Abstract
Myocardial proteasomes are comprised of 20S core particles and 19S regulatory particles, which together carry out targeted degradation of cardiac proteins. The 19S complex is unique among the regulators of proteasomes in that it affects both the capacity and specificity of protein degradation. However, a comprehensive molecular characterization of cardiac 19S complexes is lacking. In this investigation, we tailored a multidimensional chromatography-based purification strategy to isolate structurally intact and functionally viable 19S complexes from murine hearts. Two distinct subpopulations of 19S complexes were isolated based upon (1) potency of activating 20S proteolytic activity, and (2) molecular composition using a combination of immuno-detection, two-dimensional-differential gel electrophoresis, and MS-based approaches. Heat shock protein 90 (Hsp90) was identified to be characteristic to 19S subpopulation I. The physical interaction of Hsp90 with 19S complexes was demonstrated via multiple approaches. Inhibition of Hsp90 activity using geldanamycin or BIIB021 potentiated the ability of subpopulation I to activate 20S proteasomes in the murine heart, thus demonstrating functional specificity of Hsp90 in subpopulation I. This investigation has advanced our understanding of the molecular heterogeneity of cardiac proteasomes by identifying molecularly and functionally distinct cardiac 19S complexes. The preferential association of Hsp90 with 19S subpopulation I unveils novel targets for designing proteasome-based therapeutic interventions for combating cardiac disease.
The proteasome system governs protein turnover and regulates essential signaling cascades in cardiac biology (1, 2). Despite the distinct etiologies of many cardiac diseases, proteasomes are among the final effectors of ventricular remodeling during heart failure (2, 3). A decline in proteasome function occurs gradually with aging (4) and acutely with ischemia-reperfusion injury (5, 6). On the contrary, elevated proteasome function has been observed in both hypertrophic hearts (7, 8) and atrophic, diabetic hearts (9, 10). These observations highlight the enormous complexity of proteasome regulation in the heart.
The functional dynamics of proteasomes are tuned coordinately by several mechanisms. The incorporation of inducible subunits in the core particle, the 20S proteasome, leads to durable alterations in proteolytic function (11, 12). Post-translational modifications of proteasome subunits provide instantaneous adaptation of proteasome activities to receptor stimulation or stress (13, 14). The dynamic association of proteasomes with regulatory proteins, known as associating partners, serves as a key mechanism by which proteasome activities are modulated. Additionally, distinct subtypes of proteasome complexes exhibit different susceptibilities to pharmacological agents, such as competitive inhibitors (15, 16). Taken together, delineating the heterogeneity of proteasome complexes will have profound biological and clinical significance (17, 18).
The 19S complex, also known as the regulatory particle, is among the first specific regulators identified for 20S proteasomes. The 19S complexes tether ubiquitination and proteolysis by governing substrate recruitment and accessibility to the proteolytic centers. Thus, the 19S complexes serve as important modulators of targeted protein degradation (14, 19, 20). However, to date, the functional and structural dynamics of 19S complexes remain poorly defined. The current model of 19S complex biology is established primarily based on 26S proteasomes (21–23), with limited appreciation and resolution of 19S complex heterogeneity.
We previously identified heterogeneous subpopulations of cardiac 20S proteasomes, and found that they exhibited unique functional capacities and molecular constituents (24). This suggests that the protein degradation capacity of cardiomyocytes is specifically tuned to meet physiologic demands through the specialization of proteasome populations. This study led us to further dissect the diversity of cardiac proteasomes by determining whether 19S regulatory particles are heterogeneous. However, difficulties in isolating 19S complexes that retained endogenous functionality posed a major hindrance, as 19S complexes are labile and bulky in nature (25). In cardiac muscle, this is further complicated by the relatively high expression level of sarcomeric myofibrillar proteins which associate with 19S complexes (21, 26).
In this study, we developed a purification strategy (25) for endogenous cardiac 19S complexes to overcome these challenges. Consequently, two distinct subpopulations of structurally intact and functionally viable 19S complexes (19S I and 19S II) were obtained from the murine cardiac tissue. Each subpopulation exhibited different regulatory potency in stimulating 20S proteasome-dependent proteolysis, with 19S I exhibiting lower potency than 19S II. The molecular basis for this functional disparity was interrogated using quantitative proteomic tools (27) to probe for regulators of 19S complexes. A unique molecular feature observed in 19S I was the specific recruitment of heat shock protein 90 (Hsp90)1, which was subsequently confirmed with independent immunoblotting assays as well as nondenaturing native polyacrylamide gel electrophoresis. Pharmacologic inhibition of 19S I-associated Hsp90 significantly enhanced the functional potency of 19S I, thus identifying Hsp90 as an important regulator of 19S complex activity in the heart.
Studies presented herein present the first reported strategy for isolation and purification of two distinct 19S complexes from murine hearts. With functional proteomic approaches, Hsp90 was found to be uniquely associated with a specific 19S subpopulation and importantly, to restrict the regulatory potency of this subpopulation in the heart. The heterogeneity of 19S complexes identified in this study indicated that 19S complexes functionally specialize to meet the demands of heart biology. This investigation advances our understanding of basic proteasome biology, and provides avenues by which to engineer novel strategies for targeted therapeutics.
EXPERIMENTIAL PROCEDURES
All procedures were performed in accordance with the Animal Research Committee guidelines at the University of California, Los Angeles and the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health.
Purification of Murine Cardiac 19S Complexes
A protocol (25) to effectively isolate 19S complexes from murine cardiac muscle was established via numerous pilot studies. Detailed procedures can be found in the supplemental materials (supplemental Fig. S2). Briefly, 300 murine hearts (ICR stain, male, 8–9 weeks old) were homogenized. Cytosolic fraction was collected by centrifugation and then prefractionated with DE52 resin (GE Healthcare). The fraction recovered with 400 mm NaCl was further enriched with ammonium sulfate precipitation (30–40% saturation) and dialyzed overnight at 4 °C. Low molecular weight proteins were then removed via Sephacryl S-400 gel-filtration (GE Healthcare). The 19S containing fractions were pooled and resolved with DEAE Sepharose Fast Flow chromatography (GE Healthcare). Excess salts were removed via ultrafiltration, and then Mono Q 5/50GL chromatography (GE Healthcare) was used to obtain purified 19S complexes.
