Abstract
Beam-type collisional activation dissociation (HCD) offers many advantages over resonant excitation collision-activated dissociation, including improved identification of phosphorylated peptides and compatibility with isobaric tag-based quantitation (e.g. tandem mass tag (TMT) and iTRAQ). However, HCD typically requires specially designed and dedicated collision cells. Here we demonstrate that HCD can be performed in the ion injection pathway of a mass spectrometer with a standard atmospheric inlet (iHCD). Testing this method on complex peptide mixtures revealed similar identification rates to collision-activated dissociation (2883 versus 2730 IDs for iHCD/CAD, respectively) and precursor-product-conversion efficiency comparable to that achieved within a dedicated collision cell. Compared with pulsed-q dissociation, a quadrupole ion trap-based method that retains low-mass isobaric tag reporter ions, iHCD yielded isobaric tag for relative and absolute quantification reporter ions 10-fold more intense. This method involves no additional hardware and can theoretically be implemented on any mass spectrometer with an atmospheric inlet.
Beam-type collisional dissociation (HCD)1 is the primary means to effect vibronic dissociation. The approach involves energetic injection of selected precursor cations into a collision cell filled with inert gas (>1 mTorr). These dedicated collision cells are central components of numerous hybrid MS systems, e.g. Q-TOF, QqQ, Orbitrap, etc. Ion trap MS systems, however, accomplish collisional activation via resonant excitation of a trapped precursor population (ion trap collision-activated dissociation (CAD)). Rather than imparting one or two higher-energy collisions, as in HCD, CAD slowly heats precursor ions with hundreds of low energy collisions. The result is a slightly different product ion distribution (from HCD), which lacks low m/z products because of a low-mass cutoff imposed by the magnitude of the radio frequency (RF) trapping field. Yet the ubiquity of ion traps has made CAD a widely used dissociation method (1); it has resulted in some of the most comprehensive proteomic analyses to date, including the identification of the entire yeast proteome and over 30,000 phosphorylation sites in mice (2, 3).
The recent introduction of quadrupole linear ion trap (QLT) hybrids with dedicated collision cells has allowed a direct comparison of CAD to HCD (4–7). As it is able to access higher-energy fragmentation channels, HCD results in more peptide identifications than CAD, particularly for phosphorylated peptides (4, 8). Beyond that, HCD offers several advantages. First, HCD is not subject to the low-mass cutoff inherent to CAD, and is therefore compatible with isobaric tagging strategies for multiplexed quantitation (e.g. isobaric tag for relative and absolute quantification (iTRAQ) and TMT) (9–11). Second, acquisition of collision-based dissociation spectra in a standard form would greatly facilitate cross-platform comparison and data transferability. For example, downstream selected reaction monitoring experiments, which utilize HCD (i.e. QqQ), depend on prior knowledge of precursor-to-product transitions from upstream discovery data (12, 13). Third, HCD generates immonium ions and other secondary fragments that can aid both sequence determination and post-translational modification detection (14).
The dedicated collision cell required to implement HCD, however, increases instrument complexity and cost, especially for standalone QLT systems. Here we demonstrate that HCD can be performed both efficiently and quickly in the pre-existing atmospheric inlet region (inlet HCD or iHCD). iHCD requires no hardware modification and brings HCD capability to any MS system having an atmospheric pressure (AP) inlet. Typical MS operation involves ion transmission from an AP inlet to a mass analyzer, a QLT in our case, using a collection of electrostatic and electrodynamic ion optics and quadrupole ion guides (15). These elements pass ions through differentially pumped vacuum regions, the high pressure sections (>1 mTorr) of which can provide an in situ collision cell. Following injection and isolation, the resulting precursor population can be transmitted back along the ion injection pathway with kinetic energy sufficient to induce dissociation. The products of these collisions, which are trapped in that space, can be either injected into the primary mass analyzer (e.g. QLT) or sent on to a secondary analyzer (e.g. orbitrap) (16, 17).
