Abstract
Any of seven lysine residues on ubiquitin can serve as the base for chain-extension, resulting in a sizeable spectrum of ubiquitin modifications differing in chain length or linkage type. By optimizing a procedure for rapid lysis, we charted the profile of conjugated cellular ubiquitin directly from whole cell extract. Roughly half of conjugated ubiquitin (even at high molecular weights) was nonextended, consisting of monoubiquitin modifications and chain terminators (endcaps). Of extended ubiquitin, the primary linkages were via Lys48 and Lys63. All other linkages were detected, contributing a relatively small portion that increased at lower molecular weights. In vivo expression of lysineless ubiquitin (K0 Ub) perturbed the ubiquitin landscape leading to elevated levels of conjugated ubiquitin, with a higher mono-to-poly ratio. Affinity purification of these trapped conjugates identified a comprehensive list of close to 900 proteins including novel targets. Many of the proteins enriched by K0 ubiquitination were membrane-associated, or involved in cellular trafficking. Prime among them are components of the ESCRT machinery and adaptors of the Rsp5 E3 ubiquitin ligase. Ubiquitin chains associated with these substrates were enriched for Lys63 linkages over Lys48, indicating that K0 Ub is unevenly distributed throughout the ubiquitinome. Biological assays validated the interference of K0 Ub with protein trafficking and MVB sorting, minimally affecting Lys48-dependent turnover of proteasome substrates. We conclude that despite the shared use of the ubiquitin molecule, the two branches of the ubiquitin machinery—the ubiquitin-proteasome system and the ubiquitin trafficking system—were unevenly perturbed by expression of K0 ubiquitin.
Post-translational modification of cellular proteins with ubiquitin determines their fate by influencing protein-protein interactions, altering recognition, targeting to cellular compartments, or by promoting their degradation at the 26S proteasomes (1–6). In order to carry out, in parallel, such diverse cellular functions, downstream components must differentiate between ubiquitin-conjugates destined for alternative fates. This is made possible because ubiquitin polymerizes into chains and therefore does not represent a single signal embodied by a single molecule, but rather a family of polymeric signals differing in chain length, linkage type, and spatial conformation. Any of seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) on the surface of ubiquitin can be linked via a covalent amide bond to the free carboxy-terminus of a distal ubiquitin, resulting in a sizeable spectrum of configurations. Structurally distinct surfaces presented by different linkage types can be selected for by dedicated down-stream ubiquitin-binding proteins (2, 4, 7). Recent advances in mass spectrometry (MS) have provided a powerful tool for accurate, direct determination of modified lysines on ubiquitin or on the target substrate allowing for insightful analysis of ubiquitin signals and their cellular correlations (8–13). Indeed MS analysis has provided lists of ubiquitinated substrates (9, 14–17) and recognized ubiquitin chains (8, 17, 18).
An essential outcome of ubiquitination is targeting to the 26S proteasome, which is responsible for the degradation of most cytosolic, nuclear, endoplasmic reticulum lumenal or membrane proteins, and even mitochondrial proteins (5, 6, 19, 20). The majority of proteasome substrates are tagged not by a single ubiquitin (monoUb)1, but by a polyubiquitin (polyUb) chain. Lys48 is the only lysine on ubiquitin whose substitution to arginine is lethal, pointing to a unique and essential role for Lys48-linked chains (21). It is generally thought that such Lys48-linked polyUb chains longer than four ubiquitin molecules are the preferred signal for efficient recognition and degradation by 26S proteasomes (22). Once bound by proteasomes, substrate-conjugates are deubiquitinated, unfolded, and subsequently degraded.
Other biological pathways that are regulated by ubiquitination include endocytosis and intracellular trafficking (23–25), histone and transcriptional regulation (26), autophagy (27), DNA repair (28), and diverse cell signaling (29–31). Most of these nonproteolytic roles are carried out by so called “alternative” ubiquitin signals, such as embodied by conjugation of a single ubiquitin molecule (monoubiquitination), or by non-Lys48-linked polyUb chains. For instance, nonproteolytic processes associated with Lys63 chains have been documented in protein trafficking, DNA damage tolerance, the inflammatory response, and ribosomal protein function (4). In protein trafficking, Lys63-linked polyUb serves as a signal mediating the internalization of plasma membrane receptors and transporters, intracellular transport, and subsequent lysosomal and vacuolar degradation (32–39). Lys63-linked chains function also as a trafficking signal at the endosomal level for MVB sorting (40). By promoting Lys63-linked polymerization, Rsp5, an E3 ubiquitin ligase of the Nedd4 HECT family, has been shown to have a prevailing role in ubiquitination of plasma-membrane transporters in yeast cells (41–44). In most cases, this ligase does not bind its substrates directly, but is recruited to them via a subset of specific adaptors some of which were shown to undergo Rsp5-dependent ubiquitination themselves (40, 45–47). Complicating the ability to attribute specific functions to Lys63 linkages, many membrane processes associated with Lys63 linkages are also driven by monoubiquitination. Often the same substrates are documented to be targets of both Lys63 polyUb or monoUb modifications. Both signals may partially overlap, though mild discrepancies between samples may also arise from rapid deubiquitination of Lys63 chains (48, 49). Notably Ubp2, specifically deubiquitinates Lys63-linked ubiquitin chains on Rsp5-substrates leading to an increase in monoubiquitinated substrates (42). Sample preparation may thus influence efficiency of trapping of polyUb chains, in particular the relatively labile Lys63 linkages. Understanding the relative prevalence and relative efficiency of monoubiquitination versus Lys63 chains in intracellular transport and endocytosis is a subject of intense investigation and great scrutiny.
The precise biological differences between polyUb chains of various topologies have not been broadly understood yet, though sporadic observations keep coming in. Recent studies have spotlighted Lys11-linked ubiquitin chains, revealing their involvement in endoplasmic reticulum associated degradation (ERAD) as modifiers of the E2 Ubc6, which has also been proposed to participate in the synthesis of these chains (8). Other studies have related Lys11 chains to regulation of mitotic protein degradation and cell cycle control (50, 51). Lys6 chains were shown to be synthesized by the BRCA1/BARD1 complex (52, 53); Lys27 chains were described to be involved in ubiquitination of the transcription factor Jun, a modification required for its unconventional lysosomal targeting (54); Lys33 chains were recently reported to modify TCR-ζ thereby regulating cell-surface-receptor-mediated signal transduction; involvement of Lys29 chains was demonstrated in protein degradation and recruitment of a chain elongating factor E4 (55, 56). Two different substrates of AIP4/Itch were also described to be modified by K29-linked ubiquitin chains in the context of Notch trafficking and signaling (57). Adding yet another layer of complexity to the conjugated ubiquitin landscape, there are additional modifications by ubiquitin, including: mono- and multiple-mono ubiquitinations (58–60), mixed or branched ubiquitin chains (61–65), and linear chains (66). As linkages via lysines other than Lys48 are not strictly essential, these “alternative” chains may be less specific for a unique outcome allowing some overlap in signaling. Alternatively, polymerization quality control may be less rigidly enforced generating mixed signals in these pathways.
Utilization of ubiquitin mutants is a strategy used to study the role or different ubiquitin chains. Substituting lysine residues in the ubiquitin molecule with arginine blocks specific chain extensions and thereby inhibits downstream consequences associated with a specific ubiquitin linkage (67). Single-lysine ubiquitin mutants and a lysine-less, nonextendable mutant (K0 Ub) (68) are commonly used in vivo and in vitro for defining chain structure and for directing cellular ubiquitinations toward a specific ubiquitin modification. Because of the complicated dynamics of polyUb chains, when expressing ubiquitin mutants in living cells, the ratio of mutant to wild-type ubiquitin is likely to be a critical factor in the outcome of these types of experiments (67). A phenotypic analysis of all single K-to-R substitutions (excluding K48R-Ub) revealed hypersensitivity to DNA damaging agents specific for K63R mutation (69). Moreover by using the K63R mutant, the role of Lys63 ubiquitin chains in endocytosis and vacuolar targeting was unearthed (33, 70–72). Use of K48R-Ub as the sole form of cellular ubiquitin has demonstrated the essential role of this lysine residue for cell viability (21).
The large portion of proteins that are direct targets of ubiquitination, and the role that this modification has in directing their bio-stability, cellular localization and function, makes the ubiquitin system an ideal system for proteomic analysis. A large portion of ubiquitin-modified proteins have been identified by high throughput screens relying on advanced mass spectrometry (9, 14–17). However, not in all cases have high throughput screens been able correlate between groups of ubiquitinated proteins, their associated chains and biological relevance. By combining ubiquitin mutants and high throughput MS analysis we studied changes to the ubiquitin landscape and to the conjugated ubiquitinome. This approach identified new substrates and enabled to dissect use of ubiquitin in the ubiquitin-proteasome system from that in the ubiquitin trafficking system (UTS).
EXPERIMENTAL PROCEDURES
Plasmids, Yeast Strains, and Growth Conditions
Plasmids used in this study are detailed in the accompanying supplementary file. Wild-type (WT) Saccharomyces cerevisiae yeast strains were purchased from Euroscarf (genotypes detailed in accompanying supplementary file) and were transformed with the relevant plasmids by standard lithium acetate/polyethylene glycol procedure. For stable isotope labeling with amino acids in cell culture (SILAC) analysis we deleted ARG4 from MY59 strain by mating with ARG4 deletion strain (purchased from the Euroscarf collection) followed by sporulation and random spore analysis (as described in (73)). A Jen1-green fluorescent protein (GFP) expressing yeast strain was a gift from Sandra Paiva (University of Minho, Portugal). Generally, all yeast strains were grown in selective minimal yeast media or YPD at 30°, and experiments were performed when cells have reached an OD600 of 1.
Ub-AQUA Analysis
Yeast Cell Extract Preparation for AQUA Analysis
Cell cultures (5 ml) were grown to logarithmic phase; optical density was measured at 600 nm and cell density normalized. Samples were prepared by trichloroacetic acid (TCA) lysis and precipitation (except for samples described in Fig. 1B) and separated on 4–12% Tris-Bis SDS-PAGE. High-molecular-weight (MW) Gel regions were excised as indicated, and subjected to in-gel trypsin digestion. Ub-AQUA peptides were added to the digests and samples were subjected to liquid chromatography-selected reaction monitoring in a TSQuantum Ultra (ThermoElectron, San Jose, CA) (12). The Ub-AQUA method applied in yeast is detailed in (18).
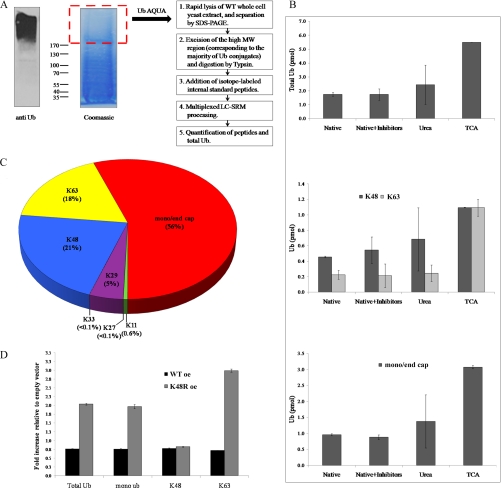
Fig. 1.
The conjugated Ub linkage profile. A, A general scheme for determining the linkage profile of conjugated ubiquitin. Logarithmically growing WT yeast cells (genotype of this and other strains used in this study are listed in supplemental Table S7) were rapidly lysed and the resulting whole-cell extract was resolved by a 4–12% gradient SDS-PAGE. The high MW region, corresponding to the majority of the Ub conjugates (as verified by immunoblotting with anti-Ub polyclonal antibody; left panel), was excised and subjected to digestion by Trypsin (right panel). Isotope-labeled Ub AQUA peptides were added as internal standards, and mixture subjected to multiplexed liquid chromatography-selected reaction monitoring processing (Ub-AQUA peptide transitions are listed in supplemental Table S9). Ubiquitin-linkage determination was performed as described (11, 18, 50, 76, 79). B, TCA lysis reproducibly generates the highest yield of ubiquitin conjugates. Logarithmic yeast cells were processed according to scheme in panel A, differing only in the lysis protocol. Top: The total amount of conjugated ubiquitin detected under the different lysis conditions is shown in pmols. TCA lysis retained the highest amount of total ubiquitin in high MW conjugates compared with other lysis conditions. Middle: Quantification of Lys48 and Lys63 ubiquitin linkages captured under different lysis conditions. TCA lysis retained the highest absolute amounts of Lys48 linkages and significantly higher amounts of Lys63 chains than those trapped in a measurable form by other lysis conditions. Bottom: Quantification of mono and endcap ubiquitin under different lysis conditions. A large portion of ubiquitin found in high MW conjugates was not modified on any lysine residue. The highest portion of such ubiquitin representing mono and endcap modifications was obtained under TCA lysis, without detracting from amounts of polyUb chains trapped in same samples (middle panel). C, Summary of ubiquitin linkage distribution obtained under TCA lysis. The relative abundance of polyUb as well as nonextended ubiquitin (mono and end cap) displayed as a percentage of total conjugated ubiquitin. In this sample, chains linked via Lys6, or N terminus are scarce (<0.03%) and are therefore not shown in this chart. D, Lys48Arg (K48R) ubiquitin mutation alters normal partitioning between polyUb linkages. Equal amounts of WT cells transformed with plasmids overexpressing (oe) WT Ub or K48R Ub were processed as in panel A, and distribution of main ubiquitin modifications was compared with that in cells transformed with empty vector (serving as background strain). Properties of these and other plasmids used in this study are listed in supplemental Table S8. Overexpression of WT Ub did not significantly alter ubiquitin conjugates in high MW relative to background strain. Upon expression of K48R Ub, Lys48-linkages remained the same as in WT background, however as other linkages increased, the relative portion of Lys48-linkages decreased.
