Abstract
Continued progress toward systematic generation of large-scale and comprehensive proteomics data in the context of biomedical research will create project-level data sets of unprecedented size and ultimately overwhelm current practices for results validation that are based on distribution of native or surrogate mass spectrometry files. Moreover, the majority of proteomics studies leverage discovery-mode MS/MS analyses, rendering associated data-reduction efforts incomplete at best, and essentially ensuring future demand for re-analysis of data as new biological and technical information become available. Based on these observations, we propose to move beyond the sharing of interpreted spectra, or even the distribution of data at the individual file or project level, to a system much like that used in high-energy physics and astronomy, whereby raw data are made programmatically accessible at the site of acquisition. Toward this end we have developed a web-based server (mzServer), which exposes our common API (mzAPI) through very intuitive (RESTful) uniform resource locators (URL) and provides remote data access and analysis capabilities to the research community. Our prototype mzServer provides a model for lab-based and community-wide data access and analysis.
A live instance of the mzServer can be accessed directly at: http://blais.dfci.harvard.edu/mzServer/
The data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash:
6g+QpUvlpxc6PM/M9t/49h0PMLwA7dTCgpwyUqfciXEyZpLun7QzPz8E+LDDJfZzBf1lGKe7t1OkXbmomzTEy70Av/kAAAAAAAAYtg==.
The effective communication of mass-spectrometry based analyses performed in support of biomedical research remains a significant challenge. As so-called discovery mode studies represent the vast majority of literature reports, it is important that data and results be available for rigorous programmatic and manual review. Experiments that focus on post-translationally modified peptides, such as phosphorylation, invite added scrutiny with respect to site of modification and sequence validation. In a companion article (Webber et al., this issue), we present mzResults, a report format and data viewer developed in-part as a response to the recently announced Philadelphia Guidelines from this journal. A substantial portion of mzResults' functionality goes beyond compliance with data reporting requirements, and enables user-driven interrogation of the underlying native mass spectrometry data files through our recently described common application programming interface (API)1 (mzAPI) (1). Consistent with current recommendations outlined in the Philadelphia Guidelines, (2, 3) authors can submit mzResults files with their manuscript and then provide the corresponding native mass spectrometry data files through the Tranche repository (4–6). Interested third parties or reviewers can use mzResults to validate claims made in the associated manuscript, or download the mass spectrometry files and interrogate additional aspects of the proteomics data.
Although the scenario described above is feasible in principle, we believe that in practice the paradigm of community-wide distribution of primary mass spectrometry data is poorly matched with the current trajectory of biomedical research. The continued march toward systematic generation of proteomic and metabolomic data sets that are intended to be “comprehensive” in some biological context (e.g. modification class, subcellular localization, tissue type, disease-specific, developmental state, organism), will likely overwhelm data sharing strategies that are based on distribution of native files from a central repository. Indeed, readers can well imagine that download of all native or surrogate mass spectrometry data files associated with recent large-scale proteomics studies will be unwieldy at best. One viable alternative would be to provide web-based, programmatic access to primary mass spectrometry data at the site of acquisition or a third party (submissions/repository) data-server. In fact this strategy is the standard in the field of astronomy, exemplified by the Virtual Astronomical Observatory (see below, and http://www.usvao.org/). Inspired by this paradigm (Fig. 1), we have developed a web-based server (mzServer), which exposes our common API (mzAPI) through very intuitive (RESTful) URLs (7) to provide efficient remote and programmatic data access and analysis capabilities to the research community.
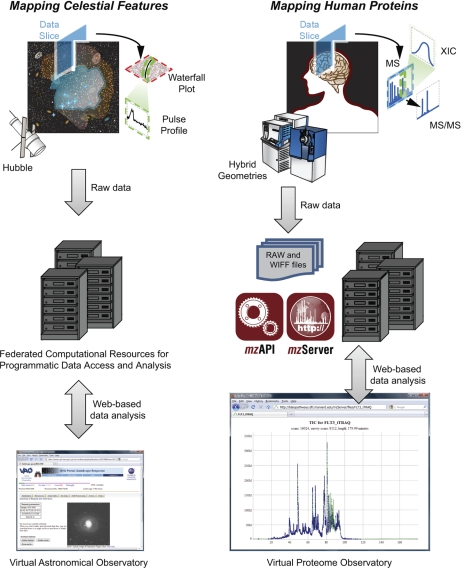
Fig. 1.