Tandem mass spectra acquired using iHCD are highly similar to MS/MS spectra acquired with a dedicated collision cell. Comparisons to existing methods revealed that iHCD identified as many or more peptides than CAD, pulsed-Q dissociation (PQD), or HCD. iHCD is robust, sensitive, and compatible with any MS system having an AP inlet.
EXPERIMENTAL PROCEDURES
Cell Growth and Lysis
Human embryonic stem cells (line H1) were maintained in a feeder-independent system, as previously described (18). Upon reaching 70% confluency, cells were passaged enzymatically using dispase (Invitrogen, Carlsbad, CA) at a 1:4 splitting ratio. Cells were harvested by individualizing for 10 min with an adequate volume of prewarmed (37 °C), 0.05% TrypLE (Invitrogen, Carlsbad, CA) to cover the culture surface. Following cell detachment, an equivalent volume of ice-cold Dulbecco's phosphate-buffered saline (DPBS) (Invitrogen, Carlsbad, CA) was added before collecting the cells. Cell pellets were subsequently washed twice in ice-cold phosphate-buffered saline and stored at −80 °C. Approximately 108 cells were collected for each analysis.
Samples were lysed via sonication in lysis buffer containing 40 mm NaCl, 50 mm Tris, 2 mm MgCl2, 50 mm NaF, 50 mm b-glyceradelhyde phosphate, 1 mm sodium orthovanadate, 10 mm sodium pyrophosphate, 1 mini EDTA-free protease inhibitor (Roche Diagnostics, Indianapolis, IN), and 1 phosSTOP phosphatase inhibitor (Roche Diagnostics, Indianapolis, IN).
Digestion and iTRAQ Labeling
Cysteine residues were reduced with dithiotreitol, alkylated using iodoacetamide, and digested in a two-step process. Proteinase Lys-C (Wako Chemicals, Richmond, VA) was added (enzyme:protein ratio = 1:100) and incubated for ∼2 h at 37 °C in lysis buffer. Samples were then diluted with 50 mm Tris pH 8 until the urea concentration was 1.5 m and digested with trypsin (Promega, Madison, WI) (enzyme:protein ratio = 1:50) at 37 °C overnight. Reactions were quenched using trifluoroacetic acid. Samples were dried to completion and purified using C18 solid phase extraction columns (SepPak, Waters, Milford, MA). iTRAQ labeling was performed according to manufacturer-supplied protocols. Once mixed, samples were dried to completion and purified by solid phase extraction.
Mass Spectrometry
Experiments were performed on three systems: LTQ, LTQ-Orbitrap, LTQ-Orbitrap XL (Thermo Fisher Scientific, San Jose, CA). Fig. 1 depicts the scan cycle. In brief, following injection and isolation of a particular precursor m/z peak (see Fig. 1A), the isolated precursor population is transmitted back along the ion injection pathway with a high degree of kinetic energy (see Fig. 1B). As the ions pass into the higher pressure regions, near the AP inlet, the ions collide with the neutral gas molecules leaking in from the atmospheric source. The precursor ions collide with the background gas molecules and fragment. The products are then trapped. Following fragmentation and trapping, product ions can either be further processed or mass analyzed (see Figs. 1C and 1D). All the associated voltages and times for this scan cycle were optimized prior to any large-scale analysis (vide infra). In these experiments, we fragmented precursor ions and trapped the resulting products in quadrupole 0 (Q0).
Fig. 1.
Instrument diagram depicting the typical events of an iHCD scan with QLT m/z analysis. Following injection and isolation of the precursor ion population (A), precursor ions are transmitted back along the ion injection pathway with a high degree of kinetic energy (B). As the ions pass into the higher-pressure regions located near the atmospheric pressure inlet, the ions collide with the neutral gas molecules leaking in from the AP inducing beam-type CAD. Generated products, trapped in that space, are then injected (C) into the QLT for m/z analysis (D).