For lysis-conditions experiment described in Fig. 1B, yeast cells were grown to log phase and then lysed using glass beads under four different conditions: (1) Nondenaturing conditions (50 mm Tris pH = 7.4, 150 mm KCl, 0.1% Triton X-100, and 2 mm dithiotreitol) without protease inhibitors (Native); (2) Nondenaturing conditions including general protease inhibitors (Roche Diagnostics) (Native+ Inhibitors); (3) Denaturing Urea lysis buffer (8 m urea, 50 mm Tris pH = 7.4) (Urea); or (4) Denaturing 20% TCA lysis buffer, which was diluted to 12% TCA buffer after breaking the cells with glass beads. After lysis, a buffering solution of 1 m Tris pH 8 was added to TCA lysed samples to neutralize any residual TCA. Laemmli buffer was added to all lysates or TCA precipitates before separation on SDS-PAGE.
Data Analysis
For each peptide, the areas under the curve were determined for the native (trypsin digested) and synthetic (isotope labeled) SRM transitions (see accompanying supplementary file for peptide transitions). The product of this ratio with the known abundance of each synthetic peptide was calculated to determine the abundance of each ubiquitin linkage type. For the K63 polyUb peptide, two distinct SRM transitions were monitored, with the reported values reflecting the average of these two measurements. To determine the total amount of ubiquitin in a sample the law of conservation of mass was applied (10) to the peptides surrounding the Lys6, Lys63, and Lys48 locus. The total amount of ubiquitin reported represents the average of the Lys6, Lys48, and Lys63 measurements. For strains expressing ubiquitin mutants (K48R and K0), the total amount of ubiquitin was calculated similarly, excluding the peptides carrying the lysine-to-arginine mutations. Nonextended ubiquitin modifications (mono and endcap Ub) were calculated by subtracting the number representing all identified modifications from the total amount of ubiquitin (detailed in (50)).
Quantitative Proteomics Analysis by SILAC
Preparation of Yeast Cell Extract and Isolation of Ubiquitin Conjugates (Pullout)
RGS-His8-WT Ub and RGS-His8-K0 Ub were each separately transformed into yeast strain auxotrophic for LYS2 and ARG4 genes. Cultures were grown in synthetic minimal media; WT Ub expressing cells in heavy medium containing 40 mg/l 13C615N2 lysine and 20 mg/l 13C615N4 arginine (Cambridge Isotope Laboratories Inc.), and K0 Ub expressing cells in light medium (containing unlabeled lysine and arginine). Both cultures were grown for at least 10 generations to ensure complete incorporation of heavy isotopes-labeled amino acids. Cultures were harvested at OD600 of 1 in a final volume of 70 ml, and lysed using glass beads in a 20% TCA solution. The supernatant was collected and the beads were washed with diluted TCA solution to retrieve the remains. The final cell lysate of each sample contained 12% TCA for efficient protein precipitation. A fraction making up 10% of the total sample volume of each heavy and light lysate was taken aside for “total extract” analysis, and all lysates were incubated at 4° for 1h, followed by centrifugation to separate precipitates from supernatants. The precipitates pH was adjusted to ∼7.5 using 1 m Tris pH = 11. Precipitates destined for “total extract” analysis were resuspended in 5% SDS solution, whereas the rest of the precipitates, destined for ubiquitin pullout, were resuspended in loading buffer (6 m GuHCl, 20 mm Tris pH = 8, 100 mm K2HPO4, 10 mm imidazole, 100 mm NaCl, 0.1%triton X-100). Protein concentration for all samples was determined using BradfordUltra (Expedeon), and then equal amounts of heavy and light samples were mixed together for either total extract or ubiquitin pullout analysis.
The combined sample destined for ubiquitin pullout (∼6 mg total protein) was loaded onto a mini NiNTA column (Qiagen, The Netherlands) and incubated overnight at 4°C. Flow-through was collected and after several column washes with loading buffer, was reloaded onto the column and incubated for 3h at room temperature. The column was subsequently washed several times with wash buffer 1 (20 mm tris pH = 8, 100 mm K2HPO4, 20 mm imidazole, 100 mm NaCl, 0.1%triton X-100) followed by washes with wash buffer 2 (20 mm tris pH = 8, 100 mm K2HPO4, 10 mm imidazole, 1 m NaCl, 0.1%triton X-100) followed by imidazole elution (20 mm tris pH = 8, 100 mm K2HPO4, 500 mm imidazole, 100 mm NaCl). The eluate was concentrated using 12% TCA and acetone precipitation and then resuspended in Laemmli buffer for separation by SDS-PAGE. Both total extract (total of 100 μg) and pullout samples (the entire concentrated eluate) were separated by a 12% SDS-PAGE, and after staining with Imperial Protein Stain (Thermo), each lane of the gel was cut into 10 to 12 slices. The above procedures were repeated twice in order to obtain two biological repeats of this experiment.
In-gel Trypsinization and Mass Spectrometry Analysis
The proteins in each gel slice were reduced (10 mm dithiotreitol), modified with 40 mm iodoacetamide (at 25 °C) and trypsinized (modified trypsin (Promega)) at a 1:100 enzyme-to-substrate ratio for 18 h at 37 °C. A similar amount of fresh trypsin was then added and samples were incubated for additional 8 h at 37 °C.
The resulting tryptic peptides from each gel slice were resolved by reverse-phase chromatography on 0.075 × 200-mm fused silica capillaries (Aligent Technologies J&W, Santa Clara, California) packed with Reprosil reversed phase material (Dr Maisch GmbH, Germany). The peptides were eluted with linear 65 min gradients of 5 to 45% and 15 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.25 μl/min. Mass spectrometry was performed by ”hybrid” mass spectrometer (Orbitrap, Thermo) in a positive mode using repetitively full MS scan followed by collision induces dissociation of the seven most dominant ion selected from the first MS scan.
Targeted quantitative MS of trypsin-derived Ub peptides (8) and of ubiquitin linkages was facilitated (when required) by a mass inclusion list (detailed in the accompanying supplementary file). In these cases, the chromatography step included a linear 34 min gradient of 5 to 40% and 10 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.25 μl/min. For inclusion list-dependent acquisition on the Orbitrap mass spectrometer, a single Orbitrap MS scan from m/z 320 to 900 at resolution 60,000 was followed by up to three MS/MS scans. Preview mode and charge state screening were enabled for selection of precursors. The m/z tolerance around targeted precursors was ±30 ppm. Dynamic exclusion was disabled. The intensity threshold for triggering of a detected peak was set to 30,000, and normalized collision energy was specified to 35.
SILAC Quantitative Data Analysis
Raw MS files from the LTQ-Orbitrap were processed in Quant.exe, the first module of MaxQuant (version 1.1.1.6) (74) combining the RAW MS files of the 2 biological repeats (i.e. Total1 and Total2 were analyzed together). The derived peak list was searched using Andromeda, MaxQuant build-in search module against the S. cerevisiae translations of all systematically named open reading frames (release date Jan 5th, 2010; Downloaded form SGD), to which the sequence of K0 Ub was added. To these 5905 sequences, known contaminants and reverse sequences were added. The search was performed using an initial precursor mass tolerance of 7 ppm and fragment mass tolerance of 0.5 Da. The following search criteria were used: tryptic/P specificity was required with two missed cleavages allowed; cysteine carbamidomethylation was set as fixed modification and oxidized-methionine, protein N-acetylation and lysine ubiquitination were set as variable modifications. In addition, the default settings were used: maximal peptide posterior error probability of 1, maximal peptide false discovery rate of 0.01, and minimal peptide length of six amino acids. For peptide and protein identification the 1% false discovery rate was determined by accumulating 1% of reverse database hits. When the identified peptide was shared by two proteins (homologues or isoforms), the two proteins were reported by MaxQuant as one protein group. Quantification was performed using MaxQuant with the following settings: minimal ratio count of two and using only unmodified peptides and peptides modified by methionine oxidation or protein N-terminal acetylation. Lists of quantified proteins including only proteins identified by two peptides or more are presented in supplementary Tables S1 and S2. Raw and normalized SILAC ratios are standard MaxQaunt output. Normalization is done at the peptide level for each of the LC-MS runs separately, allowing for different protein mixing ratios in different runs. The peptides in each LC-MS run are grouped in bins according to their intensity and for each bin the ratios are normalized by setting the median of logarithmic ratios at zero. It is done separately for lysine and arginine labeled peptides to compensate for any possible label-specific bias. Protein ratios are calculated as the median of all SILAC peptide ratios, minimizing the effect of outliers (74).
SILAC-based Targeted Quantitative MS of Ubiquitin and Ubiquitin Linkages
The intensities of the tryptic peptides derived from unmodified ubiquitin and ubiquitin linked to another ubiquitin molecule (linkage signature peptide) were manually analyzed by ion chromatograms using Xcalibur v2.0.7 software (Thermo Finnigan, San Jose, CA) and used to estimate changes to linkage profile between the two strains. The peak area of each precursor was calculated using Genesis peak algorithm with a mass tolerance of 10 ppm. The abundance ratio for each peptide was calculated by dividing for peak area of the light form (K0 Ub) of each peptide by corresponding peak area of the heavy form (WT Ub). For peptides that were identified in more than one charge or modification state (i.e. oxidized methionine or pyroglutamine) the abundance ratio of both states was averaged.
Go Enrichment Analysis
In order to characterize unique features shared by the high abundance ratio proteins found in the pullout of ubiquitin conjugates, we determined their main GO categories using AmiGO GO term enrichment tool (75) and selected the categories that showed high enrichment ratio (>2) relative to the background set.
Detection of Ectopically Expressed Ubiquitin
Cells transformed with RGS-His8 WT Ub or with RGS-His8 K0 Ub were lysed using glass beads in TCA solution and separated on SDS-PAGE. Ectopically expressed ubiquitin was visualized using mouse monoclonal anti RGS-His antibodies (Qiagen). The entire cellular ubiquitin species were visualized using rabbit polyclonal anti ubiquitin antibodies (Dako).
Cycloheximide Chase
MyoD encoding plasmid was cotransformed into WT yeast cells with either WT or K0 Ub encoding plasmids. Fresh cultures were grown to an OD600≈1 in the presence of 2% galactose, and the stability of MyoD was monitored after inhibition of protein synthesis by Cycloheximide (200 μg/ml). Cell aliquots were taken at the indicated times, lysed using glass beads in TCA solution, and separated on an SDS-PAGE. MyoD was visualized by Western blot using anti MyoD antibodies (sc-760) (Santa Cruz Biotechnology, Santa Cruz, CA). For loading control, PGK was visualized by mouse monoclonal antibodies (Invitrogen-Molecular Probes, Carlsbad, California).
The stability of Pcl5 was monitored similarly in cells cotransformed with Pcl5 encoding plasmid together with either WT or K0 Ub encoding plasmids. In this case cultures were grown in minimal medium containing 2% glucose. Pcl5 was visualized using anti Pcl5 polyclonal antibodies (a gift from Daniel Kornitzer, Rappaport Faculty of medicine, Technion).
Isolation of Ubiquitinated Syp1-GFP
Genomic Syp1-GFP yeast strain was transformed with either RGS-His8-WT Ub or RGS-His8-K0 Ub encoding plasmids. Cells from both cultures were grown in synthetic minimal medium and harvested at OD600≈1. Cells were lysed and loaded separately onto a NiNTA column as described in “Preparation of Yeast Cell Extract and isolation of ubiquitin conjugates (Pullout)” section. Total-cell-lysates and isolated ubiquitinomes were separated by SDS-PAGE and blotted with monoclonal anti GFP antibodies (clones 7.1 and 13.1, Roche).