Data from astronomy (left) and proteomics (right) have complex, multidimensional structures. The field of astronomy has successfully transitioned to a point whereby programmatic access and visualization via web-based computational resources (bottom, left) has de-coupled data analysis from the site of physical data storage (10, 11). We propose that the field of proteomics is nearing a similar threshold (bottom, right) and have therefore implemented mzServer, a web-based resource that utilizes simple, RESTful URLs to access native mass spectrometry data files via mzAPI (1).
Lessons From the Virtual Astronomical Observatory
Few scientific endeavors generate more raw data than astronomical surveys; in fact the most recent release of data from the Sloan Digital Sky Survey totals nearly 65 TB (8). Not surprisingly, practitioners in astronomy and high energy physics have been pioneers in such areas as ultra-high-bandwidth networks (9) as well as cross-platform binary data formats (10). The difficulties inherent to comprehensive distribution of data associated with these and similar studies has naturally given rise to the strategy on which we based this technical note, namely the idea of an efficient browsable resource for unconstrained data access and transformation. The most visible exemplar of this strategy is the U. S. Virtual Astronomical Observatory (11), itself a member of the International Virtual Observatory Alliance (IVOA), which is an international science consortium dedicated to providing the tools, federated systems, and logistical support necessary to enable web-based analyses of high-dimensional astronomy data generated world-wide. Within this larger framework, Spectrum Services (http://voservices.net/spectrum/) represent one example by which complex data are made available through an online resource. Here, spectra produced by the Sloan Digital Sky Survey are made available both through web-interfaces and simple object access protocol (SOAP)-based web services (12). For historical reasons, many of the data-services implemented by the IVOA (http://www.ivoa.net) are based on SOAP/web-services, although recently there has been a shift toward Resource Oriented and RESTful interfaces (see for example the most recent working draft of the VOSpace specifications (http://www.ivoa.net/Documents/VOSpace/), which are directly analogous to the information architecture underlying our prototype mzServer. We propose that, as in the field of astronomy, the long-term sustainability and success of large-scale, discovery-oriented proteomics will ultimately require an efficient mechanism for data access and re-analysis that is independent of the site of physical data storage.
Server Architecture
In order to enable efficient data browsing, it is important to store the data in a format that is optimal in terms of size and accessibility. Toward this end we chose to leverage our recently described mzAPI in the development of a prototype mzServer. The essential functionality of the mzServer is to transform valid URLs into equivalent mzAPI calls that, in turn, provide access to the original, native mass spectrometry data files. Our use of binary data files leverages the indexing scheme inherent in the manufacturers' proprietary formats and provides efficient, real-time data access and analysis, much in the same way users are accustomed to with the manufacturers' desktop data systems. This approach greatly simplifies data file storage and archiving: native files are simply copied into a predetermined directory, and the computational server is accessed through URLs which are easy to integrate with any existing web-infrastructure.
Information Architecture
As an existence proof for remote mass-informatics, we provide native data files from experiments designed to fractionate phosphopeptides (pS, pT, and pY) enriched by NTA-Fe3+ and to quantify tyrosine phosphorylated peptides isolated by immune-precipitation (13), in total spanning 42 .RAW (ThermoFisher Scientific) and 1 .WIFF (AB SCIEX) files, on a publically accessible instance of the mzServer (http://blais.dfci.harvard.edu/mzServer). Users can access data using URLs corresponding to library calls from our mzAPI (1). For example, URLs of the form: /files/filename/scans/scantime return individual scans where the scan time is provided as a floating point value representing minutes since the start of acquisition. Dropping the scantime variable yields a list of all available scans in the file, whereas dropping the scans keyword generates a total ion chromatogram of the file (Fig. 1, bottom right). Finally, dropping the filename yields a listing of all available datafiles. Similarly, users can generate extracted ion chromatograms (XIC) of any desired mass range using URLs of the form: /files/filename/xic/timeStart-timeStop/mzStart-mzStop (Fig. 2). Because the mzServer will respond to requests by issuing mzAPI calls against manufacturer data files, the response is surprisingly fast and the system is by definition, platform independent. Moreover, mzServer is readily integrated with other web-based resources. For example, we recently developed Pathway Palette (14), a freely accessible internet application that enables researchers to analyze proteomics data in the context of biological pathways. Fig. 3 illustrates the use of mzServer to provide a gateway for query and validation of mass spectrometry data that underlie protein pathways and networks. In this case (Fig. 3B) selection of the STAT3 node, color-coded based on iTRAQ ratio 114:115 (13), reveals the specific peptide sequence detected (YCRPESQEHPEADPGSAAPpYLK), selection of which creates a URL (red asterisk) that calls the corresponding tandem MS (MS/MS) spectrum. Importantly, mzServer dynamically processes peptide sequence or modification site (in this case tyrosine phosphorylation) information input through the URL, allowing users to test alternative hypotheses for peptide identifications. Modifications are specified using a combination of lowercase letters and explicit mass shifts: /files/filename/scan/time/match/i-PEPpTI[14]DE corresponds to N-terminal iTRAQ, threonine phosphorylation, and aspartic acid methylation. HyperText Markup Language (HTML5) is the default output format for mzServer, although users can specify several other common formats (supplementary Fig. S1) by simply appending a valid format-modifier to the URL: .png, .svg, .pdf, .txt, .dta, and .mgf (obviously, some combinations are invalid, e.g. an XIC in mgf format). In addition to the basic mzAPI calls, we are currently exploring various operations that would extend the capabilities of remote mass-informatics. Instrument-specific or computationally intensive functions currently under development include /files/filename/scan/time/reduced, which yields a de-isotoped and charge-reduced spectrum.
