Abstract
The growing use of mass spectrometry in the context of biomedical research has been accompanied by an increased demand for distribution of results in a format that facilitates rapid and efficient validation of claims by reviewers and other interested parties. However, the continued evolution of mass spectrometry hardware, sample preparation methods, and peptide identification algorithms complicates standardization and creates hurdles related to compliance with journal submission requirements. Moreover, the recently announced Philadelphia Guidelines (1, 2) suggest that authors provide native mass spectrometry data files in support of their peer-reviewed research articles. These trends highlight the need for data viewers and other tools that work independently of manufacturers' proprietary data systems and seamlessly connect proteomics results with original data files to support user-driven data validation and review. Based upon our recently described API1-based framework for mass spectrometry data analysis (3, 4), we created an interactive viewer (mzResults) that is built on established database standards and enables efficient distribution and interrogation of results associated with proteomics experiments, while also providing a convenient mechanism for authors to comply with data submission standards as described in the Philadelphia Guidelines. In addition, the architecture of mzResults supports in-depth queries of the native mass spectrometry files through our multiplierz software environment. We use phosphoproteomics data to illustrate the features and capabilities of mzResults.
Burgeoning demand for systematic generation of mass spectrometry data in support of biomedical studies continues to drive the development of proteomics technologies at a rapid pace. The concomitant expansion of proteomics data that accompany scientific reports, often as supplementary materials, has catalyzed numerous and somewhat disparate efforts to standardize results reporting (5–11). However, as an emerging field of endeavor, proteomics faces a unique set of challenges that complicate efforts to establish open and portable formats for sharing results; specific obstacles include:
Technology innovation: mass spectrometry in particular continues to evolve rapidly, leading to multiple hardware configurations, scan functions, and proprietary file formats.
Discovery mode experiments: the majority of studies are performed in discovery mode with the goal of maximizing new information about the protein content of each sample. As a result, methods are in a state of flux with correspondingly little standardization.
Unbounded measurement space: genetic alterations (alternate splicing, translocations, etc.) and post-translational processing (modifications, enzymatic cleavage, etc.) significantly amplify the number of chemically distinct protein products relative to that predicted by the genetic code. As a result, a large fraction of MS/MS spectra go unassigned in typical database search strategies. However, increased recognition that the vast repertoire of gene- and protein-level modifications are correlated with biological function ensures that archived proteomics data, particularly unassigned MS/MS spectra, will be frequently revisited as new information becomes available.
Uncertainty in sequence assignment: Search algorithms (Mascot, Sequest, Protein Pilot, etc.) may assign different peptide sequences to the same MS/MS spectrum. In addition, a large fraction of peptides cannot be uniquely assigned to a single protein, leading to an ambiguous relationship between a subset of the major claims in proteomics experiments (e.g. protein ID/quantification) and the underlying primary measurements (e.g. peptide sequences).
Collectively, these phenomena create a difficult environment in which to simultaneously standardize the reporting of results and enable interested third parties to browse relevant data and test alternative hypotheses with respect to peptide identification or other claims. These problems are exacerbated as the scale of proteomics studies increases.
Several groups (12–15) have developed powerful and flexible pipelines to facilitate sample tracking, data acquisition, archiving, and analysis of proteomics experiments. Although these systems play a critical role in overall project organization and systematic generation of mass spectrometry data in the context of large-scale biomedical studies, they do not provide scalable, interactive, and readily distributable viewers that enable in-depth validation of proteomics results or other related data browsing by third-parties. Similarly, submission of native mass spectrometry data files to an open-access repository such as Tranche, (8, 9, 16) as suggested by the Philadelphia Guidelines (1, 2), may circumvent potential technical limitations associated with surrogate data files (3, 4, 17), but does not provide a mechanism for browsing results or otherwise validating the claims associated with a given study. In fact, informaticians or other interested third parties who do not have access to the corresponding mass spectrometry data systems (Analyst, X-Calibur, MassLynx, etc.) may have no means to interrogate files retrieved from Tranche.
