Abstract
Human embryonic stem (hES) cells are renewable cell sources that have potential applications in regenerative medicine. The development of technologies to produce permanent and site-specific genome modifications is in demand to achieve future medical implementation of hES cells. We report herein that a baculoviral vector (BV) system carrying zinc-finger nucleases (ZFNs) can successfully modify the hES cell genome. BV-mediated transient expression of ZFNs specifically disrupted the CCR5 locus in transduced cells and the modified cells exhibited resistance to HIV-1 transduction. To convert the BV to a gene targeting vector, a DNA donor template and ZFNs were incorporated into the vector. These hybrid vectors yielded permanent site-specific gene addition in both immortalized human cell lines (10%) and hES cells (5%). Modified hES cells were both karyotypically normal and pluripotent. These results suggest that this baculoviral delivery system can be engineered for site-specific genetic manipulation in hES cells.
Introduction
Permanent transgene expression in human embryonic stem (hES) cells could potentially be beneficial for basic biological studies and regenerative medicine. One possible application involves the expression of intracellular factors to provide additional stimulus to control the differentiation of hES cells. Numerous studies on gene transfer to hES cells have been reported.1,2 Currently, the most efficient methodology to genetically engineer hES cells involves using a viral vector to introduce transgenes into the host genome. However, integrating vectors such as retroviral vectors pose the risk of insertional mutagenesis and oncogene activation.3 The development of a targeting vector that is capable of integrating into predetermined genome sites can be a safer and more desirable approach.
The insect baculovirus Autographa californica multiple nucleopolyhedrovirus has emerged as a promising gene delivery vector in recent years. This DNA virus is capable of entering mammalian cells and expressing transgenes under the control of mammalian promoters.4,5,6 Transduction by baculovirus neither causes observable cytotoxicity at high multiplicity of infection (MOI), nor does it replicate inside mammalian cells, thereby reducing the safety risk.5,6,7,8 Another significant advantage of this double-stranded DNA virus as a vector is the large Autographa californica multiple nucleopolyhedrovirus genome (130 kb), which has been shown to accommodate transgenes of up to 38 kb.9 Recently, baculoviral vectors (BVs) have been shown to be able to transduce human mesenchymal stem cells and hES cells.10,11 These data revealed that BV is a promising and safe alternative gene therapy vehicle as compared to other pathogenic viral vectors.
Zinc-finger nucleases (ZFNs) have been shown to enhance the frequency of gene correction.12,13,14,15,16,17,18 ZFNs are engineered DNA-specific zinc-finger binding proteins fused to a nonspecific DNA endonuclease domain (FokI).14,15,18 Each zinc-finger domain recognizes three base pairs of DNA and a four-finger ZFN can then recognize a DNA sequence of 12 base pairs. The endonuclease domain of FokI must dimerize to cleave the double-stranded DNA sequence, thus ZFNs are designed in pairs that bind to the target sequence in the opposite orientation with the correct spacer. The double-strand break mediated by ZFNs is then repaired by either nonhomologous end joining, an error-prone repairing process, or homologous recombination (HR), an event that copies the homologous sequence from an adjacent DNA sequence.19 ZFN-induced genetic modification has been applied to a variety of cell lines and several different species.20,21,22,23,24,25 Most notably, several groups have used ZFNs to enhance gene targeting efficiency in hES cells.12,13,17 A recent report using integrase-defective lentiviral vectors (IDLVs) delivering ZFNs and a DNA donor template showed a high level of gene targeting events in hES cells.17 Although the use of IDLVs greatly reduced the risk associated with the integrating event, some residual integration events remained.26,27 A follow-up study addressed this risk by using a virus-free system to perform ZFN-mediated gene targeting in hES cells.13 However, the gene editing efficiency was 20-fold lower than that of the IDLV system. To combine the strengths of both systems, we engineered a BV system to co-deliver both ZFNs and the DNA donor template to induce ZFN-mediated gene targeting. We report herein that the gene editing efficiency was comparable to that of the IDLVs system, and that no random integration events were observed in hES cells under our experimental conditions.