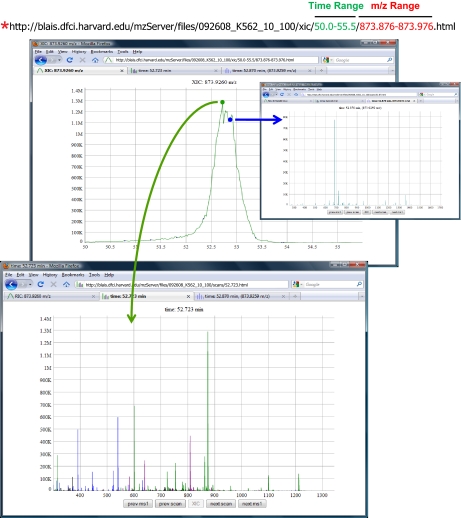
Fig. 2.
Extracted ion chromatograms (XICs) are generated from human-readable URLs (red asterisk, top) that can be modified manually or programmatically. The numbers in green represent the LC elution time in minutes and the numbers in red represent the mass range. XICs are decorated with circles that indicate associated MS (green) and MS/MS (blue) scans. A left mouse click displays the associated MS/MS spectrum in a new browser tab (blue arrow and right inset panel). Clicking on all other datapoints in the XIC displays the associated MS scan (green arrow and bottom panel), in which precursors are color-coded by charge state.
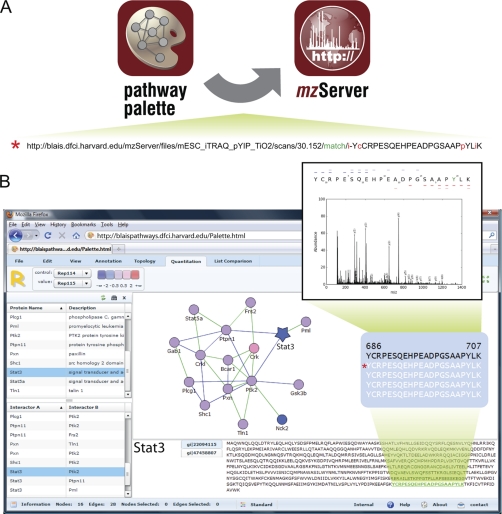
Fig. 3.
Integration of mzServer URLs in other bioinformatic resources. (A) mzServer is readily integrated with other web-based resources such as Pathway Palette (14). (B) Detected proteins are represented as nodes and arranged based on previously reported protein-protein interactions to yield a biological pathway. Nodes are color-coded based on iTRAQ ratios that represent dynamic regulation of phosphorylation resulting from cytokine withdrawal (13). Selection of a protein (STAT3, blue star) shows the sequence and detected peptide evidence (YCRPESQEHPEADPGSAAPpYLK). A left mouse click on an identified peptide sends a URL (red asterisk) to the mzServer to reveal the associated MS/MS spectrum with the amino acid sequence annotated according to the b- (blue dashes) and y- (red dashes) type fragment ions detected (lighter shades of blue and red correspond to doubly-charged fragment ions). Users can easily test alternative hypotheses for peptide sequence assignment and modification state by editing the associated URL.