The ideal solution would combine a compact format for distribution with a viewer that supports interactive browsing of results, and furthermore provide a dynamic link between peptide identification/quantification and the underlying mass spectrometry data files. Building upon our recent work in API-based tools for mass spectrometry data analysis (3, 4), and driven in part by the recently announced Philadelphia Guidelines (1, 2), we developed mzResults, a results viewer that leverages established database standards to provide:
A highly annotated, condensed version of peptide and protein ID/quantification, accessible via a user-friendly GUI.
User-driven interrogation of results through SQLite queries and Python scripts.
In-depth and dynamic interrogation of original native data files within the multiplierz environment (4).
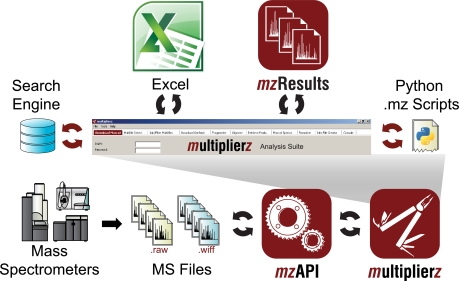
Fig. 1 illustrates the latest deployment of our open-source multiplierz data analytic environment (version 0.8.2). The mzResults viewer provides a scalable solution for distribution of results as compared with multiplierz spreadsheet-based reports, and is designed primarily to support typical discovery-mode identification/quantification experiments, but is also amenable to customization by end-users. In this brief report, we demonstrate the performance and features of mzResults based on phosphoproteomics data generated in our lab.
Fig. 1.
An overview of the multiplierz environment. The multiplierz data analytic environment provides a central point for user interaction with proprietary data files, protein/peptide identification algorithms, and other publicly available databases that contain annotation for biological pathways, mechanisms, and function. Multiple reporting formats provide a scalable solution for distribution of proteomics data. mzResults includes an interactive viewer to enable validation of peptide and protein claims, and supports user-driven data browsing via mzAPI and multiplierz.
IMPLEMENTATION
Architecture
Our overall design philosophy was to create an interactive reporting format that was well-integrated and extensible within our multiplierz environment, (3, 4) as well as readily accessible via commonly used programming languages. Toward this end we implemented the mzResults file as a SQLite database (18) with two primary tables, “PeptideData” and “ImageData.” We chose SQLite because it combines robust database capabilities with a simple, single-file design; moreover SQLite support is embedded in numerous languages including Python, our primary coding environment. The table “PeptideData” contains all data associated with each peptide identification in a given experiment. Because these are stored as text, they are readily available outside of our multiplierz environment using any SQLite database viewer (supplementary Fig. S1). The data for image generation are stored in “ImageData” using Python's built-in binary format, and thus can be extracted programmatically without the use of third party libraries.
This architecture allows mzResults to store information efficiently and provides an interactive data viewer (described below) that supports rapid and in-depth validation of proteomics results. Additional fields and tables are easily added to any mzResults file; these can be used to provide metadata in support of the MIAPE standard (10, 19) or other results such as consensus scoring from multiple peptide search algorithms. As noted above, SQLite is widely supported with libraries available for many commonly used programming languages, allowing researchers to view, use, and modify mzResults files in their preferred programming environment.
