can be critical for dissecting the individual and combined contributions of enzymes, such as UbcH5A and UbE2S (Fig. 7). An obvious benefit of the gel-based approach is that it effectively denatures the proteins and removes MS-incompatible salts and chaotropes, promoting efficient proteolysis and MS analysis. Ub is a notoriously stable protein structure and can be difficult to digest in solution without the presence of detergents or other MS-incompatible denaturing agents. Our in-gel digestion protocol includes 5% ACN in the digestion buffers to increase the activity of trypsin and minimize the loss of hydrophobic peptides. Soaking gel pieces in trypsin for >2 h further enhances digestion efficiency as described previously (47). One drawback of using a gel-based approach is that imprecise gel band excision can result in errors, such as the introduction of small amounts of Ub from adjacent lanes. As with any sample fractionation, separating Ub increases analysis time by severalfold and can complicate data analysis. As a practice, we do not sum the abundances of individual analytes across a series of consecutive gel regions because of concern over differential peptide extraction efficiency along a gradient gel.
As in most laboratories, limitations in the available instrument time force a choice between the acquisition of replicate data points for individual samples and profiling additional biological conditions. Data shown here demonstrate that AUCs for reinjections vary by ∼5%, suggesting that we benefit most by collecting data from additional gel regions, time points, and biological control samples at the expense of replicate injections. Minimizing variability between peptide ratios in biological samples relies in part on adding internal standards to digested samples as early as possible during sample preparation. Because digestion of miscleaved Ub internal standards (IQ-EG, LI-QL, and TL-ES) by residual trypsin activity can systematically alter peptide ratios, we add internal standard mixtures immediately after the digests have been quenched. To maximize the number of analytes within the linear range, we find it helpful to use total Ub Western blots for estimating the amount of Ub in a sample so that internal standard peptides can be added at an appropriate concentration relative to the digested analyte peptides. The process of adding AQUA peptides to individual samples remains a source of variability that can only be managed by careful sample handling, although preparing single use aliquots of AQUA peptide mixtures has decreased experimental variability. This batch approach also permits us to test AQUA mixtures against polyUb chains of defined lengths and linkages to correct peptide concentrations on a batch by batch basis.
The instrument platforms used in our group have been chosen because together they enable efficient characterization of Ub signals even when low end sensitivity is required. The limits of detection vary from peptide to peptide, but we have been able to successfully measure polyUb linkages across more than 4 orders of linear range from <0.5 fmol for the K48 -GG signature peptide on the Orbitrap to >5 pmol for the entire battery of peptides on our QTRAP system. Similar low end limits of detection have recently been reported for Ub peptides on a QTRAP instrument when chromatographic separation is performed at 0.20 μl/min (34). In our current configurations, the UPLC-Orbitrap setup is more sensitive than the capLC-QTRAP for most peptides by severalfold. The exceptions are the hydrophilic K29 and QLE peptides, which are not resolved on the 1 μl/min UPLC gradient but yield sharp early eluting peaks at 200 μl/min on the capLC. The Orbitrap configuration is most often used when trying to identify Ub substrates and profile Ub linkages in a single analysis. For the majority of Ub linkage profiling studies, such as the ones described in Figs. 7 and 8, we find that SRM analysis on the QTRAP at 200 μl/min offers an ideal balance of sensitivity and robustness. On this system, peptides can be loaded on the column in ∼10-fold higher abundance, permitting detection of low femtomole amounts of Ub in highly complex mixtures by simply injecting more material (Fig. 6). We found that unit resolution represented an ideal balance between absolute signal intensity and signal to noise. We focus as much dwell time as possible toward detecting signal from the most sensitive transition for each peptide rather than dividing the duty cycle among additional less sensitive transitions. Utilizing a segmented method has allowed us to further extend the dwell times for these transitions. The risk of relying on a single transition is that under certain instrument conditions (deteriorating performance) or in certain complex mixture samples quantitation could be compromised by artifactual signal. One such situation would be a sample where a peptide with precursor m/z, fragment m/z, and retention time similar to those of a Ub peptide (heavy or light) is present within the protein mixture, leading to increased signal for a monitored transition. Recently, additional transitions for characterizing Ub peptides have been reported for both yeast and human forms of Ub that may be useful in resolving such situations (34). Although monitoring multiple transitions per peptide may improve confidence and specificity in discovery-themed experiments and complex mixture quantitative studies, we find that monitoring the single best transition is effective for quantitation of Ub across a range of conditions, including certain highly complex mixtures.