Use Cases for the mzServer
In the following section we briefly describe the use of mzServers in the context of individual labs, and as a resource for the broader research community.
mzServer as a Local Laboratory Resource
The most immediate need for user-driven or custom analysis of results is typically within the confines of the laboratory that generated the data. A compelling example of the mismatch between the needs that arise ad hoc during research projects and the standard capabilities offered in manufacturers' software is illustrated by recent work in our lab focused on the development of online multidimensional phosphopeptide fractionation. Preliminary experiments in this study spanned a depth of between four and 73, three-dimensional fractions, yielding more than 20,000 unique phosphopeptide sequences in total. Optimization of separation peak capacity for our fractionation platform required calculation of XICs for all peptides detected, across the entire fractionation space. Readers can well imagine the difficulty and tedium of this analysis performed within the constraints of a commercial data system. As an effective alternative, we developed Chromatoplot, a Javascript application that uses the mzServer API to dynamically generate XICs and display them through the browser. Importantly the multi-dimensional nature of our separation platform required calculation of XICs across a 3 × 3 grid to accurately reflect relative peptide abundances in neighboring fractions. Fig. 4 shows the primary Chromatoplot webpage in which XICs from nine different mass spectrometry data files, corresponding to adjacent fractions, are simultaneously displayed. In fact, the interactive web interface (http://blais.dfci.harvard.edu/mzServer/apps/chromatoplot.html) allows users to explore XICs throughout the separation space with minimal latency (determined largely by the bandwidth available at the user's point of internet access), without ever leaving the browser.
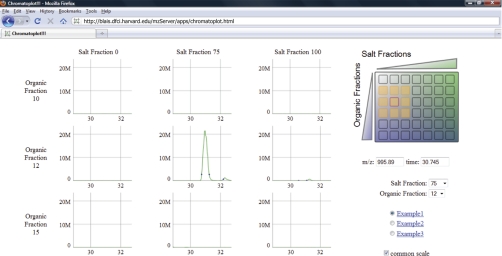
Fig. 4.
mzServer provides an intuitive URL-based API for mass-informatics that enables rapid creation of interactive data viewers. Chromatoplot allows rapid and convenient evaluation of separation peak capacity for multi-dimension fractionation schemes. A 7 × 6 grid (top, right) encompasses 42 fractions separated in three dimensions. Selection of one fraction (red outline) along with a precursor m/z value and retention time (middle, right) displays XICs for the designated mass-to-charge range in the selected and neighboring fractions.
Installation and Set-up of a Local Instance of mzServer
In order to set up a local or lab-based mzServer, users need only download a single zipped directory available from the main mzServer website (http://blais.dfci.harvard.edu/mzServer/mzServer.zip), the server is started by running the mzServer.mz multiplierz script located in the main directory. Consequently, the only software components necessary to run an mzServer instance are multipierz (15) (available for downloaded from http://sourceforge.net/projects/multiplierz/) and the vendor software for the file types of interest to the user. For example, in the case of instruments from ThermoFisher Scientific users can choose between the XCalibur XDK (typically provided as part of the instrument data system) or, alternatively, the MSFileReader installer, freely available from http://sjsupport.thermofinnigan.com/public/detail.asp?id=586. In the case of AB SCIEX instruments, the user will need to install ProteinPilot (the freely available viewer mode is sufficient: http://www.absciex.com/Products/Software/ProteinPilot-Software).We are currently working to incorporate data system libraries from other manufacturers based on our mzAPI framework (1). Users who encounter difficulties during installation or use, or who wish to discuss additional features or support of other instrument platforms are encouraged to contact us via the E-mail listed on the mzServer web site (http://blais.dfci.harvard.edu/mzServer) or under the help menu of the multiplierz application.
mzServer as a Community Resource
Beyond use within individual laboratories, mzServers can be deployed as federated resources to support sharing and publication of large scale mass spectrometry data. For example, during peer review data can be made available on a neutral mzServer configured to allow restricted and anonymized access to reviewers. Because the mzServer provides unencumbered access to individual MS and MS/MS scans in addition to associated XICs, along with the ability to overlay fragment ion assignments for putative peptide identifications, reviewers can validate claims and iteratively explore alternative sequences or sites of modification. In fact, authors can easily insert hyperlinks to the data described in their manuscript, so as to provide reviewers, and ultimately readers, with direct links to the data and spectra. Following publication the mzServer can support in silico “conversations” between the authors and interested third parties regarding virtually any aspect of the mass spectrometry data. Finally, it is worth noting that from a pedagogical perspective, the availability of large-scale datasets amenable to remote mass-informatics enables the development of educational curricula and research programs with essentially no investment in computational infrastructure beyond simple web-access. Although there will be inevitable latencies associated with network communication, they will be mitigated by increasingly powerful API primitives running server-side (spectral dot-products, statistics, spectral manipulations, etc.) and the universality of the resulting platform will constitute an ideal environment for the development and communication of mass-informatic demonstrations and prototypes.