Installation and Setup
The most convenient way to create and view mzResults files is with the multiplierz toolkit, an open-source Python application (4), available as a free (63.7 MB) download at SourceForge (http://sourceforge.net/projects/multiplierz) and our web site (http://blais.dfci.harvard.edu/multiplierz). The multiplierz installer contains Python, along with its standard libraries (including SQLite), and is compatible with the Windows XP, Vista, and 7 operating systems. Importantly this single installation includes everything required for users to generate mzResults files from Mascot or Protein Pilot search results, interactively view annotated MS/MS spectra for all peptide identifications, and use SQLite queries to further explore peptide- and protein-level data. With access to the accompanying raw data (current support includes Thermo .RAW and ABSCIEX .WIFF formats) multiplierz provides direct access to MS and MS/MS scans, extracted ion chromatograms (XICs), and other data features. Furthermore, multiplierz includes the open source search tool X!Tandem (20), allowing users to go all the way from raw data to peptide and protein identifications, without the use of commercial search algorithms. A tutorial introducing users to the mzResults format is included in the supplementary materials. Users who encounter difficulties during installation or use, or who wish to discuss additional features, are encouraged to contact us via the E-mail listed on our web site (http://blais.dfci.harvard.edu/index.php?id=63) or under the help menu of the multiplierz application.
FUNCTIONALITY
In this section we demonstrate the functionality of mzResults with brief examples based on data derived from fractionation of phosphopeptides (pS, pT, and pY) enriched by NTA-Fe3+ (unpublished data) and tyrosine phosphorylated peptides isolated by immune-precipitation (21). Based on very conservative criteria we identified 10,408 unique phosphopeptide sequences across these two experiments performed on different mass spectrometry platforms.
Compact Format for Distribution and Interactive Validation of MS/MS Spectra
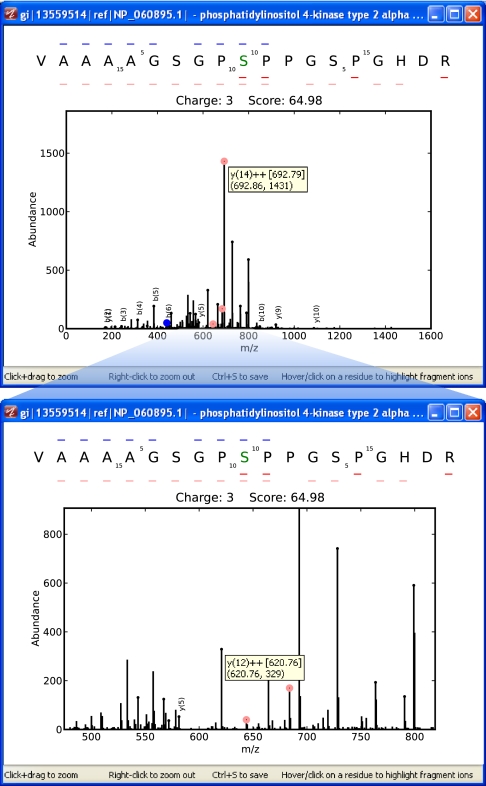
The recently released Philadelphia Guidelines (1, 2) encompass updated criteria for submission of results in support of proteomics studies. Fig. 2 illustrates how mzResults supports the requirements directly related to peptide identification. To date, the distribution and validation of associated MS/MS spectra has been problematic because static images do not allow detailed inspection of restricted m/z ranges or testing of alternative hypotheses. Interrogation of phosphopeptide spectra is particularly critical given the evidence that facile neutral losses (22, 23) and even putative side-chain rearrangements (24) can lead to a high degree of ambiguity in sequence assignments. The architecture of mzResults provides well-annotated and interactive MS/MS spectra for all reported peptides (Fig. 3). Mouse-over of a labeled fragment peak reveals a tooltip with b- or y-ion series assignment and charge state, along with measured mass, predicted mass, and observed intensity. Peptide sequence is displayed across the top, with putative site of phosphorylation highlighted in green (S, T, or Y), with small dashes above (blue) and below (red) to indicate detection of b- and y-type ions, respectively. Hovering over a sequence residue highlights the associated b/y complimentary ion pair in the MS/MS spectra, whereas a left-click freezes the colored indicators to facilitate detailed inspection of fragment ions.