Results
Construction of site-specific vectors
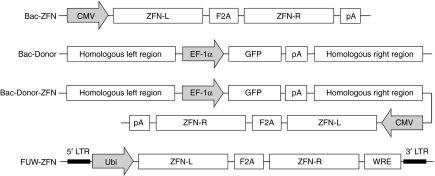
Site-specific integration to a desirable site has been a long-standing goal in the field of gene therapy. The CCR5 gene was chosen in this study as a site-specific target to introduce a foreign gene because the homozygous null mutation is prevalent in a small population of individuals28,29 and disruption of this gene is well tolerated.30 The C2H2 ZFN protein was generated by fusing the CCR5-specific zinc-finger proteins to engineered obligate heterodimers of the endonuclease domain of the FokI enzyme, which would minimize the nonspecific cleavage.31,32 The Bac-ZFN construct consists of both the right and left ZFNs linked by a F2A sequence driven by the cytomegalovirus (CMV) internal promoter. ZFNs (ZFN-R: AAA CTG CAA AAG; ZFN-L: GAT GAG GAT GAC) (Figure 2a) can induce a double-strand break at the CCR5 locus. Then, with the delivery of a suitable DNA donor template, an HR event can occur and the donor sequence can be introduced into the CCR5 locus. The DNA donor template used in this study contains a green fluorescent protein (GFP) expression cassette driven by the human elongation factor-1α promoter flanked by CCR5 homology arms to initiate HR. The human elongation factor-1α promoter has been shown to efficiently drive the expression of the GFP reporter gene in hES cells.11 Using the large transgene capacity of BV, we generated a Bac-ZFN-Donor construct by inserting the ZFN cassette directly into the Bac-Donor construct to facilitate both the double-strand break and transgene integration. We therefore constructed three different versions of BVs to deliver either ZFNs (Bac-ZFN), DNA donor template (Bac-Donor), or ZFNs and DNA donor template together (Bac-ZFN-Donor) (Figure 1).
Figure 1.
Schematic representation of key constructs in this study. These constructs include three different vectors (baculoviral vectors) carrying either ZFNs, DNA donor template, or ZFNs and DNA donor template, and a lentiviral vector encoding ZFNs. CMV, human cytomegalovirus immediate-early gene promoter; EF-1α, human elongation factor-1α promoter; F2A, a 2A sequence derived from the foot and mouth disease virus; GFP, enhanced green fluorescence protein; Homologous region, homologous DNA sequence to the exon 3 of the CCR5 gene; LTR, long terminal repeats; pA, polyadenylation signal; Ubi, the human ubiquitin-C promoter; WRE, woodchuck responsive element; ZFN-L, zinc-finger nuclease targeting the left flank of the CCR5 gene; ZFNR, zinc-finger nuclease targeting the right flank of the CCR5 gene.
Previously, IDLVs have been shown to successfully achieve gene modification in hES cells.17 The gene editing process was achieved by co-delivery of both ZFNs and a DNA donor template to the target cells. To co-deliver both ZFNs, we constructed a lentiviral vector (FUW-ZFN) encoding two ZFNs linked by F2A and driven by the human ubiquitin-C promoter;33 the woodchuck hepatitis virus posttranscriptional regulatory element was included downstream of the ZFNs to increase the level of transcription (Figure 1).
ZFN-mediated target disruption by the BV
To determine whether CCR5 ZFNs could be efficiently delivered by a BV, we transduced the Ghost-CCR5 cell line, a human cell line that highly expresses autologous CCR5 genes, with the Bac-ZFN vector. The transduced cells were maintained in culture for 5 days. Flow cytometry analysis on the Bac-ZFN-transduced cells showed several populations with various levels of CCR5 expression, indicating the success of targeted gene disruption (Figure 2b). These resulting cells can be roughly divided into three populations and are referred to as the CCR5 low (CCR5low), CCR5 intermediate (CCR5med), and CCR5 high (CCR5high) populations. In addition, we transduced both wild-type and ZFN-treated Ghost-CCR5 cells with either a lentiviral vector enveloped with a CCR5-tropsim HIV-1 glycoprotein (FUGW/GP160), or a control vector (FUGW/vesicular stomatitis virus glycoprotein (VSVG)), to validate the disruption of the CCR5 gene. Consistent with previous data, a decrease in transduction efficiency was observed for the ZFN-treated cells exposed to FUGW/GP160 (Figure 2c; Supplementary Figure S2). In the control experiment where ZFN-treated cells were transduced by FUGW/VSVG, transduction efficiency was unaltered, indicating that the decreased transduction was CCR5-dependent. To confirm the ZFN-induced cleavage at the target site, we performed a mismatch-sensitive Surveyor nuclease assay. The DNA analysis of this assay revealed a disruption rate of 36% for the targeted gene in the ZFN-treated population, as compared to <1% gene disruption in the untreated Ghost-CCR5 cell line (Figure 2d). We further sorted out the treated cells with low CCR5 expression (CCR5low). Flow cytometry analysis showed that these sorted cells contained ~70% CCR5-null cells. When these cells were subjected to the Surveyor assay, up to 60% of CCR5 alleles exhibited disruption (Figure 2d). These results demonstrate that ZFNs can be efficiently delivered by BV to induce double-strand breaks at the CCR5 target site and disable its expression. We made the attempt but failed to sort out pure CCR5-null cells by gating on the lowest corner of CCR5low cells; flow cytometry analysis of cells cultured from these sorted cells always contained 20–30% CCR5-positive cells (data not shown).