The enrichment of 19S complexes was monitored by using immunoblotting (28). Antibodies against 19S complex subunits Rpt4, Rpn7, Rpn2 (Biomol, PA) and 20S proteasome subunit α3 were elected to assess the intactness of 19S complexes and to distinguish 19S complexes from 26S proteasomes.
High-Throughput 19S Complex Functional Assay
The function of 19S complexes was assessed by measuring the potency by which 19S complexes stimulated 20S proteasome-dependent proteolysis (29). Murine cardiac 19S complexes were titrated with buffer (20 mm Tris, pH 7.6, 1 mm MgCl2, 0.1 mm EDTA, 0.5 mm dithiotreitol (DTT), 10% Glycerol, 50 mm NaCl) and then incubated with purified murine cardiac 20S proteasomes in assay buffer (25 mm Tris, 10 mm MgCl2, 0.5 mm DTT, 200 μm ATP, pH 8.0) for 45min at 37 °C. A fluorophore-tagged substrate (Suc-LLVY-AMC, Bachem, CA) was subsequently applied to signal proteolytic activity; the reaction proceeded for 1 h at 37 °C. The release of the free AMC group was measured with a Fluoroskan Ascent Fluorometer (Thermo Fischer) (excitation: 390 nm; emission: 460 nm) (30). The proteasome-specific inhibitor, epoxomicin was employed in negative control. All activity assays were performed in solution with the 96-well micro-titer plates. 19S-Independent 20S proteasome activity was measured in the same plate in parallel, which was then subtracted from the 19S stimulated proteolytic activities.
Two-Dimensional Differential Gel Electrophoresis (2-D DIGE) Analysis of Cardiac 19S Complexes
2-D DIGE was conducted based on a previously published protocol (31). 19S I and 19S II were labeled with Cy3 and Cy5, respectively, and mixed. Labeled proteins were diluted in 350 μl IPG strip rehydration buffer (6 m Urea, 2 m Thiourea, 1% CHAPS, 1% Pharmalytes). An 18 cm, pH 3–11NL IPG strip (GE Healthcare) was run with the following 6-step program: 12h at 30 V (active rehydration), 2 h at 200 V, 2 h at 600 V, 2 h at 2000 V, 1 h gradient from 2000 V to 8000 V, and held at 8000 V until the total Vhr reached 15,000. The IPG strip was incubated first with 1% DTT in equilibration buffer (50 mm Tris, pH 8.8, 2% SDS, 6 m Urea, 30% Glycerol), and then with 2.5% iodoacetamide in equilibration buffer. For the second dimension, proteins were resolved by 12% SDS-PAGE at 65 V for 15 h. Gel images were acquired using a Typhoon 9410 imager (GE Healthcare). Three independent analyses were conducted. The gel was then silver-stained and the gel spots were excised, digested with trypsin (1:50, enzyme: protein; Promega, WI), and extracted for liquid chromatography tandem MS (LC-MS/MS) analysis.
Two-dimensional Nondenaturing Native PAGE/SDS-PAGE of Proteasome Complexes
Purified 19S complexes were mixed with native electrophoresis sample-loading buffer (20% glycerol, 0.2% bromphenol blue), incubated at 4 °C for 1 h, and resolved on a nondenaturing gel (4.0%T) at 60V for 10 h at 4 °C (32). The gel lane containing 19S complexes was excised and laid atop an SDS-PAGE gel (4.0%T stacking and 10%T resolving) for denaturing electrophoresis in the second dimension. Proteins were transferred to nitrocellulose membranes and probed with antibodies to Rpn2, Rpt4 (Biomol), and Hsp90 (Stressgen, San Diego, CA). Silver staining of a parallel set of gels illustrated the overall pattern of protein separation.
LC-MS/MS Characterization of Murine Cardiac 19S Complexes
LC-MS/MS of cardiac 19S complexes was performed on an LTQ-Orbitrap and an LTQ-XL (Thermo Fischer) mass spectrometers integrated with a C18, 300A, 5 μm nanoLC column (New Objective, MA). A linear gradient of 5% buffer B to 50% buffer B was applied over 180 min for effective peptide separation (Buffer A: 0.1% formic acid, 2% acetonitrile (ACN); buffer B: 0.1% formic acid, 80% ACN). The data-dependent acquisition was set as one full MS scan followed by MS2 fragmentation of the five most abundant ions. Spectra were searched against the IPI mouse database (v.3.47, 55,298 entries) using SEQUEST (Bioworks v.3.3.1 was used for the . dta generation, default setting). The search parameters were set as: trypsin digestion: partial; missed cleavages: 2; precursor tolerance: 20ppm/Orbitrap or 2Da/LTQ; fragment tolerance: 0.5 amu; fixed mods: C 57.0215 Da; variable mods: M 15.9949 Da and N terminus 210.1984 Da. Identified peptides were filtered: Xcorr > 2.7 (+2), 3.5 (+3), ΔCn > 0.1 (30, 33), then Scaffold (v.2.0) was used to ensure reported proteins were identified with a confidence greater than or equal to 99%.
18O Labeling-Assisted Quantitative LC-MS/MS Analysis
The postdigest peptide 18O-labeling procedure was performed as previously described (26). Briefly, immobilized trypsin beads (AB SCIEX, Foster City,CA) were washed with water and mixed with trypsin-digested peptides. The sample mixture was then dried using a Speedvac concentrator (Thermo Fischer). Either 18O-enriched water (95%, Sigma Aldrich) or regular 16O water containing 20% ACN were added to samples and incubated for 18 h at 37 °C with gentle agitation. The use of immobilized trypsin beads facilitates the removal of trypsin postlabeling; thus nonspecific exchange of C-terminal oxygen atoms postlabeling can be avoided. The removal of immobilized trypsin beads was achieved with a microspin column. The 16O or 18O labeled samples were then combined in preparation for analysis on an LTQ-Orbitrap. Three independent analyses were conducted.
Statistical Analyses
All data are presented as mean ± S.E. Student's t test was used for unpaired data analysis. p < 0.05 was considered statistically significant.
RESULTS
Molecular characterization of 19S complexes has been a challenging task, especially in mammals, largely because of the labile nature of these complexes. In this investigation, we implemented a multidimensional chromatography platform to extract 19S complexes in their native state from murine hearts. Heterogeneous subpopulations of 19S complexes were isolated, and each exhibited a distinct ability to activate purified 20S proteasomes. Hsp90 was identified as a unique associating partner of 19S subpopulation I, which we further demonstrated to be a negative regulator of 19S I-induced activation of 20S (supplemental Fig. S1).