Data from these experiments, such as values calculated for percent total Ub (the portion of the Ub population represented by a single form; e.g. K48/total Ub), depend on reliably determining the amount of total Ub across a series of samples. This can be a challenge given the various structural isomers generated within cells and the permutations of Ub that may be used within experimental systems. Although it is cost-effective to determine total Ub by relying on peptides covering a single locus, using peptides from multiple loci, as described here, offers a distinct advantage by providing additional data that can be considered. We have encountered cases where results from a single locus misreport the amount of Ub because of sample composition, unexpected instrument issues, amino acid modifications, digestion inefficiencies, or alterations in stock peptide concentrations. The multiple locus approach is particularly helpful for identifying instances where results are influenced by incomplete digestion, such as during the analysis of long, homogenous polyUb chains or in cases where unfractionated mixtures of substrates are analyzed in batch (34). Built into these methods are several quality controls that recognize errors arising from incomplete digestion and other systematic effects. The miscleaved peptides, such as IQ-EG, TL-ES, and LI-QL, can be used directly to diagnose incomplete digestion, whereas TLS:EST and LIF:QLE ratios are robust numerical indicators of digestion efficiency. Peptides stemming from the same locus (such as TLS and EST) should be of equivalent abundance following complete proteolytic digestion. Past experience suggests that TLS:EST ratios systematically >1.3 are an indication of incomplete digestion because trypsin digestion produces the TLS peptide more efficiently than the EST peptide from the same locus (data not shown). The implicit assumptions that Ub peptides are not chemically or post-translationally modified (besides polyUb) or that forked polyUb chains (e.g. K6-K11, K27-K29, and K29-K33) (26, 46) are of negligible abundance can be evaluated for each experiment based on the data collected. For example, in cases where the N-terminal glutamine residue of the QLE peptide is cyclized to form pyroglutamate, the K48 locus can be excluded from total Ub calculations. When considering the N terminus, it is possible to confirm that the analyte and internal standards for each Met-containing peptide are in the same oxidation state by monitoring each of these transitions. Given these considerations, we believe that total Ub is best reported as the average of the maximum number of loci held in common between the various samples. In cases where lysineless Ub (UbK0) or several lysine mutant forms (e.g. UbK48R and UbK63R) are considered concurrently, we chose to determine total Ub based on loci for which direct measurements could be made using internal standards toward the wild-type and mutant forms (Figs. 7 and 8).
It has been shown that individual protein substrates can be modified by Ub on multiple lysines and may carry more than one form of polyUb, although the effect of these complex Ub signals on interactions with Ub-binding proteins, protein complexes, and the proteasome are not yet understood. A number of fundamental questions remain to be addressed, such as whether substrates with complex Ub signals are effectively recognized and processed by the degradation machinery or rather act as dominant negative inhibitors of the proteasome as has been suggested (46). What effects do the presence of mixed linkage substrates have on substrate-specific and broadly acting DUB enzymes, and what are the consequences of mixed linkage substrates within the cellular Ub pool? It is possible that mixed linkage substrates promote specific biological processes, although no such activity has been proposed. Whereas little is known about the effect of generating mixed linkages, there is a growing appreciation that assembly of a Ub signal frequently involves coordination of multiple chain assembly factors. The results presented here provide a framework for using mass spectrometry to study complex Ub signals in reconstituted systems with multiple competing or complementary enzymatic activities. Combining these mass spectrometry methods with linkage-specific antibody enrichment will further contribute to understanding of the mechanisms and biological consequences of the complex Ub signals in vivo.
Acknowledgments
We acknowledge Nobuhiko Kayagaki, Clifford Quan, Wendy Sandoval, Erin Dueber, Christine Yu, Dan Yansura, and Racquel Corpuz for providing reagents and technical support. We acknowledge Allison Bruce for assisting in preparation of the figures. We also thank Sarah Hymowitz, Ivan Bosanac, Jianjun Zhang, Jennie Lill, and members of the Microchemistry and Proteomics Lab and the Department of Protein Chemistry for advice and insightful discussions. All authors are employees of Genentech, Inc.
Footnotes
1 The abbreviations used are:
- Ub
- ubiquitin
- FA
- formic acid
- AMBIC
- ammonium bicarbonate
- H2O2
- hydrogen peroxide
- AUC
- area under the curve
- SRM
- selected reaction monitoring
- CV
- coefficient of variation
- AQUA
- absolute quantification
- DUB
- deubiquitinating
- polyUb
- polyubiquitin
- monoUb
- monomeric Ub
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- UPLC
- ultraperformance LC
- ox
- sulfoxide
- (ox)2
- sulfone
- APC
- anaphase-promoting complex
- IP
- immunoprecipitation.