Immunoprecipitation of Sna3–6HA
Sna3–6HA encoding plasmid was cotransformed into WT yeast cells with either WT or K0 Ub encoding plasmids. Exponentially growing cells (40 ml OD600≈1) were harvested and washed with cold TNE buffer (100 mm Tris-HCl pH 7.5, 150 mm NaCl, 5 mm EDTA). After resuspension in lysis buffer (TNE supplemented with a protease inhibitor mixture (Complete, EDTA-free mixture, Roche Diagnostics) and freshly prepared N-ethylmaleimide (5 mm final)) cells were broken using the glass beads method. Cell debris and unbroken cells were removed by centrifugation. Proteins from the lysate were precipitated by addition of 10% TCA. The protein pellet was then resuspended in Laemmli's sample buffer without β-mercaptoethanol, followed by incubation at 95 °C. TNET buffer (TNE supplemented with 1% Triton X-100) was then added to the sample before a centrifugation to get rid of unsolubilized proteins. The supernatant was incubated with a rabbit polyclonal anti-HA (Santa Cruz, sc-805). After addition of freshly prepared Protein G Sepharose beads (Gamma Bind G Sepharose, Amersham Biosciences Pharmacia), incubation was prolonged over night. The Protein G beads were collected by centrifugation, and then washed with TNET buffer. Elution of the immune complex was made by addition to the beads of an equal volume of sample buffer with 2% β-mercaptoethanol and incubation at 95 °C. Immunoprecipitated Sna3–6HA and its ubiquitin conjugates were detected by Western blot using a monoclonal anti-HA (Santa Cruz, sc-7392) or anti-RGS-His(Qiagen), respectively.
Canavanine and Nickel Phenotypic Analysis
WT yeast strains transformed with either WT or K0 Ub encoding plasmids were grown in synthetic minimal medium to a same optical density (1600 nm). Ten-fold serial dilutions in sterile double distilled water (DDW) were spotted (5 μl) onto a control, canavanine (1 μg/ml), or NiCl2 (1.5 mm) containing agar plates. Plates were incubated at 30 °C.
Cellular Localization Analysis of GFP-Phm5, Sit1-GFP, and Jen1-GFP
PRS416-GFP-Phm5 plasmid was cotransformed into WT yeast cells with either WT or K0 Ub encoding plasmids. Cells were grown in synthetic minimal medium to mid-exponential Phase and then examined by fluorescence microscopy.
Sit1-GFP encoding plasmid was cotransformed into WT yeast cells with either WT or K0 Ub encoding plasmids. Cells were grown in 2% Raffinose-containing minimal medium to mid-exponential Phase and then the expression of Sit1-GFP was induced by addition of 2% galactose. After 1 h induction cells were examined by fluorescence microscopy.
Genomically tagged Jen1-GFP yeast strain was transformed with either WT or K0 Ub encoding plasmids. Glucose (2%)-containing medium was used for growth under repression conditions. For expression conditions, glucose-grown cells in exponential growth phase were collected by centrifugation, and grown in fresh YNB medium containing lactic acid (0.5%, v/v, pH 5.0) for 4h. Cells were then examined by fluorescence microscopy (t = 0′) and 2% glucose was added for induction of endocytosis. Cells were reexamined microscopically after 30min (t = 30′).
For fluorescence microscopy, cells were concentrated by a factor of 10 by mild centrifugation. Cells were viewed immediately, without fixation, under a fluorescence microscope (type BY61, Olympus, Tokyo, Japan) using Chroma GFP II filter (excitation wavelength 440–470 nm) for detection of Green fluorescent protein (GFP)-tagged proteins.
RESULTS
Ubiquitin-linkage Profile of High MW Conjugates
A large portion of cellular ubiquitin (detected by immunoblotting) accumulated as slowly migrating species at the top of the gel when whole cell extracts (WCE) were resolved by standard SDS-PAGE (Fig. 1A left). These same high MW regions were less abundant in total protein content, reflecting a high ubiquitin-to-total protein ratio (Fig. 1A, middle). Ubiquitin is an extendable covalent modification forming polyUb chains, which may explain the migration of ubiquitin in extremely high-MW regions. Given that the ratio of ubiquitin to total protein content was very favorable in the highest MW regions we initially chose this region to assess the conjugated ubiquitin linkage profile directly from whole cell extract. The efficiency of ubiquitin extraction from whole cell extract lysed under a variety of conditions (Fig. 1B) was evaluated by comparing to isotopically labeled peptide standards (Ub-AQUA), similarly to what has been employed before for ubiquitin (18, 50, 76) or for other components of the ubiquitin system (77).
Ubiquitin conjugates are subject to deubiquitination and/or degradation, therefore lysis under native conditions may underestimate in vivo ubiquitin levels. Addition of commonly used protease inhibitors did not improve ubiquitin extraction by much (Fig. 1B; top). Urea—often used for denaturing lysis (8, 17, 76)—increased efficiency of ubiquitin capture directly from whole cell extract, though with inconsistent reproducibility. The most efficient protocol that we found for extraction of ubiquitin from whole-cell-lysate was rapid TCA lysis followed by SDS resolublization. Not only were total ubiquitin levels highest, but all linkage types were consistently recovered. Specifically we note that Lys63 linkages were better represented without detracting from identification of any other linkage type (Fig. 1B; middle), suggesting that some modifications may be particularly prone to postlysis editing. Likewise, high levels of unmodified ubiquitin were correlated with efficiency of total ubiquitin recovery (Fig. 1B; bottom). Consequently, all subsequent data in this study was collected using the protocol of TCA lysis.
The most prevalent high MW ubiquitin linkages detected in whole cell yeast extract were via Lys48 or Lys63, which together made up between 80% to 90% of polyUb. The next abundant linkage was Lys29, and all other linkage possibilities detected at trace amounts (Lys6, Lys11, Lys27, Lys33, and even N-terminal linear Ub). In total, roughly half of the ubiquitin in the high MW region was modified on at least one lysine (Fig. 1C) with all remaining ubiquitin serving either as a monoUb modification or as a chain terminator (endcap). Although the majority of ubiquitin is often thought to be tied up in long polyUb chains each made up of multiple ubiquitin molecules, it was surprising to find that roughly 50% of ubiquitin in the high MW region was unmodified on any lysine residue. This result indicated that average chain-length is rather short and that mono ubiquitination is a prevalent modification even in high MW conjugates likely representing multiple-mono ubiquitin molecules per target. Although our study focuses on high MW conjugates, analysis of lower MW regions revealed an expected increase in the mono-to-poly ratio with decreasing MW (supplemental Fig. S1). Lys48 and Lys63 remain prevalent throughout, yet a mild increase in other linkages—including Lys11—was observed with decreasing MW (supplemental Fig. S2).
Using a common building block (ubiquitin) to assemble into different orientations (linkages), may provide cells with the capability to adapt signals according to conditions. For instance, it was shown that induction of ubiquitin-binding proteins can increase the ratio of Lys48-to-Lys63 linkages (18). One way to stress the linkage profile is to limit the ability of ubiquitin to form specific linkages. We evaluated changes to the ubiquitin landscape upon overexpression of a single-site K48R mutated ubiquitin in a WT background (Fig. 1D). In this case the data is presented as relative changes from WT. With Lys48 representing a significant portion of ubiquitin conjugates one may expect that partial blocking of Lys48-chain synthesis would lead to a decrease in total ubiquitination. Interestingly, although the relative portion of Lys48 linkages did decrease, it was accompanied by a general increase of total ubiquitination. The end result was that absolute levels of Lys48 modifications (generated by endogenous WT ubiquitin) remained robust next to the elevated levels of other modifications (Fig. 1D). As a control, similar induction of a WT ubiquitin gene over the endogenous background did not alter the normal distribution between modifications.
Perturbation of the Ub-landscape by Altering the Ratio of Mono-to-Poly Ubiquitin
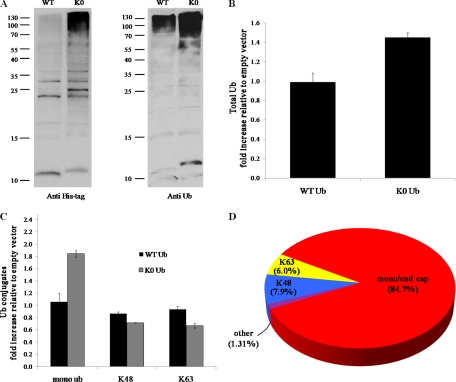
A prominent feature of the conjugated Ub landscape was that about half of the Ub molecules in WCE were in the form of nonextended Ub (Fig. 1). Such a high proportion of monoUb modifications was somewhat unexpected, because the primary ubiquitin signals are often thought to be in the form of polyUb chains (of various linkage types) (2, 7, 78). Therefore, we evaluated the perturbation upon increasing the ratio of mono-to-poly modifications using non extendable, lysineless, ubiquitin (K0 Ub). This mutant has been previously used as a chain terminator to decrease average chain length and increase modifications by monoUb (53, 79). We transformed WT yeast cells with WT [His8-Ub] or mutant [His8-K0-Ub]. In either strain, the pool of free (unconjugated) ubiquitin consisted of a mixture of both endogenous untagged WT Ub and the tagged version (Fig. 2A). Immunoblotting indicated that the mutated K0 Ub was fully usable and was incorporated into the ubiquitinome at all levels to generate a ubiquitination pattern similar to that of WT Ub, albeit with an even higher level of accumulated high MW conjugates (Fig. 2A). Likewise, mass-spectrometric analysis of ubiquitin conjugates in either strain relative to a nontransformed WT strain (empty vector) confirmed the accumulation of high MW ubiquitin conjugates with expression of the mutant ubiquitin (Fig. 2B).
Fig. 2.
Lysineless (K0) Ub penetrates into the conjugated Ub pool increasing mono/poly Ub ratio. A, WT yeast cells were transformed with plasmids for expression of either RGS-His8 tagged lysineless Ub (K0 Ub) or with tagged WT Ub as control. Rapidly lysed whole cell extracts were resolved by 18% SDS-PAGE, transferred and immunoblotted with anti RGS-His antibodies (left panel) or with anti Ub antibodies (right panel). B–D, Equal amounts of cells expressing WT Ub, K0 Ub, or empty vector (serving as background strain) were lysed and whole-cell extracts resolved by gradient SDS-PAGE. The high MW region was excised and subjected to Ub-AQUA analysis as described in Fig. 1. B, As in Fig. 1, expression of WT Ub from a plasmid did not significantly alter total conjugates relative to background strain (empty vector). Ubiquitin levels in both strains were normalized using common internal peptide standards. K0 Ub expression resulted in a roughly 50% increase in total ubiquitin conjugates. C, Increase in mono/end cap modifications accounts for bulk of ubiquitin conjugates accumulating in K0 Ub expressing cells relative to nontransformed cells. D, The relative abundance of different Ub modifications in cells expressing K0 Ub is displayed as a percentage of total cellular conjugated Ub.
The increase in total Ub conjugates (Fig. 2A, B) was due largely to a significant increase in monoubiquitination without altering the relative distribution of major linkage types (Fig. 2C). Incorporation of the nonextendable ubiquitin as a modifier (estimated at roughly 60% of total conjugates; supplemental Fig. S3) can explain the predominance of monoubiquitin modifications (Fig. 3C). Such a penetration into the ubiquitinome resulted in a perturbed landscape in which a remarkable 85% of total modifications were mono and endcaps (Fig. 3D). Remarkably, these cells were viable under standard growth conditions, possibly because of robust levels of polyubiquitin chains maintained by the endogenous WT ubiquitin. Shutting down expression of all endogenous WT Ub eventually leads to cell death, although we note that K0 Ub remained abundant in high MW conjugates even when making up nearly 100% of all available ubiquitin (supplemental Fig. S4).
Fig. 3.