Purification of Murine Cardiac 19S Proteasome Complexes
Significant efforts were made to extract structurally intact and functionally viable 19S complexes from the murine heart (supplemental Fig. S2) (25). The purification procedure included five key steps. The bulk of proteins in heart tissue homogenates were first removed by absorption with DE52 resin and selective precipitation with ammonium sulfate. 19S Complexes were collected in the fraction corresponding to 30–40% ammonium sulfate saturation. Further enrichment based on protein molecular mass and surface-to-charge ratio was conducted via size-exclusion chromatography and DEAE Sepharose chromatography, respectively. High resolution, analytical-scale MonoQ ion-exchange chromatography constituted the final refinement step for the purification of murine cardiac 19S complexes (Fig. 1A).
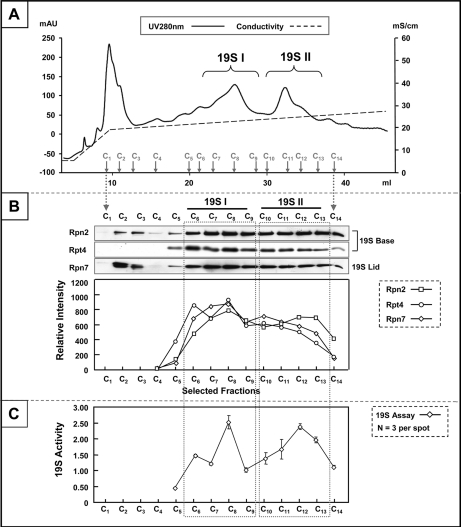
Fig. 1.
Purification of distinct 19S subpopulations from the murine heart. Panel. A, MonoQ chromatography was applied as the final step of murine 19S complex purification. With a slow ramping of salt concentration (shown in the form of conductivity: mS/cm), two distinct subpopulations of 19S complexes (19S I and 19S II) were collected (UV280: mAU). B, The structural integrity of 19S complexes was probed with a panel of antibodies targeting subunits from the “base” (Rpn2, Rpt4) and the “lid” (Rpn7) substructures. Both 19S species were shown to be structurally intact. Densitometry measurement showed a consistent enrichment pattern for 19S subunits in support of the UV280 nm absorption traces (A). C, The biological function of murine cardiac 19S complexes was evaluated as their ability to stimulate 20S proteasome-dependent proteolysis. With ATP as a necessary cofactor, the activity of 19S complexes in selected fractions (C5-C14) was assayed. The relative activities among these fractions were in agreement with both the UV280 nm profile (A) and densitometry trace (B). All analyses were made with equal volume from each chromatographic fraction.
Two distinguishable subpopulations of 19S complexes were collected from murine hearts, further referred to as 19S subpopulation I (19S I) and 19S subpopulation II (19S II) according to the sequential elution and UV 280 nm trace on the MonoQ chromatogram (Fig. 1A). The molecular identity as well as the structural intactness of 19S complexes in each subpopulation was first interrogated with immunoblotting. Each subpopulation contained components from both the 19S base and lid subcomplexes as evidenced by the detection of Rpn2, Rpt4, and Rpn7 (Fig. 1B). The possibility that 26S proteasomes constituted one of these subpopulations was ruled out with parallel immunoblotting of 20S proteasome subunit α3 (data not shown). Thus, the observed heterogeneity was attributed to the different constituents of 19S complexes. The relative abundance of conventional 19S complexes in each fraction was carefully evaluated via image densitometry measurement postimmunoblotting. The enrichment profiles of all test subunits, Rpn2, Rpt4, and Rpn7, were in agreement with UV 280 nm trace (Fig. 1B), supporting the existence of two distinct subpopulations of 19S complexes in the murine heart. With the four independent replicates analyzed, the relative abundance of 19S II over 19S I was 0.54 ± 0.08 (average ± standard deviation).
To further validate the structural intactness of 19S complexes, an unrestricted molecular characterization was conducted with LC-MS/MS. 19S Subunits identified using this approach were complimentary to immunoblotting experiments, including 19S base subunit Rpt6 (supplemental Fig. S3A) and 19S lid subunit Rpn12 (supplemental Fig. S3B). Collectively, all essential subunits of cardiac 19S complexes were identified in both subpopulations (Table I). In addition, the functional potency of 19S subpopulations, defined as the ability to stimulate 20S proteasome-mediated proteolysis in vitro, was assessed (Fig. 1C). The relative activities among individual fractions reflected profiles of the MonoQ UV 280 nm traces (Fig. 1A) and immunoblots (Fig. 1B), thus further supporting the existence of two distinct and heterogeneous subpopulations of murine cardiac 19S complexes.
Table I. Conventional subunits of cardiac 19S subpopulations identified by LC-MS/MS.
Proteomic characterization of cardiac 19S complexes. The molecular components of 19S I and 19S II were identified with an LC-MS/MS-based analysis after one dimensional SDS-PAGE separation. The number of unique peptides identified and sequence coverage percentage for each subunit are shown. The identified 19S subunits are labeled with both common nomenclature (32) and gene name.