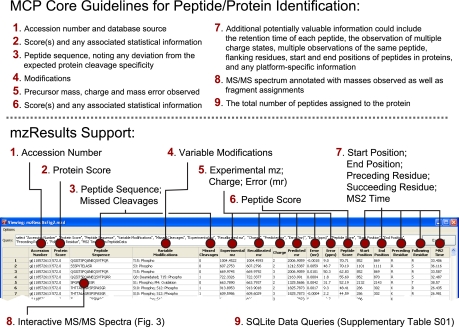
Fig. 2.
The mzResults format supports core MCP guidelines for peptide and protein identification. (Top) Information required to support peptide and protein identifications from MS/MS spectra. (Bottom) mzResults default peptide view, keyed to illustrate how various entries address manuscript submission guidelines for Molecular and Cellular Proteomics.
Fig. 3.
mzResults provides interactive validation of MS/MS spectra. (Top) Peaks with primary b- or y-type ion assignments are labeled in the spectrum with blue and red circles, respectively. The glyphs surrounding the amino acid sequence at the top indicate those fragments detected, with doubly charged ions represented as lighter shades. Mouse-over or left-click on specific residues will highlight the corresponding complimentary b- and y-ions in the spectrum. (Bottom) Users can zoom-in on restricted mass-to-charge ranges for in-depth interrogation of fragment ion assignments.
All native mass spectrometry data files and the mzResults files associated with the examples described herein may be downloaded from the ProteomeCommons.org Tranche Network using the following hash:
KEMZBJjmmT7wJtR4K3TZvn1YW+xYgRu5+mLop7gt/XENDV9acypZVYHMWAnDRCyzv5OeMKbIE72Tk++QBQ6vOKaJ8ecAAAAAAAApTQ==
Table I illustrates that the total size for combined .RAW (Thermo Orbitrap XL) and .WIFF (AB SCIEX QSTAR Elite) files was nearly 14.5 gigabytes. However, the corresponding mzResults files total less than 511 MB, representing more than a 28-fold reduction in size as compared with the native files, while providing the information necessary for data validation and manuscript submission.
Table I. Comparison of file sizes for mzResults and the corresponding native mass spectrometry data.
| Experiment | mzResults Files |
Native Mass Spectrometry Data Files |
||
|---|---|---|---|---|
| File Name | Size (MB) | Number of Files | Total Size (MB) | |
| K562 Global Phosphorylation, 68 Frxn | mzResultsFig2.mzd | 502.8 | 68 | 14,410 |
| mESC Tyrosine Phosphorylation, iTRAQ | mzResultsFig4.mzd | 8.1 | 1 | 50.5 |
| Total | 511.0 | 69 | 14,460.5 | |
Scripting Capability for User-driven Results Queries
The use of SQLite as the underlying format for mzResults enables users to explore proteomics results in more detail and customize the corresponding reports through simple database queries and Python scripts. For example, in our fractionation data (Table I), we used a very conservative strategy to account for phosphopeptides, whereby all occurrences of a given peptide sequence with the same number of phosphorylation sites were collapsed and “counted” as a single phosphopeptide, regardless of S/T/Y location or the presence of other modifications (oxidized methionine and Q/N deamidation). Based on this approach, we identified 10,251 phosphopeptides across 68 fractions acquired on a Thermo Orbitrap XL instrument. These scripts were easily extended (supplementary Table S1) to also consider peptides with an equivalent degree of phosphorylation but different modification sites as distinct sequences (11,460 total phosphopeptides), and to simply include all modification patterns as unique identifications (14,522 total phosphopeptides). These or similarly structured queries provide a convenient mechanism for comparison of results across studies and labs that may use disparate strategies to group or count modified peptides. Importantly the SQLite database library supports queries across multiple files and requires minimal computational resources. These capabilities enable researchers to perform complex data analyses through simple human-readable scripts.