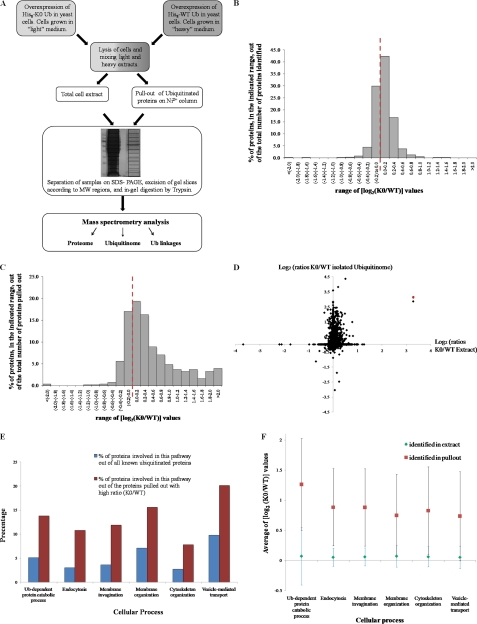
Impact of K0 Ub expression on proteome and ubiquitinome. A, Schematic description of SILAC approach used in this study. Cells expressing RGS-His8 tagged WT or RGS-His8 tagged K0 Ub were differentially isotopically labeled, mixed, lysed under denaturing conditions, and split for MW resolution of total proteome or for affinity purification of Ub-conjugates. MS/MS analysis was performed at three levels: proteins in total cell extract (proteome; panel B); proteins in pullout of ubiquitin-conjugates (ubiquitinome; panel C); and analysis of ubiquitin linkages (Fig. 4). B, 2196 different proteins were identified at high confidence in whole cell extracts of K0 Ub and WT Ub expressing cells. Enrichment factors were calculated from intensity of MS signals (log2(K0/WT); supplemental Table S2). Bars represent frequency of proteins (%) in each enrichment factor range. The red dotted line represents a ratio of unity (log21 = 0); i.e. proteins whose ratio was unaltered by expression of K0 Ub. C, A total of 856 different proteins were identified by Ub-affinity pullout (supplemental Table S1). Enrichment factors were calculated and displayed as in panel B. D, The enrichment factor for each protein in the Ub pullout (y axis) was plotted against its enrichment factor in whole cell extract (x axis). The two dimensional plot indicates the K0 Ub-induced perturbation on ubiquitination status of each substrate relative to changes in its cellular abundance. A red asterisk represents the K0 Ub protein, which was added to the database of all possible yeast translated open reading frames. E, Biological pathways significantly enriched with ubiquitinated proteins upon K0 Ub expression. Identified proteins were classified into biological pathways using the AmiGO program. Ubiquitinated proteins with a greater than twofold increase in K0 Ub expressing cells were categorized according to biological pathways. The relative portion that each category makes up out of total ubiquitinated proteins is shown in red bars. Blue bars represent the portion that these pathways make up of known ubiquitinated targets (taken from supplemental Table S3). F, To emphasize the enrichment of ubiquitinated proteins in specific biological categories the span of enrichment factors for all proteins identified in the Ub pullout is displayed for each category (surrounding the average enrichment ratio marked as red squares). For comparison, the enrichment factors of these same proteins in whole cell extract are also shown along with the average enrichment in extract of proteins belonging to each category (green diamonds). The comparison highlights that proteins belonging to these categories accumulate as ubiquitinated conjugates in K0 Ub expressing cells.
To summarize, the ubiquitin landscape is malleable and responsive to imbalances in linkage-specific or general polymerization capacity. This flexibility may channel ubiquitin into differential pathways, differing in linkage type and length, according to cellular needs.
K0 Mutant Ub Induces Perturbations in Ubiquitinome of Select Biological Pathways
Accumulation of K0 Ub-modified proteins should facilitate trapping and identification of ubiquitinated substrates with possible broader effects on the composition of the cellular proteome. Interference with the synthesis of polyUb chains as a degradation signal may directly stabilize target proteins thereby trapping normally transient ubiquitin-conjugates in a monoUb state. In order to assess the interference of K0 Ub with the steady-state population of ubiquitinated substrates, we carried out a comparative proteomic screen. The background strain used to analyze ubiquitin linkage profiles (described in Figs. 1 and 2) was genetically manipulated to be compatible with SILAC experiments using labeled arginine and lysine in media. These cells expressing either tagged WT [His8-Ub] or mutant [His8-K0-Ub] ubiquitin were grown to mid log phase in heavy or light media respectively. Cell pellets were lysed, equal amounts of extracts (total protein) were mixed, and the mixture was split for parallel analysis of proteins in extract and for affinity-purification of ubiquitin-conjugates (Fig. 3A). After resolution of samples by SDS-PAGE, gel slices were excised, trypsinized and subjected to LC-MS/MS analysis. Intensity ratios of identical peptides (Light-to-heavy) were obtained by running the entire data set through MaxQuant software (74).
Of affinity-purified ubiquitin-conjugates, light/heavy isotopic abundance ratios were obtained for 856 proteins including 186 new substrates uniquely identified here (supplemental Table S1). In total cell extract, the raw MS/MS data identified 2654 proteins (supplemental Table S2 sheet 2; ZivSTS2B), of which 2196 were positively identified by at least two different peptides with signals contributed from both light and heavy cells, thus allowing for a ratio to be calculated by MaxQuant (supplemental Table S2 sheet 1; ZivSTS2A). The validated lists were used for all subsequent analysis of data. Compiling our targets together with available published data bases of potentially ubiquitinated proteins, generates a comprehensive list of high confidence substrates in yeast (supplemental Table S3). For some of these targets, Ub-modified lysine residues (“GG-signature peptides”) were identified (supplemental Table S4 and supplemental Spectra S10).
The majority of proteins were equally abundant in total extracts from both cell types (light/heavy ratio ≈1; Fig. 3B) although the distribution was slightly off-centered toward enrichment of proteins in cells expressing K0 Ub (Fig. 3B). Among proteins most dramatically enriched with K0 Ub expression we found Orm2, Cla4, Mic17, and Ubc6 (supplemental Table S2 sheet 1; ZivSTS2A). Orm2 and Orm1 control membrane biogenesis by coordinating lipid homeostasis with protein quality control required for resistance to induced unfolded protein response (80). Cla4 is part of a complex involved in septin ring assembly, vacuole inheritance, cytokinesis, and sterol uptake (81). Mic17 is a mitochondrial intermembrane space protein, required for normal oxygen consumption (82). Ubc6 is an E2 Ub conjugating enzyme in ERAD and membrane processes (8). In the other extreme, diminished levels of Vps28, were found in K0 Ub expressing cells. Vps28 is a component of the ESCRT-I complex for ubiquitin-dependent sorting of proteins into the endosome (83). As mentioned, the overall effect of K0 Ub expression on the proteome was mild, nonetheless, of proteins whose levels did change, a larger number increased upon expression of K0 Ub. Accumulation may reflect direct stabilization of proteins modified by K0 Ub, or an indirect effect because of alteration of synthesis and degradation pathways.
Ubiquitin conjugates display a nonGaussian distribution of abundance ratios, lopsided toward enrichment of proteins tagged with K0 Ub over WT Ub (Fig. 3C). More than one third of proteins were enriched by a factor greater than 1.5 (ratio of K0 Ub-conjugates relative to conjugates of WT Ub; Fig. 3C). No correlation was found between preferred targets of K0 ubiquitination and high cellular abundance (supplemental Fig. S5; based on predicted number of copies in the cell (84)). The paucity of proteins with an abundance ratio lower than unity is probably a reflection of general protein stabilization upon interference of K0 Ub with ubiquitin polymerization. Plotting enrichment factors of all identified ubiquitin conjugates (“ubiquitinome”) against their enrichment factors in whole cell extract highlights that increased ubiquitination leads to small changes in overall protein levels, therefore implying that the ubiquitinated portion of most proteins is probably small (Fig. 3D). Nevertheless, the elevated levels of ubiquitin conjugates upon K0 Ub perturbation may serve as a means to trap targets in a relatively stable ubiquitination state.
What are the ubiquitinated proteins trapped as K0 Ub conjugates? Among the “very high scores” (ratio >4.5) listed in supplemental Table S1, one can find several membrane-associated proteins involved in stress response. Some notable examples include Dre2 that may prevent mitochondria induced cell death by shielding Tah18 in absence of stress (85), proteins involved in bud selection, cell fusion or vesicular transport (Bud1, Bud24, Gcs1), and membrane-associated proteins involved in signaling of stress responses (Opy2, Gis4, YML131W). Several proteasome subunits such as β3 (Pup3) or α3 (Pre9), and ubiquitin conjugating enzymes such as Cdc34 were also found heavily enriched with K0 Ub (though why they accumulate in ubiquitinated form in response to K0 Ub induction is somewhat of an enigma). At the other extreme, some proteins that are severely down regulated upon expression of K0 Ub include the putative RING finger E3 ligase Pep3, a component of CORVET tethering complex that promotes vesicular docking/fusion reactions in conjunction with SNARE proteins (86), She2, an RNA-binding protein that regulates mRNA localization (87), and RFC2, a subunit of the replication factor that loads the sliding clamp PCNA onto DNA templates (88).
Naturally, the next question we addressed was whether these targets represent a random selection of naturally ubiquitinated substrates, or does K0 Ub preferentially perturb certain biological pathways (reflected in the preferred targets such as those listed above). Using the AmiGo program (75) to curate the list of proteins enriched by ubiquitination of K0 Ub, we found that a substantial number of high ratio targets cluster in a limited number of biological pathways, many of which pertain to protein sorting or trafficking, or to ubiquitin-dependent proteolysis (supplemental Table S5). Of proteins associated with K0 Ub, the relative portion that these categories make up is higher than the portion they account for among all known ubiquitinated proteins (Fig. 3E, supplemental Table S6). Members of these categories were enriched specifically in their ubiquitinated state indicating that these proteins are particularly prone to ubiquitination by K0 Ub (Fig. 3F). We conclude that K0 Ub differentiates between targets and leads to increased ubiquitination of a subset of proteins associated with membrane processes, vesicle mediated transport, actin cytoskeleton, and ubiquitin-proteasome dependent proteolysis (Tables I, II).
Table I. UPS or UTS associated targets identified as preferentially ubiquitinated with K0 Ub. Ubiquitinated proteins enriched in K0 Ub expressing cells from supplemental Table S1 that are documented to be associated with ubiquitin, either as ubiquitin proteasome system components and substrates, or as ubiquitin trafficking system components. Light-to-Heavy (L/H) ratio represents abundance of each target in K0 Ub-expressing cells relative to WT. Protein description and links to protein trafficking or Rsp5 are based on SGD http://www.yeastgenome.org/.
| Systematic name | Protein | Ratio L/H | Protein description | Role in protein trafficking/sorting | Rsp5 association |
|---|---|---|---|---|---|
| YER094C | PUP3 | 14.1 | Beta subunit of the 20S proteasome involved in ubiquitin-dependent catabolism | ||
| YJL151C | SNA3a | 8.4 | Integral membrane protein has a possible role in either cell wall synthesis or protein-vacuolar targeting | + | + |
| YDR054C | CDC34 | 4.8 | Ubiquitin-conjugating enzyme (E2) and catalytic subunit of SCF ubiquitin-protein ligase complex | ||
| YPL084W | BRO1 | 4.7 | Class E vacuolar protein sorting factor that coordinates deubiquitination | + | + |
| YBL058W | SHP1 | 4.6 | UBX domain-containing protein that regulates Glc7p phosphatase activity | + | |
| YNR006W | VPS27 | 4.4 | Endosomal protein that forms a complex with Hse1p; required for recycling Golgi proteins | + | + |
| YML097C | VPS9 | 4.3 | A guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport | + | |
| YOR042W | CUE5 | 4.2 | Protein containing a CUE domain that binds ubiquitin, which may facilitate intramolecular monoubiquitination | + | + |
| YDL126C | CDC48 | 4.1 | ATPase in ER, nuclear membrane and cytosol | + | |
| YMR275C | BUL1 | 3.9 | Ubiquitin-binding component of the Rsp5p E3-ubiquitin ligase complex | + | + |
| YFL009W | CDC4 | 3.9 | F-box protein required for G1/S and G2/M transition | ||
| YDR320C | SWA2 | 3.9 | Auxilin-like protein involved in vesicular transport | + | |
| YOL038W | PRE6 | 3.9 | 20S proteasome alpha-type subunit | ||
| YEL037C | RAD23 | 3.9 | Protein with ubiquitin-like N terminus, recognizes and binds damaged DNA | ||
| YDR177W | UBC1 | 3.7 | Ubiquitin-conjugating enzyme (E2) | ||
| YOR261C | RPN8 | 3.7 | Essential, non-ATPase regulatory subunit of the 26S proteasome | ||
| YDR092W | UBC13 | 3.7 | Ubiquitin-conjugating enzyme (E2) | ||
| YDL161W | ENT1 | 3.4 | Epsin-like protein involved in endocytosis and actin patch assembly | + | + |
| YLR421C | RPN13 | 3.3 | Subunit of the 19S regulatory particle of the 26S proteasome lid | ||
| YEL012W | UBC8 | 3.2 | Ubiquitin-conjugating enzyme (E2) | ||
| YJL172W | CPS1 | 3.2 | Vacuolar carboxypeptidase yscS | + | + |
| YGR135W | PRE9 | 3.1 | 20S proteasome beta-type subunit | ||
| YOR018W | ROD1/ART4 | 3.0 | Membrane protein; contains 2 PY motifs, which are required for Rod1p interaction with Rsp5p | + | + |
| YCR030C | SYP1 | 2.8 | Protein with a potential role in actin cytoskeletal organization | + | |
| YOR322C | LDB19/ART1/CVS7 | 2.8 | Protein involved in regulating the endocytosis of plasma membrane proteins | + | + |
| YKL213C | DOA1 | 2.7 | WD repeat protein required for ubiquitin-mediated protein degradation | ||
| YBL101C | ECM21/ART2 | 2.7 | Protein involved in regulating the endocytosis of plasma membrane proteins | + | + |
| YLR392C | ART10 | 2.7 | Protein of unknown function that contains 2 PY motifs and is ubiquinated by Rsp5p | + | + |
| YLR206W | ENT2 | 2.7 | Epsin-like protein required for endocytosis and actin patch assembly | + | + |
| YFR010W | UBP6 | 2.6 | Ubiquitin-specific protease situated in the base subcomplex of the 26S proteasome | ||
| YGR068C | ART5 | 2.6 | Protein proposed to regulate the endocytosis of plasma membrane proteins | + | + |
| YER125W | RSP5 | 2.6 | Ubiquitin-protein ligase (E3) | + | + |
| YJL084C | ALY2/ART3 | 2.5 | Protein proposed to regulate the endocytosis of plasma membrane proteins | + | + |
| YKL145W | RPT1 | 2.4 | One of six ATPases of the 19S regulatory particle of the 26S proteasome | ||
| YBR082C | UBC4 | 2.4 | Ubiquitin-conjugating enzyme (E2) | ||
| YHR200W | RPN10 | 2.4 | Non-ATPase base subunit of the 19S regulatory particle of the 26S proteasome | + | |
| YBL057C | PTH2 | 2.2 | Mitochondrially-localized peptidyl-tRNA hydrolases; negatively regulates the ubiquitin-proteasome pathway | ||
| YDL007W | RPT2 | 2.2 | One of six ATPases of the 19S regulatory particle of the 26S proteasome | ||
| YBR058C | UBP14 | 2.2 | Ubiquitin-specific protease that specifically disassembles unanchored ubiquitin chains | ||
| YOR124C | UBP2 | 2.2 | Ubiquitin-specific protease that removes ubiquitin from ubiquitinated proteins | + | |
| YNL243W | SLA2 | 2.1 | Transmembrane actin-binding protein involved in membrane cytoskeleton assembly and cell polarization | + | + |
| YDR388W | RVS167 | 2.1 | Actin-associated protein, interacts with Rvs161p to regulate actin cytoskeleton, endocytosis, and viability | + | + |
| YGL045W | RIM8/ART9 | 2.1 | Protein involved in proteolytic activation of Rim101p in response to alkaline pH | + | + |
| YMR304W | UBP15 | 2.0 | Ubiquitin-specific protease that may play a role in ubiquitin precursor processing | ||
| YHR108W | GGA2 | 2.0 | Golgi-localized protein with homology to gamma-adaptin | + | |
| YML111W | BUL2 | 2.0 | Component of the Rsp5p E3-ubiquitin ligase complex, involved in intracellular amino acid permease sorting | + | + |
| YGR048W | UFD1 | 2.0 | Involved in recognition of polyubiquitinated proteins and their presentation to the 26S proteasome | ||
| YKL010C | UFD4 | 1.9 | Ubiquitin-protein ligase (E3) | + | |
| YBL047C | EDE1 | 1.9 | Key endocytic protein involved in a network of interactions with other endocytic proteins | + | + |
| YML013W | UBX2/SEL1 | 1.9 | Protein involved in ER-associated protein degradation | + | |
| YHR027C | RPN1 | 1.6 | Non-ATPase base subunit of the 19S regulatory particle of the 26S proteasome | ||
| YPR103W | PRE2 | 1.6 | 20S proteasome beta-type subunit, responsible for the chymotryptic activity of the proteasome | ||
| YDL190C | UFD2 | 1.6 | Ubiquitin chain assembly factor (E4) | + | |
| YDL097C | RPN6 | 1.6 | Essential, non-ATPase regulatory subunit of the 26S proteasome | ||
| YIL075C | RPN2 | 1.5 | Subunit of the 26S proteasome | ||
| YDL140C | RPB1 | 1.5 | RNA polymerase II largest subunit B220, part of central core | + |
a Identified by one peptide only.