| Subunit/Gene | MonoQ Peak I |
MonoQ Peak II |
||
|---|---|---|---|---|
| Unique Peptides | Coverage | Unique Peptides | Coverage | |
| 19S Proteasome ATPase Regulatory Subunits | ||||
| Rpt2/PSMC1 | 26 | 49.3% | 19 | 40.5% |
| Rpt1/PSMC2 | 24 | 51.7% | 30 | 56.2% |
| Rpt5/PSMC3 | 21 | 59.8% | 18 | 52.5% |
| Rpt3/PSMC4 | 26 | 51.7% | 16 | 45.6% |
| Rpt6/PSMC5 | 29 | 52.5% | 16 | 44.1% |
| Rpt4/PSMC6 | 24 | 54.5% | 16 | 43.4% |
| 19S Proteasome Non-ATPase Regulatory Subunits | ||||
| Rpn2/PSMD1 | 31 | 46.8% | 20 | 37.7% |
| Rpn1/PSMD2 | 31 | 46.5% | 28 | 47.3% |
| Rpn3/PSMD3 | 21 | 34.4% | 15 | 36.6% |
| Rpn10/PSMD4 | 18 | 44.3% | 12 | 41.7% |
| S5b/PSMD5 | 9 | 26.6% | 13 | 37.9% |
| Rpn7/PSMD6 | 21 | 52.7% | 15 | 47.8% |
| Rpn8/PSMD7 | 12 | 36.8% | 9 | 40.2% |
| Rpn12/PSMD8 | 6 | 37.4% | 4 | 19.8% |
| Rpn6/PSMD11 | 21 | 37.4% | 16 | 46.7% |
| Rpn5/PSMD12 | 31 | 43.1% | 17 | 43.4% |
| Rpn9/PSMD13 | 19 | 39.6% | 13 | 37.8% |
| Rpn11/PSMD14 | 5 | 31.3% | 7 | 40.3% |
| Rpn13/ADRM1 | 1 | 3.9% | 2 | 8.8% |
The Molecular Basis of the Functional Variance between 19S Subpopulations
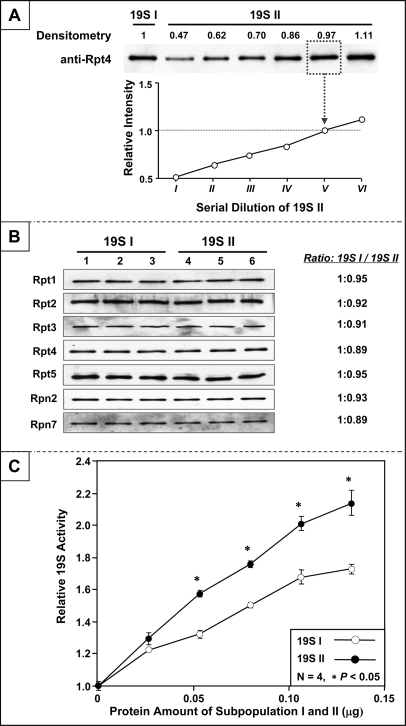
The two distinct 19S complex subpopulations were pooled separately and their functional potencies were characterized in parallel. The content of 19S complexes in each subpopulation was normalized based on results of quantitative immunoblotting using an antibody specific to 19S subunit Rpt4 (Fig. 2A). A standard curve was established with a serial dilution of 19S II to match the abundance of protein complex in 19S I. The accuracy of the normalization assay was verified with additional antibodies targeting Rpt1, Rpt2, Rpn2, and Rpn7, respectively (Fig. 2B). No significant differences in assembly stoichiometry of these subunits were observed between the two subpopulations.
Fig. 2.
Variation in functional dynamics between distinct 19S subpopulations. A, The content of 19S complexes in two pooled subpopulations was normalized based on the results of quantitative immunoblotting. The content of 19S II was titrated to match the content of 19S I. All subsequent functional comparisons were made following careful normalization of the protein. B, Postnormalization, the content of 19S complexes in the two normalized pools was verified with a panel of antibodies (Rpt1, Rpt2, Rpt3, Rpt4, Rpt5, Rpn2 and Rpn7) via quantitative immunoblotting. C, The biological potency of 19S complexes was evaluated as the ability of each to stimulate 20S proteasome-dependent proteolysis. 19S II showed significantly higher (up to 40%) activity over that of 19S I.
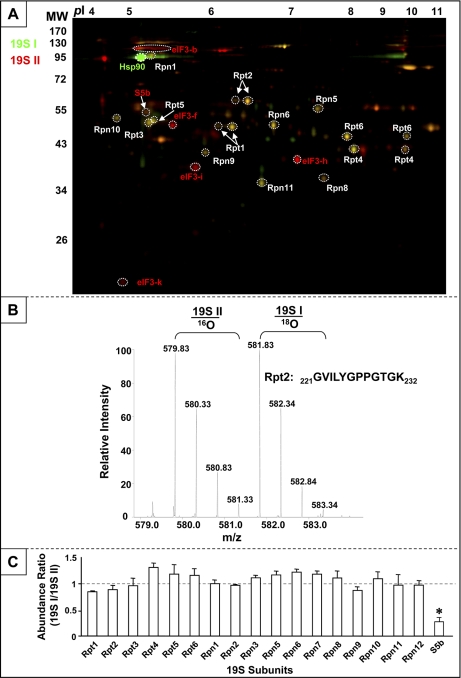
However, the two subpopulations of 19S complexes demonstrated distinct biological potency. Postnormalization, 19S II activated 20S proteasomes up to 40% greater than 19S I (Fig. 2C). Accordingly, the molecular basis for this functional variance was investigated with parallel quantitative proteomic platforms—2-D DIGE and 18O-labeling prior to LC-MS/MS. The 2-D DIGE approach showed similar molecular stoichiometries of conventional 19S subunits between subpopulations, with the exception of S5b (Fig. 3A). In previous reports, it has been suggested that S5b is an interacting partner, rather than a subunit of 19S complex (21, 34). In addition, Hsp90 was significantly enriched in 19S I, whereas eIF3 subunits were enriched in 19S II. An LC-MS/MS-based quantitative survey was conducted in parallel. Intact 19S complexes from each subpopulation were resolved on nondenaturing native PAGE gels and excised for proteomic analysis. Post-trypsin digestion, peptides derived from 19S I and 19S II were C-terminally labeled with 16O or 18O isotopes, respectively, and combined for quantitative LC-MS/MS using an LTQ-Orbitrap (Fig. 3B). In agreement with immunoblotting (Fig. 2B) and 2-D DIGE (Fig. 3A), quantitative LC-MS/MS revealed that S5b was the only conventional subunit differing between the two subpopulations (Fig. 3C). A complete list of 19S associating proteins consistently identified in both subpopulations is provided in supplemental Table S2.
Fig. 3.
Molecular composition of 19S subpopulations evaluated with quantitative proteomics. A, 2-D DIGE was applied for quantitative analysis of 19S proteasome heterogeneity. 19S I was labeled with Cy3 (green) and 19S II with Cy5 (red). The molecular identity of each spot was acquired via LC-MS/MS. Hsp90 was enriched in 19S I (green); S5b and eIF3 subunits were enriched in 19S II (red). B, The murine cardiac 19S complex subpopulations 19S I and 19S II were displayed by nondenaturing native PAGE, trypsin-digested and labeled with 18O/16O, respectively, at their C termini. The labeled peptides were then combined and quantitatively analyzed via LTQ-Orbitrap LC-MS/MS. C, The intensity ratio (19S I over 19S II) was indicative of relative protein abundance between the two subpopulations. S5b was the only conventional 19S subunit that was significantly different in abundance between the two subpopulations, with an enrichment in 19S II.