Figure 2.
Zinc-finger nuclease (ZFN)-mediated disruption of CCR5 by baculoviral vector. (a) Schematic representation of the strategy for targeted gene disruption by Bac-ZFN. The CCR5 locus is shown and exons are indicated by arrows. The enlarged views depict the binding sites for the CCR5-ZFN pairs. DSB, double-strand break; NHEJ, nonhomologous end joining. (b) Ghost-CCR5 cells were transduced by the Bac-ZFN vector and the decreased level of CCR5 expression was measured by flow cytometry at day 5 post-treatment. The untreated cells were included as a control. Left: histogram plot of CCR5 expression on treated (open line) and untreated (shaded area) cells. Right: mean fluorescence intensity of Bac-ZFN-treated and untreated cells. (c) Entry efficiency of lentiviral vectors enveloped with an CCR5-tropsim HIV-1 glycoprotein (FUGW/GP160) toward Bac-ZFN-treated Ghost-CCR5 cells. Untreated cells and lentiviral vectors enveloped with a CCR5-independent vesicular stomatitis virus glycoprotein (FUGW/VSVG) were included as controls. Green fluorescent protein expression as an indication of successful entry was measured by flow cytometry and relative transduction efficiency was shown. (d) The level of targeted gene disruption in Bac-ZFN-treated Ghost-CCR5 cells was assessed by the Surveyor assay. The untreated control cells are included as a control. In addition to Bac-ZFN-treated cell sample, we also include the sample with enriched CCR5-low/negative population (~70%) that was sorted from the Bac-ZFN-treated cells and labeled it as Bac-ZFN (sorted). The lower migrating bands indicate the ZFN-mediated gene disruption. The percentage of disruption was quantified by a phosphorimager and calculated using a formula (1−SQRT (parental fraction)) × 100.
ZFN-associated toxicity for vector-producing cells
Previously, Lombardo et al. reported the utilization of IDLVs to deliver ZFNs to disrupt the CCR5 gene.17 However, their approach requires the target cells to be cotransduced by two vectors for a gene disruption. We hypothesized that incorporating both ZFNs linked by a 2A sequence in a single IDLV vector could improve the efficiency of gene targeting. To determine whether FUW-ZFN could mediate enhanced gene disruption, we generated a ZFN-containing vector FUW-ZFN/VSVG and a ZFN-lacking vector FUW/VSVG. However, the lentiviral vector containing both ZFNs was not efficiently produced. A p24 assay revealed that the production level of FUW-ZFN/VSVG was <half of that of FUW/VSVG (Figure 3a). Immunostaining to detect the protein 53BP1, which tends to localize at the DNA damage sites and form foci, revealed that the FUW-ZFN-transfected 293T cells yielded a higher amount of 53BP1-stained foci than the FUW-transfected cells, suggesting that the decreased production of FUW-ZFN/VSVG was due to the ZFN-mediated DNA cleavage (Figure 3b). We further performed immunostaining on mammalian 293T and insect SF9 cells transduced with Bac-ZFN and found that 53BP1-stained foci were only observed in 293T cells. Thus, we suspect that the toxicity was due to both the presence of ZFNs that were only effectively translated by the mammalian-active CMV promoter in 293T cells and the presence of the target sequence (CCR5) in the 293T genome (Figure 3c). These sets of results indicate that the reduction in virus production was due to ZFN-mediated toxicity associated with the lentiviral vector-producing cells (293T), whereas no such toxicity was detected in the BV producing cells (SF9).
Figure 3.