Table II. Additional trafficking or membrane associated proteins preferentially ubiquitinated with K0 Ub. Ubiquitinated proteins enriched in K0 Ub expressing cells from supplementary Table S1 that are associated with protein sorting, trafficking, or membrane-associated processes, but not included in Table I. Light-to-Heavy (L/H) ratio represents abundance of each target in K0 Ub-expressing cells relative to WT. Protein descriptions are based on SGD http://www.yeastgenome.org.
| Systematic name | Protein | Ratio L/H | Protein description |
|---|---|---|---|
| YMR316W | DIA1 | 12.2 | Protein of unknown function, involved in invasive and pseudohyphal growth, target of calcineurin signaling |
| YDL226C | GCS1 | 6.1 | ADP-ribosylation factor GTPase activating protein (ARF GAP), involved in ER-Golgi transpor |
| YDR425W | SNX41 | 5.1 | Sorting nexin, involved in the retrieval of late-Golgi SNAREs |
| YLR219W | MSC3 | 3.9 | Protein of unknown function; may be a component of eisosomes |
| YDL203C | ACK1 | 3.5 | Protein that functions upstream of Pkc1p in the cell wall integrity pathway |
| YOR171C | LCB4 | 3.5 | Sphingoid long-chain base kinase, responsible for synthesis of long-chain base phosphates |
| YJR125C | ENT3 | 3.0 | Protein containing an N-terminal epsin-like domain involved in clathrin recruitment and traffic |
| YGR086C | PIL1 | 2.9 | Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis |
| YOR109W | INP53 | 2.8 | Polyphosphatidylinositol phosphatase, dephosphorylates multiple phosphatidylinositols |
| YMR079W | SEC14 | 2.7 | Phosphatidylinositol/phosphatidylcholine transfer protein |
| YNL054W | VAC7 | 2.6 | Integral vacuolar membrane protein involved in vacuole inheritance and morphology |
| YAL002W | VPS8 | 2.6 | Membrane-associated protein that interacts with Vps21p to facilitate soluble vacuolar protein localization |
| YGR136W | LSB1 | 2.4 | Protein containing an N-terminal SH3 domain |
| YGR130C | YGR130C | 2.4 | Putative protein of unknown function; possible component of the eisosome |
| YIL041W | GVP36 | 2.1 | BAR domain-containing protein that localizes to both early and late Golgi vesicles |
| YDL029W | ARP2 | 2.1 | Essential component of the Arp2/3 complex, which is a highly conserved actin nucleation center |
| YPL195W | APL5 | 2.0 | Delta adaptin-like subunit of the clathrin associated protein complex (AP-3) |
| YJL154C | VPS35 | 1.9 | Endosomal subunit of membrane-associated retromer complex required for retrograde transport |
| YKL212W | SAC1 | 1.8 | Phosphatidylinositol phosphate (PtdInsP) phosphatase involved in hydrolysis of PtdIns[4]P |
| YCR088W | ABP1 | 1.8 | Actin-binding protein of the cortical actin cytoskeleton |
| YDR129C | SAC6 | 1.8 | Fimbrin, actin-bundling protein; cooperates with Scp1p in the organization of the actin cytoskeleton |
| YNL044W | YIP3 | 1.7 | Protein localized to COPII vesicles, proposed to be involved in ER to Golgi transport |
| YGR261C | APL6 | 1.7 | Beta3-like subunit of the yeast AP-3 complex |
| YEL013W | VAC8 | 1.7 | Phosphorylated and palmitoylated vacuolar membrane protein, required for cytoplasm-to-vacuole targeting |
| YPR139C | VPS66 | 1.6 | Cytoplasmic protein of unknown function involved in vacuolar protein sorting |
| YFR051C | RET2 | 1.6 | Delta subunit of the coatomer complex (COPI), which coats Golgi-derived transport vesicles |
| YFL039C | ACT1 | 1.6 | Actin, structural protein involved in cell polarization, endocytosis, and other cytoskeletal functions |
| YER143W | DDI1 | 1.5 | DNA damage-inducible v-SNARE binding protein, contains a ubiquitin-associated (UBA) domain |
| YBR080C | SEC18 | 1.5 | ATPase required for vesicular transport between ER and Golgi |
| YML048W | GSF2 | 1.5 | ER localized integral membrane protein that may promote secretion of certain hexose transporters |
| YKR001C | VPS1 | 1.5 | Dynamin-like GTPase required for vacuolar sorting; also involved in actin cytoskeleton organization |
| YCR009C | RVS161 | 1.5 | Amphiphysin-like lipid raft protein; regulates polarization of the actin cytoskeleton, endocytosis, cell polarity |
| YPR029C | APL4 | 1.5 | Gamma-adaptin, large subunit of the clathrin-associated protein (AP-1) complex |
One of the interesting targets is Rsp5, an E3 ubiquitin ligase whose substrates include many membrane proteins (41), components of vesicular trafficking machinery (89), and proteins in connection with the actin cytoskeleton (90, 91). Rsp5 function is mediated via a series of substrate specific adaptors such as Art1–Art10 and Bul1, Bul2 (45–47, 92, 93). Rsp5-associated proteins—adaptors as well as substrates—show up prominently in our list of targets preferentially ubiquitinated by K0 Ub (Tables I, II). Among Rsp5-ubiquitinated targets, our list uncovers even substrates for proteasome dependent degradation, such as Rbp1 (94). With the exception of Rsp5 related proteins, other biological categories (such as transcriptional regulation, metabolic enzymes, protein translation, and cell cycle) did not appear to be altered by K0 Ub expression. This was not a self-evident observation as K0 Ub is often thought to serve as a nondiscriminatory chain terminator, expected to lead to pervasive substrate stabilization (79). Observations presented in the current study suggest that in vivo, K0 mutant ubiquitin dissects the ubiquitin system, perturbing some branches more than others.
K0 mutant Ub is Preferentially Linked to Lys63 Chains
A nonrandom effect of K0 Ub expression on the identity and steady state levels of ubiquitinated proteins (Fig. 3) raises the question whether this is the outcome of a general effect on the nature of ubiquitin modifications (polyUb chain length, for instance), or whether K0 Ub also has a nonrandom effect on the ubiquitin landscape (by altering the ratio of modification types). Prior to analysis of chains directly linked to K0 Ub, we looked at the broader effects on the ubiquitin profile in WCE. Lys48 and Lys63 were prevalent in the high MW ubiquitin-rich region of mixed SILAC samples (>130kDa; Fig. 4A) in agreement with determination based on synthetically labeled standards (AQUA analysis; Figs. 1 and 2). Induction of K0 Ub had a similar effect on the levels of both of these linkages, suggesting that in the high MW region their relative ratio to one another remained stable. Using a mass-inclusion list for all potential tryptic peptides derived from ubiquitin (including the seven linkage-signature peptides depicted in supplemental Fig. S6) positively identified GGLys 48 and GGLys63 signals in two adjacent MW slices (67–90 and 90–130 kDa; Fig. 4A). In one slice, around 100 kDa, Lys63 linkages increased slightly upon expression of K0 Ub. This analysis also confirmed that K0 Ub induction led to a 2.5- to fourfold increase in total ubiquitin conjugates relative to WT expressing cells (Fig. 4B). This increase was mainly because of accumulation of nonextended monoubiquitin modifications (Fig. 4C), supporting independent observations by Ub AQUA (Fig. 2).
Fig. 4.
Distribution of K0 Ub with ubiquitin chain-linkages. A, The ratios of the main Ub linkages, Lys48 and Lys63 in whole cell extract (K0/WT Ub) were determined by SILAC according to MW regions. For MW regions <130 kDa, Orbitrap detection of signature peptides (supplemental Fig. S6) was performed with a mass inclusion list. B, Ratio of total conjugated Ub in cells expressing K0 Ub relative to cells expressing WT Ub based on the MS intensity ratio of the tryptic peptide EGIPPDQQR (see explanation in supplemental Fig. S6). C, Penetration of K0 Ub into the ubiquitin landscape according to MW. Ubiquitin in the SILAC cells comes from two sources: endogenous (WT) and expressed tagged ubiquitin (either WT or K0). The ratio of extendable Ub (derived from peptide TITLEVESSDTIDNVK) to total ubiquitin (derived from peptide EGIPPDQQR) between strains provides a measure of K0 Ub penetration (details in supplemental Fig. S6). D, Main ubiquitin linkages pulled out with tagged ubiquitin. Bars reflect ratio of linkages associated with tagged-K0 Ub relative to tagged-WT Ub in each MW region. K0 Ub is preferentially associated with Lys63 linkages over Lys48 in affinity purified chains.
Utilizing SILAC ratios of multiple tryptic peptides of ubiquitin allowed an estimation of K0 Ub penetration into the ubiquitin landscape over the entire MW span (Fig. 4C). The highest incorporation of K0 Ub into conjugates was detected in very high MW and in the low-mid range MW regions. Penetration of K0 Ub into these conjugates nears 75%, similar to independent estimates by Ub AQUA (supplemental Fig. S3). Because K0 Ub serves as a chain terminator, the lower MW fractions likely represent monoubiquitinated substrates or those modified by very short chains, whereas high-MW region conjugates probably reflect multiple-monoubiquitinated targets as well as K0 Ub capping long chains. K0 Ub was also enriched in the free pool of unanchored ubiquitin (<15kDa) implying possible difficulties in utilization.