As illustrated in Fig. 2, mzResults tables are displayed from a “peptide-centric” perspective, a default view appropriate for a phosphoproteomics study. However, users may prefer a protein summary as the primary view, and in fact, protein-level information is required by the Philadelphia Guidelines (1, 2). Supplementary Fig. S2 shows an mzResults protein-level view as generated by a separate SQLite query (Fig. 2, bottom right and supplementary Table S1), and reveals that in total we identified 3594 proteins, of which 3471 were phosphorylated, in the context of a liquid chromatography-tandem MS (LC-MS/MS) experiment that spanned 68 phosphopeptide fractions (Table I).
Integration with Multiplierz to Provide a Dynamic Link to Native Mass Spectrometry Files
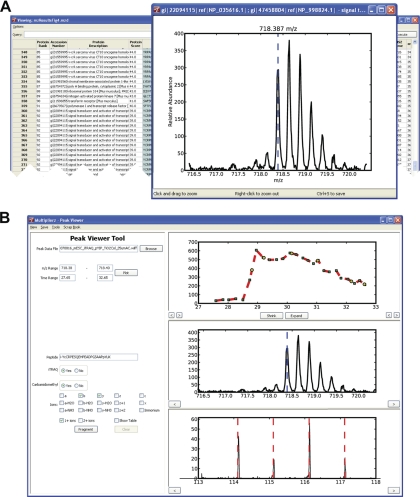
Despite the detailed requirements outlined in the Philadelphia Guidelines (1, 2), the emergent nature of the proteomics field ensures that submission criteria will be subject to continual refinement as new technologies and methods are introduced. This phenomenon is reflected in the fact that the current guidelines are more comprehensive for peptide and protein identification as compared with quantification. As a result of these observations, reviewers or other interested parties would benefit from a dynamic link between the results report and the underlying mass spectrometry data. For example, it is now well recognized that iTRAQ- (25, 26) or TMT-based (27) quantification data are susceptible to errors derived from mixed MS/MS of co-eluting peptide precursors that have similar mass-to-charge ratios (28). Unfortunately, to date there is no widely accepted standard for quantifying or reporting the degree of precursor contamination, exemplifying the need for interrogation of data beyond the peptide identification stage. The architecture of mzResults supports the use of Python scripts that access the underlying mass spectrometry data files, via mzAPI (3), and enables researchers to embed images that span the m/z region around each precursor. Fig. 4A shows the m/z region near the iTRAQ-labeled, phosphorylated peptide sequence, YCRPESQEHPEADPGSAAPpYLK, from STAT3, acquired on a QSTAR Elite mass spectrometer (21). In this way users and reviewers alike can rapidly scan identified peptides for evidence of potential precursor contamination that could skew quantification results. Moreover, use of mzResults files within the multiplierz environment (Fig. 4B) provides the opportunity to dynamically explore mass spectrometry data files in further detail, including generation of extracted ion chromatograms (XICs), inspection of all spectra (MS and MSn) including the iTRAQ reporter ion region, and testing alternative sequences for putative peptide identifications (4).
Fig. 4.
Integration of mzResults and the multiplierz Peak Viewer tool for in-depth query of MS and MS/MS data. A, Data content of mzResults files can be easily extended through simple Python scripts to include a user-defined m/z range spanning each peptide precursor to rapidly scan for potential iTRAQ interference from coeluting peptides of similar mass-to-charge ratio. B, Use of mzResults within the multiplierz environment provides unencumbered and dynamic access to the underlying mass spectrometry data files, via mzAPI, for in-depth validation of the mass spectrometry data.