Along with the significant opportunities described above, there are also real technological hurdles associated with the use of mzServer on a community-wide scale; chief among these is the difficulty in enabling secure, potentially anonymous scripting by researchers interested in the implementation of large-scale remote mass-informatics algorithms. However as with many projects ongoing in the IVOA we are confident that synergistic, multi-lab development efforts with the mzServer concept will refine a strategy to bring together data and computation, a challenge cogently expressed by the noted computer scientist Jim Gray (16), “In the future, working with large data sets will typically mean sending computations to the data, rather than copying the data to your work station. But the management of distributed computations raises new questions of security, free access to public data and cost. Few data archives address these issues today.”
Acknowledgments
We thank Eric Smith for valuable discussion and preparation of the figures.
Footnotes
* Generous support for this work was provided by the Dana-Farber Cancer Institute and the National Institutes of Health, NHGRI (P50HG004233), and NINDS (P01NS047572).
 This article contains supplemental Fig. S1.
This article contains supplemental Fig. S1.
1 The abbreviations used are:
- API
- Application Programming Interface
- URL
- Uniform Resource Locator
- HTML
- HyperText Markup Language
- REST
- Representational State Transfer
- SOAP
- Simple Object Access Protocol
- XIC
- Extracted Ion Chromatogram.
REFERENCES
- 1. Askenazi M., Parikh J. R., Marto J. A. (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat. Methods 6, 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cottingham K. (2009) MCP ups the ante by mandating raw-data deposition. J. Proteome Res. 8, 4887–4888 [DOI] [PubMed] [Google Scholar]
- 3. Chalkley R. J., Clauser K. R., Carr S. A. (2009) Updating the MCP proteomic publication guidelines. ASBMB Today, August, 16–18 [Google Scholar]
- 4. Falkner J., Andrews P. C. (2006) The DFS tool for dissemination and annotation of proteomics data. HUPO. 5th Annual World Congress p. 98, Long Beach, CA U.S.A [Google Scholar]
- 5. Mead J. A., Bianco L., Bessant C. (2009) Recent developments in public proteomic MS repositories and pipelines. Proteomics 9, 861–881 [DOI] [PubMed] [Google Scholar]
- 6. Mead J. A., Shadforth I. P., Bessant C. (2007) Public proteomic MS repositories and pipelines: available tools and biological applications. Proteomics 7, 2769–2786 [DOI] [PubMed] [Google Scholar]
- 7. Richardson L. (2007) RESTful web services, O'Reilly, Farnham [Google Scholar]
- 8. Abazajian K. N., et al. (2009) The Seventh Data Release of the Sloan Digital Sky Survey. Astrophys. J. Suppl. Series 182, 543–558 [Google Scholar]
- 9. Uose H. (2003) Application of ultrahigh-speed network to advanced science. NTT. Tech. Rev. 1, 39–47 [Google Scholar]
- 10. Shasharina S. G., Li C., Wang N., Pundaleeka R., Wade-Stein D. (2007) Distributed technologies for remote access of HDF data. IEEE, Enabling Technologies: Infrastructure for Collaborative Enterprises, 2007. WETICE 2007. 16th IEEE International Workshops on, 255–260 [Google Scholar]
- 11. Szalay A., Gray J. (2001) The world-wide telescope. Science 293, 2037–2040 [DOI] [PubMed] [Google Scholar]
- 12. Dobos L., Budavári T., Csabai I., Szalay A. S., Herczegh G. (2008) Improved search in spectrum databases. ASP Conference Series 394, 389–392 [Google Scholar]
- 13. Ficarro S. B., Zhang Y., Lu Y., Moghimi A. R., Askenazi M., Hyatt E., Smith E. D., Boyer L., Schlaeger T. M., Luckey C. J., Marto J. A. (2009) Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem. 81, 3440–3447 [DOI] [PubMed] [Google Scholar]
- 14. Askenazi M., Li S., Singh S., Marto J. A. (2010) Pathway Palette: A rich internet application for peptide-, protein- and network-oriented analysis of MS data. Proteomics 10, 1880–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parikh J. R., Askenazi M., Ficarro S. B., Cashorali T., Webber J. T., Blank N. C., Zhang Y., Marto J. A. (2009) multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10, 364–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Szalay A., Gray J. (2006) 2020 Computing: Science in an exponential world. Nature 440, 413–414 [DOI] [PubMed] [Google Scholar]