Liquid Chromatography-Tandem MS (LC-MS/MS) Analysis
LC-MS/MS analysis was performed using a NanoAcquity UPLC system (Waters, Milford, MA) coupled to a LTQ-orbitrap XL (Thermo Fisher Scientific, San Jose, CA). Peptide samples were loaded onto a precolumn (75 μm ID, packed with 5 cm C18 particles (Alltech, Deerfield, IL)) for 10 min at a flow rate of 1 μl/min. Peptides were then eluted from a reversed-phase LC column (50 μm ID, packed with 15 cm C18 particles (Alltech)) with a 120 min linear gradient from 1% to 35% acetonitrile (0.2% formic acid) at a flow rate of 300 nL/min. An additional 30 min were used for column washing and equilibration. The column making procedure was previously described (5).
MS1 spectra were acquired at a resolving power of 60,000 and an automated gain control target of 1,000,000. Following MS1 analysis, the 10 most intense precursors were selected for data-dependent activation using iHCD. Precursors having either unassigned or <2 charge states were rejected. A 60 s dynamic exclusion window was employed (repeat count of 1). MS/MS AGC targets were set to 40,000.
Database Search and False Discovery Rate (FDR) Analysis
The resulting data files were searched using the Open Mass Spectrometry Search Algorithm (OMSSA) version 2.1.4 (19). Pre- and postsearch processing was performed using the Coon OMSSA Proteomic Analysis Software Suite (COMPASS) (20). Data was searched against the International Protein Index (IPI; http://www.ebi.ac.uk/IPI/) human database version 3.57, which had been concatenated with a reversed version of the same database (21). Full enzymatic specificity was required, allowing up to three missed cleavages. Carbamidomethylation of cysteines, iTRAQ 4-plex on the N terminus, and iTRAQ 4-plex on lysines were set as fixed modifications, whereas oxidation of methionines and iTRAQ 4-plex on tyrosines were set as variable modifications. An average mass tolerance of ±4.5 Da was used for the precursor, whereas a monoisotopic mass tolerance of ±0.5 Da was used for fragments ions. The resulting peptide spectral matches were trimmed to a 1% FDR using both e-value and precursor mass accuracy (20). Peptides were grouped into proteins following the rules previously established (22). P-scores for unique peptides that were grouped together by a common protein were multiplied to obtain the protein P-scores. Proteins were filtered by this score to achieve a 1% FDR.
RESULTS AND DISCUSSION
iHCD Implementation
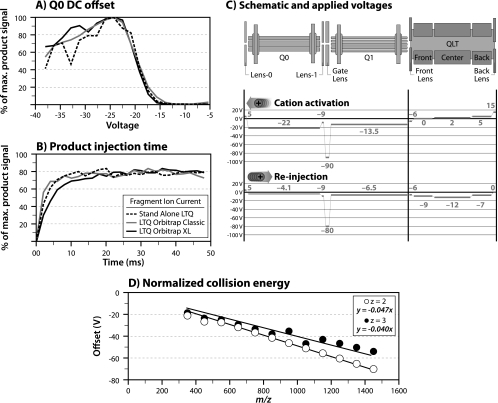
As shown in Fig. 1 we elected to implement iHCD in Q0, which operates at a pressure comparable to dedicated collision cells (∼1–5 mTorr). The iHCD scan cycle involves four major steps: (A) injection and isolation of selected precursor m/z peak (QLT), (B) activation in the high pressure region near the AP inlet (Q0), (C) injection of product ions into the system for further processing (e.g. product isolation, ion/ion reaction, etc.) and/or (D) m/z analysis in either the primary or secondary analyzer, if present (e.g. QLT or orbitrap, respectively). For maximum sensitivity steps B and C required extensive optimization (i.e. high precursor-to-product conversion efficiency). Initial evaluation of voltages and times was performed using triply charged angiotensin cations. Fig. 2 presents the results of these procedures as performed on three generations of QLT instruments (LTQ, LTQ-Orbitrap, and LTQ-Orbitrap XL). Fig. 2A details one such analysis; here the product ion current produced upon iHCD of triply protonated angiotensin cations was measured as a function of the voltage offset between the QLT and Q0. To achieve optimal results one must balance low transmission efficiency, at low voltage offsets, and ion loss, because of scattering at high offsets. A 22-V offset was optimal for this triply charged precursor. Such an offset imparts ∼66 eV of kinetic energy, a comparable amount to that used in a dedicated collision cell. Fig. 2B depicts a similar experiment in which we monitor the ejection of product ions from Q0 to the QLT. At short times not all of the product ions trapped in Q0 are ejected; however, as the time is extended to 20 ms most product ions are effectively ejected. This extended ejection time is expected as Q0 is 85 mm long and has no axial gradient. Fig. 2C shows the optimal voltages (all ion optics) for the interrogation of triply charged cations at m/z 433. With these conditions we estimate the overall precursor-to-product conversion efficiency of iHCD at ∼40%– a yield typical of collision cell HCD.