REFERENCES
- 1. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 2. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garnett M. J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A. R. (2009) UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin L., Williamson A., Banerjee S., Philipp I., Rape M. (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williamson A., Wickliffe K. E., Mellone B. G., Song L., Karpen G. H., Rape M. (2009) Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 106, 18213–18218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu T., Merbl Y., Huo Y., Gallop J. L., Tzur A., Kirschner M. W. (2010) UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 107, 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 10. Guterman A., Glickman M. H. (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279, 1729–1738 [DOI] [PubMed] [Google Scholar]
- 11. Hofmann R. M., Pickart C. M. (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276, 27936–27943 [DOI] [PubMed] [Google Scholar]
- 12. Kirkpatrick D. S., Hathaway N. A., Hanna J., Elsasser S., Rush J., Finley D., King R. W., Gygi S. P. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 [DOI] [PubMed] [Google Scholar]
- 13. Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. (2009) Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol. Cell 33, 496–504 [DOI] [PubMed] [Google Scholar]
- 14. Winget J. M., Mayor T. (2010) The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell 38, 627–635 [DOI] [PubMed] [Google Scholar]
- 15. Cheng L., Watt R., Piper P. W. (1994) Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol. Gen. Genet. 243, 358–362 [DOI] [PubMed] [Google Scholar]
- 16. Watt R., Piper P. W. (1997) UBI4, the polyubiquitin gene of Saccharomyces cerevisiae, is a heat shock gene that is also subject to catabolite derepression control. Mol. Gen. Genet. 253, 439–447 [DOI] [PubMed] [Google Scholar]
- 17. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 18. Hanna J., Leggett D. S., Finley D. (2003) Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swaminathan S., Amerik A. Y., Hochstrasser M. (1999) The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cook C., Petrucelli L. (2009) A critical evaluation of the ubiquitin-proteasome system in Parkinson's disease. Biochim. Biophys. Acta 1792, 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petroski M. D. (2008) The ubiquitin system, disease, and drug discovery. BMC Biochem. 9, Suppl. 1, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Randow F., Lehner P. J. (2009) Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 11, 527–534 [DOI] [PubMed] [Google Scholar]
- 23. Finley D., Sadis S., Monia B. P., Boucher P., Ecker D. J., Crooke S. T., Chau V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14, 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mimnaugh E. G., Chen H. Y., Davie J. R., Celis J. E., Neckers L. (1997) Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36, 14418–14429 [DOI] [PubMed] [Google Scholar]
- 25. Xu M., Skaug B., Zeng W., Chen Z. J. (2009) A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol. Cell 36, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 27. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 28. Blankenship J. W., Varfolomeev E., Goncharov T., Fedorova A. V., Kirkpatrick D. S., Izrael-Tomasevic A., Phu L., Arnott D., Aghajan M., Zobel K., Bazan J. F., Fairbrother W. J., Deshayes K., Vucic D. (2009) Ubiquitin binding modulates IAP antagonist-stimulated proteasomal degradation of c-IAP1 and c-IAP2(1). Biochem. J. 417, 149–160 [DOI] [PubMed] [Google Scholar]
- 29. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 30. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 31. Sliter D. A., Kubota K., Kirkpatrick D. S., Alzayady K. J., Gygi S. P., Wojcikiewicz R. J. (2008) Mass spectrometric analysis of type 1 inositol 1,4,5-trisphosphate receptor ubiquitination. J. Biol. Chem. 283, 35319–35328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumoto M. L., Wickliffe K. E., Dong K. C., Yu C., Bosanac I., Bustos D., Phu L., Kirkpatrick D. S., Hymowitz S. G., Rape M., Kelley R. F., Dixit V. M. (2010) K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 39, 477–484 [DOI] [PubMed] [Google Scholar]
- 34. Mirzaei H., Rogers R. S., Grimes B., Eng J., Aderem A., Aebersold R. (2010) Characterizing the connectivity of poly-ubiquitin chains by selected reaction monitoring mass spectrometry. Mol. Biosyst. 6, 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 36. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kirkpatrick D. S., Gerber S. A., Gygi S. P. (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35, 265–273 [DOI] [PubMed] [Google Scholar]
- 38. Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., Iwai K. (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pesavento J. J., Garcia B. A., Streeky J. A., Kelleher N. L., Mizzen C. A. (2007) Mild performic acid oxidation enhances chromatographic and top down mass spectrometric analyses of histones. Mol. Cell. Proteomics 6, 1510–1526 [DOI] [PubMed] [Google Scholar]
- 40. Dai J., Zhang Y., Wang J., Li X., Lu Z., Cai Y., Qian X. (2005) Identification of degradation products formed during performic oxidation of peptides and proteins by high-performance liquid chromatography with matrix-assisted laser desorption/ionization and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19, 1130–1138 [DOI] [PubMed] [Google Scholar]
- 41. Rodrigo-Brenni M. C., Morgan D. O. (2007) Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
- 42. Ye Y., Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 44. Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., Jentsch S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644 [DOI] [PubMed] [Google Scholar]
- 45. Baboshina O. V., Haas A. L. (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 46. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 47. Havlis J., Shevchenko A. (2004) Absolute quantification of proteins in solutions and in polyacrylamide gels by mass spectrometry. Anal. Chem. 76, 3029–3036 [DOI] [PubMed] [Google Scholar]
- 48. Dynek J. N., Goncharov T., Dueber E. C., Fedorova A. V., Izrael-Tomasevic A., Phu L., Helgason E., Fairbrother W. J., Deshayes K., Kirkpatrick D. S., Vucic D. (2010) c-IAPI and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 29, 4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]