Ubiquitin itself is also a substrate for ubiquitination, a property that allows trapping of chains that are physically correlated with K0 Ub, either as a chain terminator or as modifiers of the same substrate (on different lysines). SILAC-based analysis of linkage types pulled out with affinity tagged ubiquitin (K0 versus WT), revealed that Lys63 chains are two times more likely than Lys48 chains to be associated with targets of K0 Ub; this is particularly so in the low-mid range MW (Fig. 4D). At this stage we cannot distinguish between a situation in which K0 Ub caps short Lys63 chains from that whereby K0 Ub and Lys63 chains are conjugated to separate lysines on a common substrate on. As a side note, we were unable to identify any Lys63 linkages in unanchored di- or tri-ubiquitin chains (low MW fractions of Fig. 4D). In conclusion, K0 Ub seems to associate with a subset of substrates, those that are also linked to short Lys63 chains. The next question we asked was “how does modification by K0 Ub influence the fate of these substrates?”.
K0 Mutant Ub Interferes With Protein Sorting and Localization but not with Proteasome-dependent Proteolysis
Simultanous accumulation of ubiquitinated proteins alongside changes to the cellular ubiquitination pattern upon K0 Ub expression posits that both outcomes reflect the altered fate of modified targets. A logical explanation would be that proteasome substrates evade degradation and accumulate in cellulo because of the nonoptimal signal embodied by K0 Ub. To evaluate the direct effect of K0 Ub on ubiquitin-proteasome dependent degradation, we measured the degradation rates of two ubiquitin-proteasome substrates. MyoD is a short-lived transcription factor (95) whose degradation in yeast is strictly dependent on Lys48-linked polyubiquitin (96). Overexpression of lysineless ubiquitin caused no delay in MyoD turnover (Fig. 5A). Likewise, an endogenous yeast cyclin for the CDK Pho85, Pcl5, is turned over by the E3 ubiquitin ligase complex SCFGrr1 (97). Once again, overexpression of K0 Ub caused no delay in Pcl5 degradation (Fig. 5A).
Fig. 5.
Induction of K0 Ub interferes with protein sorting more than it impacts ubiquitin-proteasome dependent degradation. A, Influence of K0 Ub induction on known ubiquitin-proteasome substrates. Cellular stability of ectopically expressed MyoD or Pcl5 in the presence of either WT or K0 Ub was monitored by addition of cycloheximide to exponentially growing yeast cells. Aliquots were taken at the indicated time points after addition of cycloheximide and analyzed by immunoblotting with anti-MyoD (top panel) or anti-Pcl5 (bottom) antibodies, and anti-PGK serving as loading control. B, Expression of K0 Ub increases the ubiquitination levels of Syp1. Cells expressing Syp1-GFP together with either RGS-His8-K0 Ub or RGS-His8-WT Ub were lysed and loaded onto a Ni-NTA column for isolation of ubiquitin conjugates. Syp1 content in whole cell extracts (WCE) and eluate of isolated ubiquitin conjugates (El) were analyzed by immunoblotting with anti-GFP antibodies. C, Expression of K0 Ub alters the ubiquitination pattern of Sna3. Cells expressing Sna3–6HA together with either WT or K0 Ub were lysed, immunoprecipitated with anti-HA antibodies, and analyzed for ubiquitinated Sna3 species. Immunoblotting the immunoprecipitate with anti-HA identified unmodified Sna3 as well as modified higher MW forms (left). Immunoblotting with anti His-tag confirmed the presence of K0 Ub in high MW conjugates of ubiquitinated Sna3 (right). D, Cells expressing K0 Ub display typical phenotypes of defected endocytosis: sensitivity to canavanine but resistance to nickel ions. Ten-fold serial dilutions of cells expressing either WT or K0 Ub were spotted onto selective medium (control) or media supplemented with either canavanine (1 μg/ml) or NiCl2 (1.5 mm), and grown at 30°. E, Expression of K0 Ub partially impairs MVB sorting of GFP-Phm5 and Sit1-GFP. Cells expressing GFP-Phm5 together with K0 or WT Ub were grown to midexponential phase in selective medium (carbon source glucose). Intracellular localization of GFP-Phm5 was examined by fluorescence microscopy. GFP-Phm5 localizes to vacuolar lumen in WT cells at steady state, whereas accumulation at vacuolar membrane periphery and endosomes occurs upon K0 Ub induction. Similar monitoring of Sit1 was performed after 1 h induction with galactose for the expression of Sit1-GFP. For this target too, K0 Ub interferes with vacuolar lumen sorting. F, Expression of K0 Ub partially impairs plasma membrane internalization and MVB sorting of the Jen1 transporter. Cells encoding for Jen1-GFP were transformed with either K0 or WT Ub. Cells were induced in lactic acid for 4 h for expression and plasma membrane targeting of Jen1-GFP. GFP fluorescence was monitored before (t = 0) and 30 min after (t = 30′) the addition of 2% glucose, which triggers endocytosis and vacuolar targeting of Jen1. Within 30 min, all GFP fluorescence was detected within vacololar lumen, whereas ∼10% of K0 Ub expressing cells retained vacuolar membrane and faint plasma membrane staining.
Even though high cellular abundance of K0 Ub did not seem to interfere with ubiquitin-proteasome dependent degradation (Fig. 5A), it did lead to accumulation of a substantial number of proteins involved in membrane-associated processes (Table I, Table II, Figs. 3E, 3F). One such protein that was not reported to be modified by ubiquitin is Syp1, a conserved protein involved in regulation of endocytic pathway initiation (98–100). Tagged WT or K0 Ub were separately affinity purified from cells expressing genomically tagged Syp1 and immunoblotted for Syp1 (Fig. 5B). More monoubiquitinated-Syp1 was isolated from K0 Ub expressing cells. Thus although not documented before, ubiquitination of Syp1 is confirmed in this study by two independent methods: SILAC of affinity “Ub-pullout” samples, and by immunoblotting for ubiquitinated forms of Syp1. As mentioned above, K0 Ub emerges as a practical approach to trap otherwise difficult-to-detect ubiquitin conjugates. Another protein, Sna3, was previously identified as an MVB cargo targeted to the vacuole upon modification by Lys63 chains on a single target lysine (101). A general accumulation of ubiquitinated Sna3 was visualized (using antibodies for both anti-Ub and anti-Sna3 epitopes) upon K0 Ub expression. Interestingly, even when modified with K0 Ub, a prominent mono-ubiquitinated band of Sna3 was present at the base of a ladder extending to higher- and even much higher-MW forms. Notably, Sna3 was identified by a single peptide in the ubiquitin pullout (supplemental Table S1; Table I) and therefore was not considered as a high confidence hit in our proteomic screen (supplemental Table S3), yet immunoblotting validated the status of Sna3 as a bona fide target of ubiquitination (Fig. 5C). A focus on an individual substrate (Fig. 5C) confirms that multiple ubiquitin modifications occur per target even in presence of K0 Ub; a similar pattern is seen also in the general ubiquitinome of K0 Ub conjugates migrating as high-MW forms (Fig. 2, supplemental Fig. S4).
We may now ask whether the perturbed ubiquitination pattern of MVB cargos (such as Sna3) represents a defect in endocytosis or downstream trafficking. Many membrane-bound proteins (receptors, transporters) are internalized at the plasma membrane, sorted to early endosomes, and then to late endosomes, where they meet proteins arriving from the Golgi apparatus by the VPS (vacuolar protein sorting) pathway. Endocytosed proteins are then either sorted to internal vesicles of late endosomes/multivesicular bodies and delivered to the vacuole for degradation, or recycled back to the plasma membrane. This trafficking pathway includes two ubiquitin-dependent steps involving the E3 ubiquitin ligase, Rsp5: internalization (102), and MVB sorting (41). A defect in either of these ubiquitin-dependent events results in accumulation of plasma membrane proteins in the plasma membrane (103). One such protein is Can1, an amino acid transporter of arginine that can also import the toxic analog canavanine. As a result, cells defective in Can1 internalization, or MVB sorting, are particularly sensitive to canavanine (46, 104). Indeed, expression of K0 Ub was sufficient to cause sensitivity to canavanine (Fig. 5D), indicative of elevated levels of the Can1 transporter at the plasma membrane. However, as sensitivity to canavanine may also be an outcome of other regulatory malfunctions, we assayed for Nickel ion resistance, another phenotype displayed by many mutants in MVB sorting or endosomes-to-Golgi transport (105, 106). In this assay, expression of K0 Ub provided some resistance to Ni2+ (Fig. 5D), once again pointing to a general misregulation of protein traffic networks.
In order to follow the effect of K0 Ub on intracellular trafficking directly, we checked the fate of several GFP-tagged membrane-bound proteins that are targeted to the vacuole, either from the Golgi apparatus, or from the plasma membrane. The vacuolar resident, Phm5, is a polyphosphatase that reaches its destination via MVB sorting, apparently in an Rsp5-dependent manner (107, 108). In WT cells GFP-Phm5 accumulated inside vacuoles (Fig. 5E; left). In contrast, in some K0 Ub expressing cells, GFP-Phm5 resided at the vacuolar membrane staining the periphery instead of the lumen; this being a typical phenotype of MVB sorting defects (Fig. 5E; left). Another example is the siderophore transporter Sit1, which in the absence of its natural substrates is sorted from the Golgi apparatus to the vacuole (rather than to the plasma membrane), again upon Lys63-linked ubiquitin-modification (70). Whereas Sit1-GFP was efficiently imported into vacuolar lumen in all WT cells, it was largely localized at the vacuolar rim in some K0 Ub expressing cells (Fig. 5E; right). Mislocalization of these targets was observed in a limited number of K0 Ub expressing cells, however it was reproducible, and moreover, not a single case of protein mislocalization was observed in the control WT cells. Impaired sorting resulting in delivery of these targets to the vacuolar membrane instead of the vacuolar lumen is a typical phenotype of defects in MVB sorting machinery (the ESCRT proteins) or in ubiquitination of cargo (109).
In addition to these two examples of MVB cargoes, we also followed the fate of an endocytic cargo in cells expressing K0 Ub. Growth phenotypes associated with K0 Ub expression (Fig. 5D) may imply an interference with proper internalization or MVB sorting of such targets. The monocarboxylate transporter, Jen1, resides in the plasma membrane, yet when cells are exposed to preferred carbon sources such as glucose, Jen1 is rapidly internalized and degraded within the vacuole (33). This dynamic process can be monitored upon addition of glucose to Jen1-GFP expressing cells (Fig. 5F). In some K0 Ub expressing cells, Jen1-GFP appeared to start the internalization process but displayed defective MVB sorting as judged from vacuolar membrane and endosomes enhanced fluorescence rather than complete accumulation in lumen (Fig. 5F; right). Some K0 Ub expressing cells even still displayed residual plasma membrane staining (Fig. 5F and data not shown). K0 Ub appears to delay endocytosis.
DISCUSSION
In this study we have charted the cellular ubiquitin landscape, focusing on the various modes of ubiquitin conjugates, and ubiquitin-in-conjugates. To take a broadest possible snapshot of the landscape, we refined a protocol for rapid lysis of cells under denaturing conditions without involving further purification steps. This approach enabled us to grasp a representative spectrum of conjugated ubiquitin. The ubiquitin signal is not limited to polyUb chains, but encompasses a variety of chain lengths and linkage types distributed throughout the entire MW range, from unanchored molecules to extremely high MW conjugates (Figs. 1, 2, 4, supplemental Figs. S1, S2, S4). Of polyUb modifications, the most prevalent linkages that we detected in whole cell extract were via Lys48 and Lys63. This was particularly evident for high MW conjugates. Together these linkages make up close to half of all conjugated ubiquitin, with the bulk of the remainder going to monoUb (and chain terminating endcaps). Very few studies have charted the linkage profile in whole cell extract without including any enrichment steps. One recent study (76) identified Lys48 and Lys63 as the primary and secondary linkages in high MW conjugates of mammalian extract. A range of linkage distributions has been reported when isolating conjugates to enrich for the ubiquitin content. In one such case, Lys48 and Lys63 made up 90% of chains in conjugates isolated from Drosophila neuronal tissue (110). In another case, ubiquitin captured from mouse brain showed the highest content to be of Lys48 linkages, followed by lower levels of Lys63 and Lys11 (111). Conjugates isolated from urea-denatured yeast lysate identified Lys11 as the second most abundant linkage making up over a quarter of chains and nearing the abundance of the majority type, Lys48 (8). Lys11 of ubiquitin was shown to play a role in ERAD (8) and in cell cycle progression (51, 112), suggesting that levels of Lys11 conjugates cyclically fluctuate. The relatively higher abundance of Lys11 found in lower MW samples of unsynchronized cells (supplemental Fig. S2) or of specifically ubiquitinated samples (76) indicates that many Lys11 targets may be small proteins or modified by short chains. Considering these studies together, the ubiquitin linkage profile seems to be extremely sensitive to experimental conditions such as culture medium, growth phase, lysis conditions, and sample handling postlysis. Of the conditions tested herein, TCA-SDS lysis provided the most consistent results and highest levels of all linkages captured. Using these conditions we were able to capture high levels of Lys48 and particularly high levels of Lys63 chains (Fig. 1), which may partially explain the low portion (though not necessarily low levels) of “alternative” ubiquitin chains.