Hsp90 Is a Unique Functional Associating Partner of 19S Subpopulation I
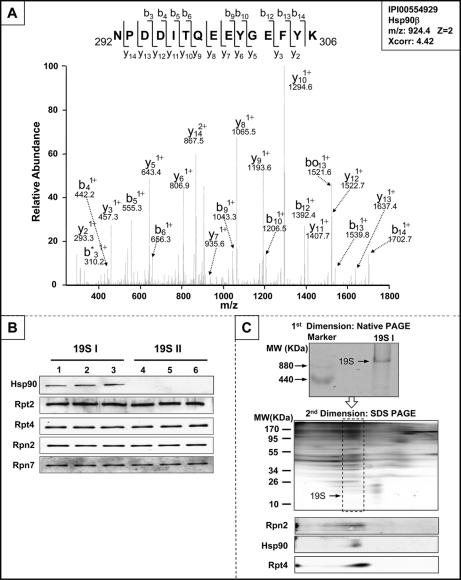
Hsp90 was identified to be a component of 19S I by native PAGE-LC-MS/MS (Fig. 4A). A total of eight distinct peptides derived from Hsp90 were identified in murine cardiac 19S I, exemplifying the substantial and preferential association of Hsp90 with 19S I complexes compared with 19S II (supplemental Fig. S4). Furthermore, immunoblotting analyses confirmed this unique molecular feature of 19S I (Fig. 4B).
Fig. 4.
Differential recruitment of Hsp90 contributes to cardiac 19S complex heterogeneity. A, The molecular identity of 19S-associated Hsp90 was confirmed with LC-MS/MS following nondenaturing native PAGE. The spectrum for Hsp90 peptide 292NPDDITQEEYGEFYK306 was shown. B, Quantitative immunoblotting assays with Hsp90 specific antibodies demonstrated that Hsp90 was unique to subpopulation 19S I. The relative contents of 19S subunits Rpt2, Rpt4, Rpn2 and Rpn7 were also probed as loading controls. C, Two-dimensional native PAGE and SDS-PAGE was performed for 19S I, confirming the interaction of Hsp90 with 19S I under a native, nondenaturing state.
Hsp90 is a molecular chaperone known to interact with 20S, 26S, and 11S proteasomes during assembly or exposure to oxidative stress (35–37). However, the direct association of Hsp90 with 19S complexes has not been previously reported. Using 2-D nondenaturing native-PAGE/SDS-PAGE coupled with immunoblotting, we demonstrated a specific interaction between Hsp90 with 19S I from murine hearts (Fig. 4C).
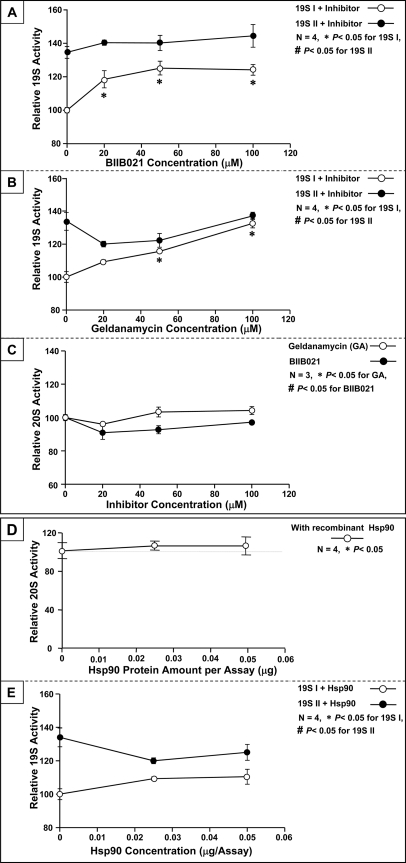
The functional impact of Hsp90 on the ability of 19S complexes to activate 20S proteasomes was examined in vitro. Because the depletion of Hsp90 from 19S I under native conditions was not feasible, we employed specific inhibitor of Hsp90 with distinct structure, BIIB021 and geldanamycin, to evaluate the influence of Hsp90 on 19S I function (Figs. 5A and 5B). Inhibition of Hsp90 led to a significant enhancement in 19S I activating potential of 20S, thus identifying Hsp90 as a negative regulator of 19S I complexes in the heart. Neither HSP90 inhibitor significantly affected the functional potency of 19S II. In addition, neither BIIB021 nor geldanamycin affects 20S activity significantly (Fig. 5C). HSP90 have been previously shown to interact with 20S proteasomes; in this assay, Hsp90 alone did not significantly affect 20S proteolytic activity (Fig. 5D); thereby confirming 19S I complexes as specific regulatory targets of Hsp90. Additionally, in vitro treatment of 19S I or 19S II with recombinant Hsp90 did not imitate a significant functional changes in 19S activities (Fig. 5E). This observation suggested 19S I contains sufficient endogenous Hsp90 and 19S population II may have adapted a conformation rejecting the association of HSP90. With the addition of recombinant Hsp90, the activity of 19S I and 19S II are still clearly distinct.
Fig. 5.
Hsp90 Suppressed the biological potency of 19S subpopulation I. A, The functional impact of Hsp90 on 19S complexes was evaluated with an Hsp90 inhibitor, BIIB021. Inhibition of 19S I-associated Hsp90 significantly enhanced the activating potency of 19S I on 20S-dependent proteolytic activity, indicating that Hsp90 is a negative regulator of 19S I (n = 4). B, The functional impact of Hsp90 on 19S complexes was validated with another Hsp90 inhibitor, geldanamycin, supporting Hsp90 as a specific negative regulator of 19S I (n = 4). C, Neither BIIB021 nor geldanamycin impacts 20S activity significantly (n = 3). D, As another negative control, the functional impact of Hsp90 on 20S proteasomes alone (without 19S) was examined. The proteolytic function of 20S proteasomes was not inhibited with exogenous Hsp90, suggesting that the Hsp90-induced functional inhibition of 20S proteolysis was transduced through 19S (n = 4). E, In vitro treatment with recombinant Hsp90 did not significantly alter the function of 19S I or 19S II (n = 4).
DISCUSSION
The 19S complex is the first recognized as well as the most intricate regulator of the proteasome system. A major proportion of 19S complexes interacts with 20S proteasomes, serving to recruit and catalyze targeted degradation of protein substrate repertoires (38). In addition, 19S complexes function independently from 20S proteasomes as molecular chaperones (39) or regulators of post-translational modifications (40). Our current knowledge of 19S complexes is primarily derived from studies of 26S proteasomes, which may have limited our appreciation of 19S heterogeneity and specialization within cardiac cells.