Analysis of zinc-finger nuclease (ZFN)-associated toxicity for producer cells to make lentiviral or baculoviral vectors. (a) Productivity of mammalian 293T producer cells for making ZFN-containing (FUW-ZFN) or ZFN-negative (FUW) lentiviral particles. p24 is a key component of the lentiviral capsid and its concentration is a direct indication of the quantity of vector particles present. Concentration of p24 was measured by enzyme-linked immunosorbent assay. (b) Representative images of transfected 293T producer cells to make lentiviral vectors (FUW-ZFN or FUW). These cells were intranuclearly stained with antibodies to 53BP1 (red). Bar = 50 µm. (c) Representative images of vector producer cells (mammalian 293T or insect SF9) treated with the DNA-damaging agent etoposide (1 µm, as a positive control) or transfected with Bac-ZFN for 5 days before staining with antibodies to 53BP1 (red). Bar = 50 µm.
ZFN-mediated targeted gene addition in cell lines
The transient nature of gene expression delivered by conventional BVs has limited the usage of this vector system in gene therapy. To extend the duration of BV-mediated gene expression, we constructed three BVs, Bac-ZFN, Bac-Donor, and Bac-Donor-ZFN, and tested their ability to achieve targeted integration into the CCR5 gene (Figure 4a). We cotransduced both 293T and U87 cell lines with Bac-ZFN and Bac-Donor and observed that the GFP reporter gene could be stably expressed at 1 month post-transduction, whereas no detectable expression was seen when Bac-Donor was used alone (Figure 4b). We then cotransduced the cell lines with Bac-ZFN and Bac-Donor at various MOI combinations to seek an optimal protocol for achieving higher efficiency of targeted gene integration. The flow cytometry analysis performed at 1 month post-transduction revealed that GFP expression culminated at a MOI of 150 for Bac-ZFN and a MOI of 500 for Bac-Donor, as evidenced by the high percentage (8–10%) of GFP expression (Figure 4c). Transduction with Bac-Donor alone resulted in <1% GFP-positive cells, indicating the necessity of the ZFN-mediated double-strand break for efficient gene insertion. Similarly, when 293T and U87 cell lines were transduced by Bac-Donor-ZFN, the baculoviral construct that delivered both ZFNs and donor DNA to the cells, up to 6% targeted gene integration was observed (Figure 4d). In all of cell types tested, targeted integration into the CCR5 gene was confirmed by PCR analysis on GFP+ cells sorted by fluorescence-activated cell sorting at both integration junctions (Figure 4e). To further characterize the double-strand break-mediated HR, we performed limiting dilution to isolate single clones. We employed a TaqMan quantitative PCR assay to determine the genome copies of the integrated GFP expressing cassette. This assay revealed that ~80% of the transduced cells do not contain randomly integrated viral vector (Supplementary Figure S1). This series of experiments confirmed that BVs can mediate specific gene addition to a predetermined target site with high efficiency.
Figure 4.
Targeted gene editing at the CCR5 gene locus in 293T and U87 cell lines. (a) Schematic diagram of the strategy to use baculoviral vectors (Bac-ZFN and Bac-Donor) for achieving the targeted gene addition to the CCR5 gene locus. The gray arrows indicate the exons in the CCR5 locus. Bac-ZFN and Bac-Donor deliver the CCR5 zinc-finger nucleases (ZFNs) and the donor DNA encoding homologous arms and a green fluorescent protein (GFP) expression cassette, respectively. Expression of a pair of CCR5-ZFNs in human cells induces a double-strand break (DSB) at the ZFNs-binding site. If the homologous recombination (HR)-mediated repair occurs, the GFP expression cassette will be added into the CCR5 locus through targeted integration. EF-1α, human elongation factor-1α promoter; pA, polyadenylation signal. (b) 293T or U87 cells were transduced with either Bac-Donor alone or Bac-Donor and Bac-ZFN. Phase contrast and fluorescence images were acquired by epifluorescence microscopy and representative images are shown. Bar = 50 µm. (c) 293T or U87 cells were treated with the indicated multiplicity of infections (MOIs) doses of baculoviral vectors (Bac-Donor and Bac-ZFN) and their GFP expression was analyzed by flow cytometry at 1 month post-transduction. (d) 293T or U87 cells were treated with the Bac-Donor-ZFN vector at the MOI of 150 or 500 and their GFP expression was analyzed by flow cytometry at 1 month post-transduction. (e) Top: schematic diagram of targeted integration into the CCR5 locus. The black arrows indicate the primers used in the analysis. Bottom: the treated cells were sorted by GFP expression and analyzed by PCR for targeted integration (TI) using two sets of primers specific for the 5′ or 3′ integration junctions. A pair of primers to amplify a nontargeted locus (NTL) region was included as a control.