Use of mzResults in Manuscript Submissions
As noted above the multiplierz installer contains all components necessary to generate mzResults files. We envision that these files will be particularly useful as supplements to journal articles in the scientific literature. In a typical scenario, researchers would download the Mascot search result(s) using multiplierz from their laboratory's server, and output a spreadsheet-based report. At this point users can further manipulate the data (FDR calculations, statistics for modification site assignment or quantification, protein grouping, etc.) depending on their experimental focus, and then create a mzResults file using the “Formatter” tool in multiplierz. Finally mzResults files can be further augmented with images such as XICs or annotated MS/MS spectra by extraction (via multiplierz) of these data from the corresponding mass spectrometer file(s) or Mascot DAT file(s), respectively. Upon submission of their manuscript for peer-review, authors would upload mzResults files as part of their supplementary materials. If the native mass spectrometry data files are also made available through Tranche or another public repository, then, as described above, reviewers and ultimately readers will have complete access to all data (MS, MS/MS, peptide sequence and modification, and quantification) underlying the proteomics-related claims of published studies. In particular, the ability of reviewers and other interested third parties to dynamically explore MS and MS/MS spectra, along with scripting capabilities that enable customization, provides a level of autonomous data review that is not, to our knowledge, currently available in any commercial or open-source software.
DISCUSSION
Although the architecture of mzResults enables integration within data analytic pipelines, widespread adoption will be facilitated by related standardization efforts. For example, in our current implementation mzResults includes fragment ion assignments as determined by Mascot (via DAT files), and the code base can be expanded to include similar functionality for SEQEUST, X!Tandem, OMSSA, and other algorithms. Ideally, however, search engine output formats, such as pepXML (12) or the forthcoming mzIdentML (http://psidev.info/index.php?q=node/40#mzIdentML), will be expanded to include fragment ions assignments as determined by the search algorithm. Once these and analogous criteria for quantification stabilize, mzResults can serve as a common platform for results distribution and viewing based on a standardized input file. Moreover, when combined with the native mass spectrometry files, users, reviewers, and other interested third parties will have unencumbered access to the data that underlie experimental claims.
Despite open questions related to standardization of search engine output, mzResults nonetheless provides an accessible and powerful desktop framework for distribution and validation of proteomics results, and in fact supports the Philadelphia Guidelines for submission of manuscripts to Molecular and Cellular Proteomics. Although the full functionality of mzResults and multiplierz requires local access to the native mass spectrometry data files, we fully appreciate the logistical difficulties and inconvenience associated with download of the mass spectrometry data files for proteomics studies of even moderate size to the user's desktop. In the course of developing the multiplierz environment (3, 4), it has not escaped us that translating our mzAPI library into self-describing URLs would decouple programmatic access from the site of physical storage, and hence provide a mechanism for web-based proteomics data analysis. A prototype server based on this design philosophy is the topic of the accompanying article.
Acknowledgments
We thank Eric Smith for preparation of figures, and members of the Marto lab for helpful discussions.
Footnotes
* Generous support for this work was provided by the Dana-Farber Cancer Institute and the National Institutes of Health, NHGRI (P50HG004233), and NINDS (P01NS047572).
 This article contains supplemental Figs. S1 and S2 and Table S1.
This article contains supplemental Figs. S1 and S2 and Table S1.
1 The abbreviations used are:
- API
- Application Programming Interface.