Purification of Murine Cardiac 19S Complexes
The labile nature of 19S complexes and their association with abundant myofibrillar proteins (21, 26) necessitate a delicate protocol for effective isolation. With significant efforts invested in preliminary experiments, we adjusted the purification procedure to accommodate the unique characteristics of cardiac tissue (25, 41). In the salt precipitation step, the window of ammonium sulfate concentration was narrowed from 0–40% to 30–40%. Together with DE52 resin absorption, the majority of contaminant proteins were efficiently removed from heart tissue homogenates. ATP was added as a supplement in between isolation steps, resulting in considerable stabilization of labile 19S complexes. High resolution MonoQ ion-exchange chromatography was applied to replace hydroxyapetite chromatography as the final step of purity refinement (25). MonoQ chromatography exhibited superior sensitivity, reproducibility and resolution for purifying murine cardiac 19S complexes. An even deeper delineation of 19S heterogeneity may be possible with additional fractionation technology.
The 19S complex is configured jointly with two substructures, the “base” and the “lid” (38, 42). A panel of antibodies specific to three distinct 19S subunits; two from the “base” (Rpn2 and Rpt4) and one from the “lid” (Rpn7), was formulated to track the correlation between these substructures along the purification procedure. Nondenaturing native electrophoresis, LC-MS/MS, and enzymatic activity assays were conducted in parallel to thoroughly interrogate the intactness of purified murine cardiac 19S complexes.
Heterogeneity of Murine Cardiac 19S Complexes
The functional dynamics of proteasome complexes, in general, vary from tissue to tissue (26, 43). In the heart, there exist an array of heterogeneous proteasome species with distinct activities, which collectively may direct targeted protein homeostasis (15, 24). The variation in molecular assembly leads to differences in biochemical activities (14, 44) as well as susceptibility to oxidative stress (45) and inhibitors (15). However, the diversity of 19S complexes and its molecular basis until now were poorly defined. This investigation reported a novel, large-scale chromatographic purification of cardiac 19S complexes, which enabled molecular and enzymatic characterization in a systematic fashion.
Heterogeneity of 19S complexes was first suggested from studies demonstrating post-translational modifications on 19S subunits (20, 21, 44, 46). In addition, 19S complexes from bovine erythrocytes were distinguished into two subpopulations with hydroxyapatite chromatography (41). Erythrocyte 19S complexes from the second-eluting subpopulation exhibited a lower biological activity than the first one; however the molecular basis underlying this difference was not elucidated. In this investigation, we isolated two distinct 19S subpopulations from the murine heart using MonoQ chromatography. Contrary to erythrocyte 19S complexes, the later-eluting subpopulation of cardiac 19S complexes (19S II) showed a higher activating capacity. One possible reason for this discrepancy is that the separation kinetic of 19S complexes via MonoQ chromatography is quite distinct from that of hydroxyapatite chromatography. Another feasible scenario is that cardiac 19S complexes exhibit significant differences in functional dynamics or molecular features from that of erythrocytes.
In this study, we took a functional proteomic approach to characterize the molecular features leading to functional heterogeneity of 19S complexes in the heart. Unique molecular constituents were observed via quantitative proteomic approaches. Among them, Hsp90 was identified to be a functional associating partner of 19S I, but not 19S II, murine cardiac complexes, and the presence of active Hsp90 significantly suppressed the functional potency of 19S I. HSP90 may influence the function of 19S in many ways. It may reduce the binding affinity between 19S and 20S proteasomes or hinder the passage of substrates toward the proteolytic center. The ATPase activities within 19S complexes may be the regulatory targets as well. Eukaryotic initiation factor 3 (eIF3) was also found to be markedly different between 19S subpopulations in 2-D DIGE analysis. This may reflect the specialization of this subpopulation for a role in protein translation (47). Experimental protocols are being implemented for further analyses of the role of eIF3 and other unique molecular features to offer additional insights toward the diverse function of cardiac 19S complexes. S5b was found to be dominantly associated with 19S II. S5b has been previously shown to assist in 19S assembly, and it also maintains a functional affiliation with mature 19S complexes (48). Our data supports the notion that S5b is interacting with mature proteasome complexes in the heart, as 19S II was not constituted with assembly intermediates. Rather, quantitative assays suggested that 19S I and 19S II have matching subunit stoichiometry (Figs. 3A and 3C). Additionally, 19S II has full functionality, which further supports that this population is mature in nature.
Implications of 19S Heterogeneity in Cardiovascular Biology
The proteasome is more than just a passive protein degradation machine. A dysregulation in proteasome function has been recognized in many forms of cardiomyopathy, including ischemia-reperfusion injury (5), hypertrophy (7), myocarditis (11), and diabetic cardiomyopathy (10). Applications of pharmacological reagents to inhibit (49, 50) or preserve proteasome function (51) have shown promising benefits in combating cardiomyopathy. However, prolonged application of high-dose proteasome inhibitors may also induce detrimental side-effects on the heart (52). Therefore, it is important to engineer targeted intervention strategies with precise specificity.
The identification and characterization of heterogeneity in the proteasome system provides knowledge for achieving such goals. Distinct populations of 19S complexes confer different functional states; the dynamic balance among them collectively defines protein degradation. The characterization of this diversity at the molecular level may foster targeted intervention strategies. The various species of proteasome complexes demonstrated distinct properties in functional dynamics (53), resistance to oxidative damage (45, 54) and susceptibility to pharmacological agents (15). Taking advantage of the distinction in molecular properties, it is feasible to develop agents which target specific subpopulations of proteasomes (55). Recently, pharmacological agents aiming at 19S complexes have also emerged (56, 57), which may enable selective adjustment of targeting ubiquitinated proteins for degradation. The comprehensive delineation of the heterogeneity in cardiac 19S subpopulations will aid in the engineering of a new generation of agents with enhanced specificity.
Footnotes
* This work was supported, in part, by NHLBI Proteomics Center Award (HHSN268201000035C), R01 HL098954, and a Theodore C. Laubisch endorsement at UCLA to Dr. Peipei Ping, as well as the R01 HL088640 to Dr. Enrico Stefani.
 This article contains supplemental material, Figs. S1 to S4, and Tables S1 and S2.
This article contains supplemental material, Figs. S1 to S4, and Tables S1 and S2.