In the high-MW region, where we find the highest ubiquitin-to-protein ratio, the most striking observation is the prevalence of nonextended ubiquitin molecules (Fig. 1). These consist of both monoubiquitin modifications and chain terminators (endcap) implying that average chain length is short (either a majority of short chains or a mixture of few long chains among multiple-monoUb modifications). Monoubiquitination on targets may represent intermediate steps in chain elongation, intermediate steps in polyUb disassembly, or a specific stand-alone signal. Additionally, there is also the option of branched chains (61, 62) which are impossible to analyze using current MS protocols and therefore may account for a portion of ubiquitin-in-conjugates (adding multiple endcaps per chain). Modifications by multiple mono-ubiquitin on a single substrate were shown to sustain proteasomal processing in vitro (113). Mono-ubiquitin has even been shown to target substrates to the 26S proteasome for degradation (albeit at lower efficiency) (114, 115), suggesting that the cell may be able to tolerate a certain level of premature chain-terminations or short chains. Involvement of monoubiquitin has also been documented as a signal for endocytosis and intracellular sorting of proteins (3, 24, 25, 116), though the relative portion of such monomeric modifications has not been quantified. Moreover, attributing a distinct, unique, signal to monoubiquitin has been tricky as many substrates for which a role of monoubiquitination has been documented, are also found modified by polyUb chains, particularly Lys63 linked chains (29, 40).
Ubiquitin mutants in which arginine has been substituted for one or more of the lysine residues on ubiquitin have been used extensively to tilt the ubiquitin landscape by preventing/promoting polymerization of certain chain linkages over others (21, 55, 117, 118). In the extreme case, nonextendable lysineless Ub (K0 Ub) has been used as a chain terminator to probe the fate of monoubiquitin conjugates (53, 64, 79, 113). Because decreasing average chain length has generally been assumed to be toxic to cells, most studies using K0 Ub were performed in vitro, or in cell free extract. In the current study we assessed the overall effect of K0 Ub induction over WT background and correlated changes to the ubiquitin signal with changes to the population of ubiquitinated proteins. We deliberately expressed K0 Ub from a mild promoter so as not to overwhelm the ubiquitin pool with the mutant ubiquitin. Interestingly, even though K0 Ub consisted of more than half of the ubiquitin molecules in the cells (supplemental Fig. S3, Fig. 4C), the resulting stress on growth rates and viability was minimal. K0 Ub was utilized by the ubiquitination machinery to modify proteins, increasing the portion of monoubiquitin from 50% to 80% of total signal in high MW conjugates, reflecting multiple modifications per target (Fig. 2). Even when K0 Ub literally replaces all available cellular ubiquitin (supplemental Fig. S4), the ubiquitination processes continued generating substrates in heavily ubiquitinated forms, which are improperly targeted and hence accumulate. Over time, the accumulation of these mutant ubiquitin-conjugates eventually leads to cell death.
Analysis of conjugates preferentially tagged with K0 Ub drew a correlation between K0 Ub and Lys63 linkages (Fig. 4). The implication being that K0 Ub was not evenly distributed throughout the ubiquitinome, but was more likely to be found on targets that were also modified with Lys63 chains (Fig. 3, 4). In particular, K0 Ub interfered more with the nonessential pathways of protein internalization and sorting rather than with the essential pathways of cell cycle, transcriptional regulation, protein synthesis or general metabolism. Such a skewing of ubiquitin between pathways and target populations was nontrivial, as we initially assumed that K0 Ub would serve as a general chain terminator, equally capping chains of all linkage types. Based on results presented in this study, we propose that K0 Ub concentrates primarily on proteins associated with membrane processes for protein sorting and trafficking, and to a lesser extent with rapidly turned-over short-lived proteasome targets (Fig. 3, 5, Tables I, II, and supplemental Tables S3 and S5).
Understanding the direct targets and relative roles of monoubiquitination versus Lys63-linked polyubiquitination in endocytosis, protein sorting, and vacuolar targeting is an ongoing endeavor. We wish to raise the possibility, subject to further investigation, that monoubiquitin (or multiple monoubiquitin) is a sufficient signal for most steps in endocytosis and vacuolar targeting but does not represent the preferred signal, particularly so for MVB sorting and late steps of vacuolar trafficking. Overexpression of K0 Ub, which covered 80% of conjugated cellular ubiquitin, triggered a slight defect on internalization of plasma membrane proteins (Fig. 5F), yet interfered to a greater extent with MVB sorting leading to mislocalized vacuolar targets failing to enter the vacuole lumen.
Partially impaired Lys63 modifications on MVB cargoes such as Sna3 (Fig. 5E) might be one reason for their altered fate. Another reason could be a change in the ubiquitinated status of ESCRT proteins. Strikingly, some ESCRT proteins were recovered in the list of enhanced ubiquitinated proteins (Tables I and II). Notable is the case of Vps27 (Hrs in mammals; ESCRT0) and Bro1, the yeast homolog of Alix (119). Ubiquitination of yeast ESCRT proteins has not yet been studied in as great a detail as their mammalian counterparts. Thus, human Hrs was described to undergo ubiquitination by AIP4, one of the human HECT domain ligases homologous to Rsp5. Ubiquitination was proposed to trigger a conformational change of Hrs, influencing its function in MVB cargo sorting (120). STAM (in yeast; Hse1), an Hrs partner, was also described to be ubiquitinated, and to be a substrate of the deubiquitinating enzyme AMSH (121) that specifically disassembles Lys63-linked ubiquitin chains (122). Accumulation of ubiquitinated STAM in AMSH deficient cells was shown to result in defective MVB sorting (121). Likewise, accumulation of ubiquitinated yeast ESCRT proteins induced by K0 Ub expression may impair correct functioning and regulation of the ESCRT machinery. Finding Vps28 as the most significantly diminished protein in the proteome upon induction of K0 Ub coincides with defective MVB sorting (supplemental Table S2). As a member of the ESCRT I complex, low levels of Vps28 would be expected to hinder sorting of proteins into the endosome (MVBs) with some cargo ending up on the outer membrane of the vacuole as seen in Fig. 5.
In a seeming contradiction to the mild effects observed on proteasome-dependent turnover, a large number of ubiquitination machinery components, including proteasome subunits, were found enriched conjugated to K0 Ub (Fig. 4). Particularly notable are Rsp5 adaptors for ubiquitination of plasma membrane proteins (Bul1/2, Arts) and Rsp5 substrates in connection with the actin cytoskeleton, and components of the 26S proteasome (Table I). Rsp5 itself was also recovered among the protein more heavily ubiquitinated in the presence of K0 Ub. This phenomenon suggests that ubiquitin-proteasome components bear the brunt of the stress imposed by nonextendable K0 ubiquitin, possibly as a ubiquitin-landscape sensing response mechanism. In this respect, finding so many Rsp5-associated proteins raises the possibility that phenotypes observed are largely an indirect result of Rsp5 malfunctioning. As detailed above, Rsp5 is heavily involved in regulating trafficking events via Lys63-ubiquitination of endocytic and MVB cargos (41, 46, 71). Actin cytoskeleton organization and Rsp5 are genetically linked via rsp5 mutant phenotypes (123, 124). Indeed, several proteins in connection with actin were already described to undergo Rsp5-dependent ubiquitination (90, 91), but the underlying function is still largely unknown. The actin cytoskeleton is known to play a crucial role in both yeast and mammals in the internalization step of endocytosis (125). Several proteins located at actin patches and/or involved in actin cytoskeleton assembly/disassembly or in actin nucleation and polymerization (Ent1/2, Sla2, Rvs167, Syp1, Arp2, and Act1 itself) were present in the list of proteins preferentially ubiquitinated upon expression of K0 Ub (Tables I, II), and GO analysis identified this category as prone to perturbation (Fig. 3). Of particular interest is the case of Syp1, a protein that interacts with Ede1 and is one of the first proteins arriving at actin patches, the sites of endocytosis (100). Our study emphasizes that ubiquitination and deubiquitination events play subtle roles in the regulation of protein/protein interactions in the process of actin-mediated endocytosis.
Taking into account ubiquitin in all its forms, the resulting ubiquitin landscape seems to be roughly dissected into two main spheres: the ubiquitin proteasome arm (dominated by Lys48-linked polyUb), and the ubiquitin trafficking arm (where Lys63 linkages and monoUb modifications prevail). The two branches of the ubiquitin system are codependent as they both share the pool of free unconjugated ubiquitin, a large portion of which is recycled from conjugates. A profound implication is that inhibition of one branch could tie up ubiquitin in conjugates leading to a shortage of available ubiquitin, and in turn inhibit other processes indirectly. In this respect, the effectiveness of K0 Ub to dissect the ubiquitin landscape into its two arms may serve as a powerful experimental tool to tweak and study specific cellular processes, which may be encouraging for drug discovery efforts.
Acknowledgments
We are indebted to Dan Finley for “pointing us in the right direction.” We warmly acknowledge Tamar Ziv, Ilana Navon (special thanks), Keren Bendelak, Yonit Horev, Sharona Schpund, Rina Zuchman, and Hila Wolf from the Smoller Proteomics Center in the Department of Biology at the Technion for dedicated operation of Mass Spectrometers and help with generation of data.
Footnotes
* This research was funded by grants from ISF Bikura (MHG), Israel-France program in Biophysics from the Israel Ministry of Science (MHG), and a USA-Israel Binational Science Foundation Grant (MHG). Oded Kleifeld was a recipient of an Aly Kaufman postdoctoral fellowship (at the Technion). RHT's laboratory is supported by the Centre National de la Recherche Scientifique (CNRS), Paris 7 University, and by grants from the Association Pour la Recherche Contre le Cancer (ARC, grant 3298), the Ligue contre le Cancer (Comité de Paris) (Ref: RS09/75–26), and from the EU 6th Framework Programme (NoE -Network of Excellence- Role of ubiquitin and ubiquitin-like modifiers in cellular regulation - RUBICON, and the Marie Curie Network Ubiregulators) Zoi Erpapazoglou is a postdoctoral fellow of the RUBICON NoE.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S10.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S10.
1 The abbreviations used are:
- monoUb
- single ubiquitin
- polyUb
- polyubiquitin
- Ub
- ubiquitin
- WCE
- whole cell extract
- SILAC
- stable isotope labeling with amino acids in cell culture
- MW
- molecular weight
- ERAD
- ER-associated degradation.