REFERENCES
- 1. Chalkley R. J., Clauser K. R., Carr S. A. (2009) Updating the MCP proteomic publication guidelines. ASBMB Today, August 16–18 [Google Scholar]
- 2. Cottingham K. (2009) MCP ups the ante by mandating raw-data deposition. J. Proteome Res. 8, 4887–4888 [DOI] [PubMed] [Google Scholar]
- 3. Askenazi M., Parikh J. R., Marto J. A. (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat. Methods 6, 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parikh J. R., Askenazi M., Ficarro S. B., Cashorali T., Webber J. T., Blank N. C., Zhang Y., Marto J. A. (2009) multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10, 364–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradshaw R. A. (2005) Revised draft guidelines for proteomic data publication. Mol. Cell Proteomics 4, 1223–1225 [PubMed] [Google Scholar]
- 6. Bradshaw R. A., Burlingame A. L., Carr S., Aebersold R. (2006) Reporting protein identification data: the next generation of guidelines. Mol. Cell Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 7. Falkner J. A., Falkner J. W., Andrews P. C. (2006) ProteomeCommons.org JAF: reference information and tools for proteomics. Bioinformatics 22, 632–633 [DOI] [PubMed] [Google Scholar]
- 8. Mead J. A., Bianco L., Bessant C. (2009) Recent developments in public proteomic MS repositories and pipelines. Proteomics 9, 861–881 [DOI] [PubMed] [Google Scholar]
- 9. Mead J. A., Shadforth I. P., Bessant C. (2007) Public proteomic MS repositories and pipelines: available tools and biological applications. Proteomics 7, 2769–2786 [DOI] [PubMed] [Google Scholar]
- 10. Orchard S., Jones P., Taylor C., Zhu W., Julian R. K., Jr., Hermjakob H., Apweiler R. (2007) Proteomic data exchange and storage: the need for common standards and public repositories. Methods Mol. Biol. 367, 261–270 [DOI] [PubMed] [Google Scholar]
- 11. Orchard S., Taylor C. F., Hermjakob H., Weimin Zhu, Julian R. K., Jr., Apweiler R. (2004) Advances in the development of common interchange standards for proteomic data. Proteomics 4, 2363–2365 [DOI] [PubMed] [Google Scholar]
- 12. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kohlbacher O., Reinert K., Gröpl C., Lange E., Pfeifer N., Schulz-Trieglaff O., Sturm M. (2007) TOPP–the OpenMS proteomics pipeline. Bioinformatics 23, e191–197 [DOI] [PubMed] [Google Scholar]
- 14. Yu K., Salomon A. R. (2010) HTAPP: high-throughput autonomous proteomic pipeline. Proteomics 10, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu K., Salomon A. R. (2009) PeptideDepot: flexible relational database for visual analysis of quantitative proteomic data and integration of existing protein information. Proteomics 9, 5350–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Falkner J., Andrews P. C. (2006) The DFS tool for dissemination and annotation of proteomics data. HUPO 5th Annual World Congress, p. 98, Long Beach, CA U.S.A [Google Scholar]
- 17. Lin S. M., Zhu L., Winter A. Q., Sasinowski M., Kibbe W. A. (2005) What is mzXML good for? Expert Rev. Proteomics 2, 839–845 [DOI] [PubMed] [Google Scholar]
- 18. Owens M., Hagar P., and Books24x7 Inc (2006) The definitive guide to SQLite. Apress, Berkeley, CA [Google Scholar]
- 19. Taylor C. F., Binz P. A., Aebersold R., Affolter M., Barkovich R., Deutsch E. W., Horn D. M., Hühmer A., Kussmann M., Lilley K., Macht M., Mann M., Müller D., Neubert T. A., Nickson J., Patterson S. D., Raso R., Resing K., Seymour S. L., Tsugita A., Xenarios I., Zeng R., Julian R. K., Jr. (2008) Guidelines for reporting the use of mass spectrometry in proteomics. Nat. Biotechnol. 26, 860–861 [DOI] [PubMed] [Google Scholar]
- 20. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 21. Ficarro S. B., Zhang Y., Lu Y., Moghimi A. R., Askenazi M., Hyatt E., Smith E. D., Boyer L., Schlaeger T. M., Luckey C. J., Marto J. A. (2009) Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem. 81, 3440–3447 [DOI] [PubMed] [Google Scholar]
- 22. Payne S. H., Yau M., Smolka M. B., Tanner S., Zhou H., Bafna V. (2008) Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J. Proteome Res. 7, 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 24. Palumbo A. M., Reid G. E. (2008) Evaluation of gas-phase rearrangement and competing fragmentation reactions on protein phosphorylation site assignment using collision induced dissociation-MS/MS and MS3. Anal. Chem. 80, 9735–9747 [DOI] [PubMed] [Google Scholar]
- 25. Choe L., D'Ascenzo M., Relkin N. R., Pappin D., Ross P., Williamson B., Guertin S., Pribil P., Lee K. H. (2007) 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics 7, 3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 27. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 28. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”. J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]