1 The abbreviations used are:
- Hsp90
- heat shock protein 90
- 20S
- proteasome proteolytic core particle
- 19S
- proteasome regulatory particle
- 2-D DIGE
- two-dimensional differential gel electrophoresis
- LC-MS/MS
- liquid chromatography tandem MS.
REFERENCES
- 1. Wang X., Su H., Ranek M. J. (2008) Protein quality control and degradation in cardiomyocytes. J. Mol. Cell Cardiol. 45, 11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patterson C., Ike C., Willis P. W., 4th, Stouffer G. A., Willis M. S. (2007) The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation 115, 1456–1463 [DOI] [PubMed] [Google Scholar]
- 3. Gomes A. V., Zong C., Edmondson R. D., Berhane B. T., Wang G. W., Le S., Young G., Zhang J., Vondriska T. M., Whitelegge J. P., Jones R. C., Joshua I. G., Thyparambil S., Pantaleon D., Qiao J., Loo J., Ping P. (2005) The murine cardiac 26S proteasome: an organelle awaiting exploration. Ann. N.Y. Acad. Sci. 1047, 197–207 [DOI] [PubMed] [Google Scholar]
- 4. Bulteau A. L., Szweda L. I., Friguet B. (2002) Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 397, 298–304 [DOI] [PubMed] [Google Scholar]
- 5. Bulteau A. L., Lundberg K. C., Humphries K. M., Sadek H. A., Szweda P. A., Friguet B., Szweda L. I. (2001) Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J. Biol. Chem. 276, 30057–30063 [DOI] [PubMed] [Google Scholar]
- 6. Das S., Powell S. R., Wang P., Divald A., Nesaretnam K., Tosaki A., Cordis G. A., Maulik N., Das D. K. (2005) Cardioprotection with palm tocotrienol: antioxidant activity of tocotrienol is linked with its ability to stabilize proteasomes. Am. J. Physiol. Heart Circ. Physiol. 289, H361–367 [DOI] [PubMed] [Google Scholar]
- 7. Depre C., Wang Q., Yan L., Hedhli N., Peter P., Chen L., Hong C., Hittinger L., Ghaleh B., Sadoshima J., Vatner D. E., Vatner S. F., Madura K. (2006) Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation 114, 1821–1828 [DOI] [PubMed] [Google Scholar]
- 8. Hedhli N., Lizano P., Hong C., Fritzky L. F., Dhar S. K., Liu H., Tian Y., Gao S., Madura K., Vatner S. F., Depre C. (2008) Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am. J. Physiol. Heart Circ. Physiol. 295, H1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zu L., Bedja D., Fox-Talbot K., Gabrielson K. L., Van Kaer L., Becker L. C., Cai Z. P. (2010) Evidence for a role of immunoproteasomes in regulating cardiac muscle mass in diabetic mice. J. Mol. Cell Cardiol. 49, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu J., Klein J. D., Du J., Wang X. H. (2008) Cardiac muscle protein catabolism in diabetes mellitus: activation of the ubiquitin-proteasome system by insulin deficiency. Endocrinology 149, 5384–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szalay G., Meiners S., Voigt A., Lauber J., Spieth C., Speer N., Sauter M., Kuckelkorn U., Zell A., Klingel K., Stangl K., Kandolf R. (2006) Ongoing coxsackievirus myocarditis is associated with increased formation and activity of myocardial immunoproteasomes. Am. J. Pathol. 168, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marfella R., Di Filippo C., Portoghese M., Siniscalchi M., Martis S., Ferraraccio F., Guastafierro S., Nicoletti G., Barbieri M., Coppola A., Rossi F., Paolisso G., D'Amico M. (2009) The ubiquitin-proteasome system contributes to the inflammatory injury in ischemic diabetic myocardium: the role of glycemic control. Cardiovasc. Pathol. 18, 332–345 [DOI] [PubMed] [Google Scholar]
- 13. Asai M., Tsukamoto O., Minamino T., Asanuma H., Fujita M., Asano Y., Takahama H., Sasaki H., Higo S., Asakura M., Takashima S., Hori M., Kitakaze M. (2009) PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J. Mol. Cell Cardiol. 46, 452–462 [DOI] [PubMed] [Google Scholar]
- 14. Zachara N. E., Hart G. W. (2004) O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell Biol. 14, 218–221 [DOI] [PubMed] [Google Scholar]
- 15. Kloss A., Meiners S., Ludwig A., Dahlmann B. (2010) Multiple cardiac proteasome subtypes differ in their susceptibility to proteasome inhibitors. Cardiovasc. Res. 85, 367–375 [DOI] [PubMed] [Google Scholar]
- 16. Busse A., Kraus M., Na I. K., Rietz A., Scheibenbogen C., Driessen C., Blau I. W., Thiel E., Keilholz U. (2008) Sensitivity of tumor cells to proteasome inhibitors is associated with expression levels and composition of proteasome subunits. Cancer 112, 659–670 [DOI] [PubMed] [Google Scholar]
- 17. Husom A. D., Peters E. A., Kolling E. A., Fugere N. A., Thompson L. V., Ferrington D. A. (2004) Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 421, 67–76 [DOI] [PubMed] [Google Scholar]
- 18. Dahlmann B., Ruppert T., Kuehn L., Merforth S., Kloetzel P. M. (2000) Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J. Mol. Biol. 303, 643–653 [DOI] [PubMed] [Google Scholar]
- 19. Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 20. Satoh K., Sasajima H., Nyoumura K. I., Yokosawa H., Sawada H. (2001) Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry 40, 314–319 [DOI] [PubMed] [Google Scholar]
- 21. Gomes A. V., Zong C., Edmondson R. D., Li X., Stefani E., Zhang J., Jones R. C., Thyparambil S., Wang G. W., Qiao X., Bardag-Gorce F., Ping P. (2006) Mapping the murine cardiac 26S proteasome complexes. Circ. Res. 99, 362–371 [DOI] [PubMed] [Google Scholar]
- 22. Wang X., Chen C. F., Baker P. R., Chen P. L., Kaiser P., Huang L. (2007) Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46, 3553–3565 [DOI] [PubMed] [Google Scholar]
- 23. Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drews O., Wildgruber R., Zong C., Sukop U., Nissum M., Weber G., Gomes A. V., Ping P. (2007) Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol. Cell Proteomics 6, 2021–2031 [DOI] [PubMed] [Google Scholar]
- 25. DeMartino G. N. (2005) Purification of PA700, the 19S regulatory complex of the 26S proteasome. Methods Enzymol. 398, 295–306 [DOI] [PubMed] [Google Scholar]