REFERENCES
- 1. Hicke L., Dunn R. (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 2. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mukhopadhyay D., Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 4. Pickart C. M., Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 5. Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 6. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li W., Ye Y. (2008) Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol. Life Sci. 65, 2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seyfried N. T., Xu P., Duong D. M., Cheng D., Hanfelt J., Peng J. (2008) Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 80, 4161–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirkpatrick D. S., Denison C., Gygi S. P. (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirkpatrick D. S., Weldon S. F., Tsaprailis G., Liebler D. C., Gandolfi A. J. (2005) Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics 5, 2104–2111 [DOI] [PubMed] [Google Scholar]
- 12. Kirkpatrick D. S., Gerber S. A., Gygi S. P. (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35, 265–273 [DOI] [PubMed] [Google Scholar]
- 13. Denison C., Kirkpatrick D. S., Gygi S. P. (2005) Proteomic insights into ubiquitin and ubiquitin-like proteins. Curr. Opin. Chem. Biol. 9, 69–75 [DOI] [PubMed] [Google Scholar]
- 14. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 15. Mayor T., Graumann J., Bryan J., MacCoss M. J., Deshaies R. J. (2007) Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol. Cell Proteomics 6, 1885–1895 [DOI] [PubMed] [Google Scholar]
- 16. Mayor T., Lipford J. R., Graumann J., Smith G. T., Deshaies R. J. (2005) Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol. Cell Proteomics 4, 741–751 [DOI] [PubMed] [Google Scholar]
- 17. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 18. Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matiuhin Y., G. M. H. (2006) Ubiquitin and Ubiquitination: an overview of the Ubiquitin-proteasome system for protein degradation. In: The UPS in the nervous system: from Physiology to Pathology Nova science publishers Inc [Google Scholar]
- 20. Livnat-Levanon N., Glickman M. H. (2010) Ubiquitin-proteasome system and mitochondria - Reciprocity. Biochim. Biophys. Acta [DOI] [PubMed] [Google Scholar]
- 21. Finley D., Sadis S., Monia B. P., Boucher P., Ecker D. J., Crooke S. T., Chau V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cellular Biol. 14, 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madshus I. H. (2006) Ubiquitin binding in endocytosis–how tight should it be and where does it happen? Traffic 7, 258–261 [DOI] [PubMed] [Google Scholar]
- 24. Mosesson Y., Yarden Y. (2006) Monoubiquitylation: a recurrent theme in membrane protein transport. Isr. Med. Assoc. J. 8, 233–237 [PubMed] [Google Scholar]
- 25. Saksena S., Sun J., Chu T., Emr S. D. (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 26. Kodadek T. (2010) No Splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J. Biol. Chem. 285, 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 28. Thomson T. M., Guerra-Rebollo M. (2010) Ubiquitin and SUMO signalling in DNA repair. Biochem. Soc. Trans. 38, 116–131 [DOI] [PubMed] [Google Scholar]
- 29. Chen Z. J., Sun L. J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 30. Kawadler H., Yang X. (2006) Lys63-linked polyubiquitin chains: linking more than just ubiquitin. Cancer Biol. Ther. 5, 1273–1274 [DOI] [PubMed] [Google Scholar]
- 31. Haglund K., Dikic I. (2005) Ubiquitylation and cell signaling. EMBO J. 24, 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duncan L. M., Piper S., Dodd R. B., Saville M. K., Sanderson C. M., Luzio J. P., Lehner P. J. (2006) Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25, 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. (2009) Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 284, 19228–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barriere H., Nemes C., Du K., Lukacs G. L. (2007) Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol. Biol. Cell 18, 3952–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 36. Kamsteeg E. J., Hendriks G., Boone M., Konings I. B., Oorschot V., van der Sluijs P., Klumperman J., Deen P. M. (2006) Short-chain ubiquitination mediates the regulated endocytosis of the aquaporin-2 water channel. Proc. Natl. Acad. Sci. U.S.A. 103, 18344–18349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geetha T., Jiang J., Wooten M. W. (2005) Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell 20, 301–312 [DOI] [PubMed] [Google Scholar]
- 38. Li J. G., Haines D. S., Liu-Chen L. Y. (2008) Agonist-promoted Lys63-linked polyubiquitination of the human kappa-opioid receptor is involved in receptor down-regulation. Mol. Pharmacol. 73, 1319–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Varghese B., Barriere H., Carbone C. J., Banerjee A., Swaminathan G., Plotnikov A., Xu P., Peng J., Goffin V., Lukacs G. L., Fuchs S. Y. (2008) Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol. Cell. Biol. 28, 5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 41. Belgareh-Touzé N., Léon S., Erpapazoglou Z., Stawiecka-Mirota M., Urban-Grimal D., Haguenauer-Tsapis R. (2008) Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 36, 791–796 [DOI] [PubMed] [Google Scholar]
- 42. Kee Y., Muñoz W., Lyon N., Huibregtse J. M. (2006) The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J. Biol. Chem. 281, 36724–36731 [DOI] [PubMed] [Google Scholar]
- 43. Kim H. C., Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., Tanaka K. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Léon S., Haguenauer-Tsapis R. (2009) Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 46. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 47. Shearwin-Whyatt L., Dalton H. E., Foot N., Kumar S. (2006) Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays 28, 617–628 [DOI] [PubMed] [Google Scholar]
- 48. Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E. (2009) K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sims J. J., Cohen R. E. (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirkpatrick D. S., Hathaway N. A., Hanna J., Elsasser S., Rush J., Finley D., King R. W., Gygi S. P. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 [DOI] [PubMed] [Google Scholar]
- 51. Matsumoto M. L., Wickliffe K. E., Dong K. C., Yu C., Bosanac I., Bustos D., Phu L., Kirkpatrick D. S., Hymowitz S. G., Rape M., Kelley R. F., Dixit V. M. (2010) K11-Linked Polyubiquitination in Cell Cycle Control Revealed by a K11 Linkage-Specific Antibody. Mol. Cell [DOI] [PubMed] [Google Scholar]
- 52. Nishikawa H., Ooka S., Sato K., Arima K., Okamoto J., Klevit R. E., Fukuda M., Ohta T. (2004) Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279, 3916–3924 [DOI] [PubMed] [Google Scholar]
- 53. Wu-Baer F., Lagrazon K., Yuan W., Baer R. (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743–34746 [DOI] [PubMed] [Google Scholar]
- 54. Ikeda H., Kerppola T. K. (2008) Lysosomal localization of ubiquitinated Jun requires multiple determinants in a lysine-27-linked polyubiquitin conjugate. Mol. Biol. Cell 19, 4588–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456 [DOI] [PubMed] [Google Scholar]
- 56. Mastrandrea L. D., You J., Niles E. G., Pickart C. M. (1999) E2/E3-mediated assembly of lysine 29-linked polyubiquitin chains. J. Biol. Chem. 274, 27299–27306 [DOI] [PubMed] [Google Scholar]
- 57. Chastagner P., Israël A., Brou C. (2008) AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One 3, e2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5, 461–466 [DOI] [PubMed] [Google Scholar]
- 59. Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 60. Hicke L. (2001) A New Ticket for Entry into Budding Vesicles–Ubiquitin. Cell 106, 527–530 [DOI] [PubMed] [Google Scholar]
- 61. Ziv I., Kleifeld O., Glickman M. (2009) Nonconformity in ubiquitin compliance. EMBO J. 28, 1825–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 63. Uchiki T., Kim H. T., Zhai B., Gygi S. P., Johnston J. A., O'Bryan J. P., Goldberg A. L. (2009) The ubiquitin-interacting motif protein, S5a, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. J. Biol. Chem. 284, 12622–12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 65. Boname J. M., Thomas M., Stagg H. R., Xu P., Peng J., Lehner P. J. (2010) Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic 11, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 67. Volk S., Wang M., Pickart C. M. (2005) Chemical and genetic strategies for manipulating polyubiquitin chain structure. Methods Enzymol. 399, 3–20 [DOI] [PubMed] [Google Scholar]
- 68. Arnason T., Ellison M. J. (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14, 7876–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spence J., Sadis S., Haas A. L., Finley D. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., Belgareh-Touzé N. (2008) Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic 9, 1372–1391 [DOI] [PubMed] [Google Scholar]
- 71. Galan J. M., Haguenauer-Tsapis R. (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16, 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lauwers E., Jacob C., André B. (2009) K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 74. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 75. Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B., Lewis S. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Phu L., Izrael-Tomasevic A., Matsumoto M. L., Bustos D. J., Dynek J. N., Fedorova A. V., Bakalarski C. E., Arnott D., Deshayes K., Dixit V. M., Kelley R. F., Vucic D., Kirkpatrick D. S. (2010) Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010) Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell 143, 951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Winget J. M., Mayor T. (2010) The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell 38, 627–635 [DOI] [PubMed] [Google Scholar]
- 79. Tan J. M., Wong E. S., Kirkpatrick D. S., Pletnikova O., Ko H. S., Tay S. P., Ho M. W., Troncoso J., Gygi S. P., Lee M. K., Dawson V. L., Dawson T. M., Lim K. L. (2008) Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17, 431–439 [DOI] [PubMed] [Google Scholar]
- 80. Han S., Lone M. A., Schneiter R., Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Caviston J. P., Longtine M., Pringle J. R., Bi E. (2003) The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gabriel K., Milenkovic D., Chacinska A., Müller J., Guiard B., Pfanner N., Meisinger C. (2007) Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 365, 612–620 [DOI] [PubMed] [Google Scholar]
- 83. Katzmann D. J., Babst M., Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 84. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 85. Vernis L., Facca C., Delagoutte E., Soler N., Chanet R., Guiard B., Faye G., Baldacci G. (2009) A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS One 4, e4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Peterson M. R., Emr S. D. (2001) The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2, 476–486 [DOI] [PubMed] [Google Scholar]
- 87. Shen Z., St-Denis A., Chartrand P. (2010) Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 24, 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Majka J., Burgers P. M. (2004) The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acids Res. Mol Biol 78, 227–260 [DOI] [PubMed] [Google Scholar]
- 89. Shih S. C., Prag G., Francis S. A., Sutanto M. A., Hurley J. H., Hicke L. (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 22, 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dores M. R., Schnell J. D., Maldonado-Baez L., Wendland B., Hicke L. (2010) The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic 11, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Stamenova S. D., Dunn R., Adler A. S., Hicke L. (2004) The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J. Biol. Chem. 279, 16017–16025 [DOI] [PubMed] [Google Scholar]
- 92. Nikko E., Sullivan J. A., Pelham H. R. (2008) Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep 9, 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nikko E., Pelham H. R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Beaudenon S. L., Huacani M. R., Wang G., McDonnell D. P., Huibregtse J. M. (1999) Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6972–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17, 5964–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sadeh R., Breitschopf K., Bercovich B., Zoabi M., Kravtsova-Ivantsiv Y., Kornitzer D., Schwartz A., Ciechanover A. (2008) The N-terminal domain of MyoD is necessary and sufficient for its nuclear localization-dependent degradation by the ubiquitin system. Proc. Natl. Acad. Sci. U.S.A. 105, 15690–15695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Aviram S., Simon E., Gildor T., Glaser F., Kornitzer D. (2008) Autophosphorylation-induced degradation of the Pho85 cyclin Pcl5 is essential for response to amino acid limitation. Mol. Cell. Biol. 28, 6858–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Reider A., Barker S. L., Mishra S. K., Im Y. J., Maldonado-Báez L., Hurley J. H., Traub L. M., Wendland B. (2009) Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 28, 3103–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Boettner D. R., D'Agostino J. L., Torres O. T., Daugherty-Clarke K., Uygur A., Reider A., Wendland B., Lemmon S. K., Goode B. L. (2009) The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr. Biol. 19, 1979–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Stimpson H. E., Toret C. P., Cheng A. T., Pauly B. S., Drubin D. G. (2009) Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol. Biol. Cell 20, 4640–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Stawiecka-Mirota M., Pokrzywa W., Morvan J., Zoladek T., Haguenauer-Tsapis R., Urban-Grimal D., Morsomme P. (2007) Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic 8, 1280–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Dupré S., Urban-Grimal D., Haguenauer-Tsapis R. (2004) Ubiquitin and endocytic internalization in yeast and animal cells. Biochim. Biophys. Acta 1695, 89–111 [DOI] [PubMed] [Google Scholar]
- 103. Bugnicourt A., Froissard M., Sereti K., Ulrich H. D., Haguenauer-Tsapis R., Galan J. M. (2004) Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins. Mol. Biol. Cell 15, 4203–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Grenson M., Mousset M., Wiame J. M., Bechet J. (1966) Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta 127, 325–338 [DOI] [PubMed] [Google Scholar]
- 105. Ruotolo R., Marchini G., Ottonello S. (2008) Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 9, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Arita A., Zhou X., Ellen T. P., Liu X., Bai J., Rooney J. P., Kurtz A., Klein C. B., Dai W., Begley T. J., Costa M. (2009) A genome-wide deletion mutant screen identifies pathways affected by nickel sulfate in Saccharomyces cerevisiae. BMC Genomics 10, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Reggiori F., Pelham H. R. (2001) Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20, 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hettema E. H., Valdez-Taubas J., Pelham H. R. (2004) Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Katzmann D. J., Odorizzi G., Emr S. D. (2002) Receptor down regulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3, 893–905 [DOI] [PubMed] [Google Scholar]
- 110. Franco M., Seyfried N. T., Brand A. H., Peng J., Mayor U. (2010) A novel strategy to isolate ubiquitin conjugates reveals wide role of ubiquitination during neural development. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 112. Wu T., Merbl Y., Huo Y., Gallop J. L., Tzur A., Kirschner M. W. (2010) UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 107, 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. (2009) Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol. Cell 33, 496–504 [DOI] [PubMed] [Google Scholar]
- 114. Guterman A., Glickman M. H. (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279, 1729–1738 [DOI] [PubMed] [Google Scholar]
- 115. Hershko A., Heller H. (1985) Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 116. Haglund K., Di Fiore P. P., Dikic I. (2003) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 117. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 118. Spence J., Gali R. R., Dittmar G., Sherman F., Karin M., Finley D. (2000) Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102, 67–76 [DOI] [PubMed] [Google Scholar]
- 119. Nikko E., André B. (2007) Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot. Cell 6, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Malik R., Marchese A. (2010) Arrestin-2 interacts with the endosomal sorting complex required for transport machinery to modulate endosomal sorting of CXCR4. Mol. Biol. Cell 21, 2529–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sierra M. I., Wright M. H., Nash P. D. (2010) AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J. Biol. Chem. 285, 13990–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. McCullough J., Clague M. J., Urbé S. (2004) AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kamińska J., Gajewska B., Hopper A. K., Zoladek T. (2002) Rsp5p, a new link between the actin cytoskeleton and endocytosis in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 6946–6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kamińska J., Kwapisz M., Grabińska K., Orlowski J., Boguta M., Palamarczyk G., Zoladek T. (2005) Rsp5 ubiquitin ligase affects isoprenoid pathway and cell wall organization in S. cerevisiae. Acta Biochim. Pol. 52, 207–220 [PubMed] [Google Scholar]
- 125. Galletta B. J., Mooren O. L., Cooper J. A. (2010) Actin dynamics and endocytosis in yeast and mammals. Curr. Opin. Biotechnol [DOI] [PMC free article] [PubMed] [Google Scholar]