Fig. 2.
iHCD parametric evaluation. A, the relationship between the maximum product ion signal and the voltage offset between Q0 and the QLT. B, the time required for the ejection of product ions out of Q0 and into the QLT. As judged by data from three platforms, a standalone LTQ, an LTQ-Orbitrap Classic, and an LTQ-Orbitrap XL, the optimal conditions are identical. A schematic overview of the conditions necessary to effect iHCD on triply protonated angiotensin ions is presented in C. Finally, plots relating the optimal offset voltages between Q0 and the QLT (i.e. the optimal collision energy) for producing a sequence-informative MS/MS spectrum as a function of precursor m/z and charge state are shown in D.
Ideal activation parameters, of course, vary with precursor charge (z) and mass (m); more specifically, the optimal voltage offset between the QLT and Q0 is precursor z and m dependent. To counter this we developed a normalized collision energy algorithm that automatically tailors iHCD parameters to precursor characteristics. To generate the algorithm we repeatedly interrogated a yeast whole-cell lysate that was enzymatically digested with LysC by nLC-MS/MS. During each experiment we varied one instrument parameter (e.g. the voltage offset between Q0 and the QLT), producing over 20,000 peptide spectral matches. Next, we empirically determined what instrument settings produce the most identifications for a given z and m/z range– e.g. for precursors between 300 and 400 m/z and with a charge state of 2 the offset that produced the maximal number of peptide spectral matches was ∼20V (Fig. 2D). This process was repeated for each m/z range to generate a line of best fit. The trend shows an increasing amount of kinetic energy must be applied with increasing m/z. Trends very similar to those depicted in Fig. 2D have been described for other collision-activated dissociation methods (23–28). We then incorporated the normalized collision energy algorithm into the instrument control code to automatically calculate optimal iHCD voltage offsets for any selected precursor.
Comparison to Existing Fragmentation Methods
To compare iHCD to existing collision-based methods we infused angiotensin and activated the +3 precursor using CAD, PQD, HCD, and iHCD. Because they result from beam-type fragmentation, iHCD and HCD closely resemble one another (Fig. 3). For example, both display pronounced histidine immonium ion m/z peaks and have an appreciable amount of lower m/z secondary fragment ions, which tend to accumulate in beam-type CAD MS/MS spectra. A notable difference between iHCD and CAD is the complete absence of the histidine immonium ion (m/z 100) in the CAD spectrum, because of the inherent low mass cutoff of the QLT. PQD, a variant of CAD designed to overcome low mass cutoff, yields intermediate immonium ion levels.
Fig. 3.
MS/MS spectra resulting from either iHCD (A), CAD (B), PQD (C), or HCD (D) of triply protonated angiotensin cations.
We next evaluated the ability of iHCD to identify peptides from a complex mixture (LysC digest) as compared with CAD and HCD. Under method-specific optimal conditions (e.g. preferred AGC targets, NCE values, etc.), iHCD outperformed HCD and was comparable to CAD (1% FDR; Fig. 4A). We conclude this performance gap between iHCD and HCD is mainly analyzer driven. A separate comparison of iHCD to HCD using the same analyzer yielded comparable identifications (2883 versus 2538 PSMs, respectively). Despite the reduced number of iHCD scans (9556, avg. rate 280 ms versus 9933, avg. rate 260 ms for CAD), iHCD returned slightly more identifications (2883 versus 2730). These results are typical of other comparisons we have performed (data not shown). We conclude that though somewhat less efficient and slightly slower, iHCD generates more sequence-informative MS/MS spectra than CAD.