- 26. Gomes A. V., Young G. W., Wang Y., Zong C., Eghbali M., Drews O., Lu H., Stefani E., Ping P. (2009) Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Mol. Cell Proteomics 8, 302–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berhane B. T., Zong C., Liem D. A., Huang A., Le S., Edmondson R. D., Jones R. C., Qiao X., Whitelegge J. P., Ping P., Vondriska T. M. (2005) Cardiovascular-related proteins identified in human plasma by the HUPO Plasma Proteome Project pilot phase. Proteomics 5, 3520–3530 [DOI] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. Leggett D. S., Glickman M. H., Finley D. (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 [DOI] [PubMed] [Google Scholar]
- 30. Zong C., Gomes A. V., Drews O., Li X., Young G. W., Berhane B., Qiao X., French S. W., Bardag-Gorce F., Ping P. (2006) Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ. Res. 99, 372–380 [DOI] [PubMed] [Google Scholar]
- 31. Scruggs S. B., Hinken A. C., Thawornkaiwong A., Robbins J., Walker L. A., de Tombe P. P., Geenen D. L., Buttrick P. M., Solaro R. J. (2009) Ablation of Ventricular Myosin Regulatory Light Chain Phosphorylation in Mice Causes Cardiac Dysfunction In Situ and Affects Neighboring Myofilament Protein Phosphorylation. J. Biol. Chem. 284, 5097–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glickman M. H., Rubin D. M., Fried V. A., Finley D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu H., Zong C., Wang Y., Young G. W., Deng N., Souda P., Li X., Whitelegge J., Drews O., Yang P. Y., Ping P. (2008) Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Mol. Cell Proteomics 7, 2073–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guerrero C., Tagwerker C., Kaiser P., Huang L. (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 35. Imai J., Maruya M., Yashiroda H., Yahara I., Tanaka K. (2003) The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 22, 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conconi M., Djavadi-Ohaniance L., Uerkvitz W., Hendil K. B., Friguet B. (1999) Conformational changes in the 20S proteasome upon macromolecular ligand binding analyzed with monoclonal antibodies. Arch. Biochem. Biophys. 362, 325–328 [DOI] [PubMed] [Google Scholar]
- 37. Whittier J. E., Xiong Y., Rechsteiner M. C., Squier T. C. (2004) Hsp90 enhances degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 279, 46135–46142 [DOI] [PubMed] [Google Scholar]
- 38. Coux O., Tanaka K., Goldberg A. L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 39. Liu C. W., Strickland E., Demartino G. N., Thomas P. J. (2005) Recognition and processing of misfolded proteins by PA700, the 19S regulatory complex of the 26S proteasome. Methods Mol. Biol. 301, 71–81 [DOI] [PubMed] [Google Scholar]
- 40. Koues O. I., Dudley R. K., Mehta N. T., Greer S. F. (2009) The 19S proteasome positively regulates histone methylation at cytokine inducible genes. Biochim. Biophys. Acta 1789, 691–701 [DOI] [PubMed] [Google Scholar]
- 41. Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. (1994) Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20 S proteasome. J. Biol. Chem. 269, 3539–3547 [PubMed] [Google Scholar]
- 42. Yoshimura T., Kameyama K., Takagi T., Ikai A., Tokunaga F., Koide T., Tanahashi N., Tamura T., Cejka Z., Baumeister W., et al. (1993) Molecular characterization of the “26S” proteasome complex from rat liver. J. Struct. Biol. 111, 200–211 [DOI] [PubMed] [Google Scholar]
- 43. Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. (2008) Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell Proteomics 7, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Isasa M., Katz E. J., Kim W., Yugo V., González S., Kirkpatrick D. S., Thomson T. M., Finley D., Gygi S. P., Crosas B. (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 38, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gurusamy N., Goswami S., Malik G., Das D. K. (2008) Oxidative injury induces selective rather than global inhibition of proteasomal activity. J. Mol. Cell Cardiol. 44, 419–428 [DOI] [PubMed] [Google Scholar]
- 46. Ullrich O., Reinheckel T., Sitte N., Hass R., Grune T., Davies K. J. (1999) Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc. Natl. Acad. Sci. U.S.A. 96, 6223–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baugh J. M., Pilipenko E. V. (2004) 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell 16, 575–586 [DOI] [PubMed] [Google Scholar]
- 48. Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 49. Pye J., Ardeshirpour F., McCain A., Bellinger D. A., Merricks E., Adams J., Elliott P. J., Pien C., Fischer T. H., Baldwin A. S., Jr., Nichols T. C. (2003) Proteasome inhibition ablates activation of NF-kappa B in myocardial reperfusion and reduces reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 284, H919–926 [DOI] [PubMed] [Google Scholar]
- 50. Meiners S., Dreger H., Fechner M., Bieler S., Rother W., Günther C., Baumann G., Stangl V., Stangl K. (2008) Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension 51, 302–308 [DOI] [PubMed] [Google Scholar]
- 51. Das S., Lekli I., Das M., Szabo G., Varadi J., Juhasz B., Bak I., Nesaretam K., Tosaki A., Powell S. R., Das D. K. (2008) Cardioprotection with palm oil tocotrienols: comparision of different isomers. Am. J. Physiol. Heart Circ. Physiol. 294, H970–978 [DOI] [PubMed] [Google Scholar]
- 52. Voortman J., Giaccone G. (2006) Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer 6, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cai Z. P., Shen Z., Van Kaer L., Becker L. C. (2008) Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. FASEB J. 22, 4248–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Farout L., Mary J., Vinh J., Szweda L. I., Friguet B. (2006) Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch. Biochem. Biophys. 453, 135–142 [DOI] [PubMed] [Google Scholar]
- 55. Ho Y. K., Bargagna-Mohan P., Wehenkel M., Mohan R., Kim K. B. (2007) LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem. Biol. 14, 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lim H. S., Archer C. T., Kim Y. C., Hutchens T., Kodadek T. (2008) Rapid identification of the pharmacophore in a peptoid inhibitor of the proteasome regulatory particle. Chem. Commun. 1064–1066 [DOI] [PubMed] [Google Scholar]
- 57. Lim H. S., Archer C. T., Kodadek T. (2007) Identification of a peptoid inhibitor of the proteasome 19S regulatory particle. J. Am. Chem. Soc. 129, 7750–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]