Fig. 4.
The number of peptides identified from a yeast whole-cell lysate that was enzymatically digested with Lys-C and interrogated by HCD (FTMS), CAD (ITMS), and iHCD (ITMS) (A). B, the number of peptides identifications from a human embryonic stem cell lysate that was enzymatically digested with LysC and labeled with isobaric tags (iTRAQ).
Because compatibility with isobaric tagging strategies is one of the major differences between HCD and CAD, we next sought to characterize the ability of iHCD to identify and generate quantitative information from isobarically tagged peptides (e.g. iTRAQ/TMT). iTRAQ-labeled peptides, generated from a Lys-C digest of human ES proteins, were analyzed by PQD, HCD, and iHCD. We substituted PQD for CAD, which cannot retain the quantitative reporter ions. iHCD produced more than twice as many identifications compared with either PQD or HCD (Fig. 4B). As we have previously shown, reporter ion intensity directly correlates with quantitative accuracy (4, 29). To assess the ability of iHCD to produce iTRAQ reporter ions, we performed one analysis in which both PQD and iHCD were executed on every precursor. On average, reporter ions were ∼10-fold more intense when interrogated by iHCD than by PQD (Fig. 5). iHCD and HCD generate comparable amounts of iTRAQ reporter signal. Taken together, these identification and quantification data indicate that iHCD is an excellent method for isobaric tag-based quantification.
Fig. 5.
MS/MS spectra following dissociation of an iTRAQ-labeled peptide cation interrogated using either PQD or iHCD. In both cases, the resulting MS/MS spectra easily identified the corresponding peptide; however, the iTRAQ reporter ions are ∼10-fold more intense in the iHCD spectrum. Additionally, secondary fragment ions are more prominent in the iHCD spectrum.
CONCLUSION
We present a method for performing HCD in the mass spectrometer AP inlet region (iHCD). iHCD provides similar or better rates of identification compared with CAD and carries with it all the benefits associated with beam-type dissociation: (1) compatibility with isobaric tags for multiplexed quantitation, (2) direct transferability of precursor-to-product transitions from discovery to targeted analysis, (3) elimination of low-mass cutoff inherent to CAD, (4) improved sequencing capabilities, especially for post-translational modification (PTM)-containing peptides. iHCD eliminates the need for a dedicated collision cell, thereby reducing instrument complexity and cost. By doing so, iHCD brings beam-type fragmentation to virtually any MS system having an AP inlet (e.g. ion traps, single quadrupoles, etc.). Here we describe implementation of iHCD on an LTQ quadrupole linear ion trap mass spectrometer. Recent work has extended iHCD to the newer generation dual cell QLT (Velos) with similar success (30). This result is significant because that system's quite different AP inlet utilizes a stacked ring ion guide. In sum, iHCD is an effective, simple to implement technology that delivers beam-type CAD to any system having an AP inlet.
Acknowledgments
We are grateful to Derek Bailey for experimental assistance.
Footnotes
* This work was supported by the University of Wisconsin, the Beckman Foundation, and National Institutes of Health (NIH) Grant R01GM080148 (to J.J.C.). D.H.P. acknowledges support from an NIH predoctoral traineeship—the Genomic Sciences Training Program, NIH 5T32HG002760.
1 The abbreviations used are:
- HCD
- beam-type collisional activation dissociation
- CAD
- collision-activated dissociation
- QLT
- quadrupole liear ion trap
- AP
- atmospheric pressure
- PQD
- pulsed-Q dissociation
- iTRAQ
- isobaric tag for relative and absolute quantification.
REFERENCES
- 1. Hunt D. F., Buko A. M., Ballard J. M., Shabanowitz J., Giordani A. B. (1981) Sequence-analysis of polypeptides by collision activated dissociation on a triply quadrupole mass-spectrometer. Biomed. Mass Spectrom. 8, 397–408 [DOI] [PubMed] [Google Scholar]
- 2. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 3. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A Tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McAlister G. C., Phanstiel D., Wenger C. D., Lee M. V., Coon J. J. (2010) Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal. Chem. 82, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y., Ficarro S. B., Li S., Marto J. A. (2009) Optimized Orbitrap HCD for Quantitative Analysis of Phosphopeptides. J. Am. Soc. Mass Spectrom. 20, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 6. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 7. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagaraj N., D'Souza R. C., Cox J., Olsen J. V., Mann M. (2010) Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 [DOI] [PubMed] [Google Scholar]
- 9. Bantscheff M., Boesche M., Eberhard D., Matthieson T., Sweetman G., Kuster B. (2008) Robust and sensitive iTRAQ quantification on an LTQ orbitrap mass spectrometer. Mol. Cell. Proteomics 7, 1702–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 11. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 12. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U. S. A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 14. Falick A. M., Hines W. M., Medzihradszky K. F., Baldwin M. A., Gibson B. W. (1993) Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass-spectrometry. J. Am. Soc. Mass Spectrom. 4, 882–893 [DOI] [PubMed] [Google Scholar]
- 15. Schwartz J. C., Senko M. W., Syka J. E. P. (2002) A two-dimensional quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 13, 659–669 [DOI] [PubMed] [Google Scholar]
- 16. Syka J. E., Marto J. A., Bai D. L., Horning S., Senko M. W., Schwartz J. C., Ueberheide B., Garcia B., Busby S., Muratore T., Shabanowitz J., Hunt D. F. (2004) Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 [DOI] [PubMed] [Google Scholar]
- 17. Makarov A., Denisov E., Kholomeev A., Balschun W., Lange O., Strupat K., Horning S. (2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 18. Ludwig T. E., Levenstein M. E., Jones J. M., Berggren W. T., Mitchen E. R., Frane J. L., Crandall L. J., Daigh C. A., Conard K. R., Piekarczyk M. S., Llanas R. A., Thomson J. A. (2006) Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185–187 [DOI] [PubMed] [Google Scholar]
- 19. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 20. Wenger C. D., Phanstiel D. H., Lee M. V., Bailey D. J., Coon J. J. (2011) COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: An integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 22. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 23. Neta P., Simon-Manso Y., Yang X., Stein S. E. (2009) Collisional energy dependence of peptide ion fragmentation. J. Am. Soc. Mass Spectrom. 20, 469–476 [DOI] [PubMed] [Google Scholar]
- 24. Cox K. A., Gaskell S. J., Morris M., Whiting A. (1996) Role of the site of protonation in the low-energy decompositions of gas phase peptide ions. J. Am. Soc. Mass Spectrom. 7, 759. [DOI] [PubMed] [Google Scholar]
- 25. Dongre A. R., Jones J. L., Somogyi A., Wysocki V. H. (1996) Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J. Am. Chem. Soc. 118, 8365–8374 [Google Scholar]
- 26. Haller I., Mirza U. A., Chait B. T. (1996) Collision induced decomposition of peptides. Choice of collision parameters. J. Am. Soc. Mass Spectrom. 7, 677–681 [DOI] [PubMed] [Google Scholar]
- 27. Summerfield S. G., Gaskell S. J. (1997) Fragmentation efficiencies of peptide ions following low energy collisional activation. Int. J. Mass Spectrom. 165, 509–521 [Google Scholar]
- 28. Wysocki V. H., Tsaprailis G., Smith L. L., Breci L. A. (2000) Special feature: Commentary - Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrom. 35, 1399–1406 [DOI] [PubMed] [Google Scholar]
- 29. Phanstiel D., Zhang Y, Marto J. A., Coon J. J. (2008) Peptide and protein quantification using iTRAQ with electron transfer dissociation. J. Am. Soc. Mass Spectrom. 19, 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwartz J. C. Personal Communication. Thermo Fisher Scientific, 2011 [Google Scholar]