ZFN-mediated targeted gene addition in hES cells
Stable gene expression is an important tool for future implementation of hES cells for regenerative medicine. To explore the feasibility of using ZFNs for targeted gene addition, we dissociated hES cells (H9) into single cells and incubated them with BVs (Bac-ZFN and Bac-Donor) at a MOI of 500 for 2 hours. We found that up to 4.38% of the hES cells were modified at 2 weeks post-transduction, whereas only a background level GFP was seen in hES cells modified with the Bac-Donor vector alone (Figure 5a). The TaqMan quantitative PCR revealed that no random integration event was observed in the modified hES cells (Supplementary Figure S1d). To confirm that the integration event was targeted to the desired CCR5 gene, we performed PCR amplifications for the junctions of the integration sites and found that the GFP reporter cassette had undergone the predicted HR event at the CCR5 locus (Figure 5b).
Figure 5.
Targeted gene editing at the CCR5 gene locus in human embryonic stem cells. (a) H9 human embryonic stem (hES) cells were dissociated into single cells and treated with either Bac-ZFN and Bac-Donor or Bac-Donor alone at a multiplicity of infection (MOI) of 500. Green fluorescent protein (GFP) expression was detected by flow cytometry at week 3 post-transduction. (b) The treated cells were analyzed by PCR to confirm the targeted integration (TI) of the GFP cassette into the CCR5 gene using primers specific for the 5′ and 3′ integration junctions. A pair of primers to amplify a nontargeted locus (NTL) region was included as a control. (c) Karyotyping analysis on baculoviral vector (BV)-transduced hES cells after 20 passages demonstrates a normal karyotype (46, XX). (d) Representative confocal microscopy images of H9 1 month post-transduction for GFP (green), the embryonic stem cell markers, OCT4, NANOG, GCTM2, and SSEA4 (red), and nuclear DNA (Hoescht, blue), showing the undifferentiated stem cells. (e) Left: BV-transduced hES cells were sorted based on GFP expression. Right: expression of selected markers of embryonic bodies (EBs) (MAP2, NEUROD1, AFP, DCN, HAND1, IGF2) was examined by reverse transcriptase-PCR.
Transduction by BVs caused no microscopically observed cytotoxic effects or morphological changes on the hES cells as compared to nontransduced control hES cells and the cells retained a normal karyotype (Figure 5c). The transduced cells were immunostained to confirm the undifferentiated state of these modified hES cells. Transduced hES cells stably expressed the GFP reporter gene and remained undifferentiated, as indicated by the expression of the hES markers OCT4, NANOG, GCTM2, and SSEA4 (Figure 5d). To confirm the pluripotency of the modified hES cells, transduced hES cells were sorted based on GFP expression, and these sorted hES cells were employed to form embryoid bodies (EBs). Reverse transcriptase-PCR analysis showed that the EBs were able to form all three germ layers, as the expression of germ layer–specific markers were detected in EBs but absent in undifferentiated hES cells, indicating the pluripotency of the modified hES cells (Figure 5e). Collectively, these data demonstrate that the ZFN-based BV approach allows for stable and efficient integration of transgenes in hES cells without any apparent detrimental effects to the self-renewal and pluripotent state of the modified cells.
Discussion
Baculoviruses have been shown to be a promising transient gene delivery vehicles in many mammalian cell types.34 The high-level transduction, low cytotoxicity, lack of replication in mammalian cells, and large transgene capacity make the baculovirus a useful tool for delivering genes of interest. However, one major limitation in the broad range of applications of the BV system is the lack of permanent, site-specific transgene expression. To address this challenge, others have generated a baculovirus/adeno-associated virus hybrid vector system containing a gene cassette flanked by the inverted terminal repeats of adeno-associated virus to prolong the expression of the transgene.11,35 The limitation of this strategy is that the location of the integration site is restricted to the adeno-associated virus-preferred integration site and cannot be used to target-specific genes. Permanent modification of the genome via site-specific integration has been a long-standing goal in the field, and has significant advantages over other viral integration approaches. Traditionally, the HR pathway has been exploited to edit a specific locus. However, this approach is both labor-intensive and time-consuming as the efficiency of site-specific HR is very low. To overcome the low HR efficiency, ZFNs have been used along with a donor template to enhance the gene targeting efficiency. The double-strand break induced by ZFNs triggers two downstream repair mechanisms. One involves the nonhomologous end joining of the recessed strands at the double-strand break and results in localized deletion or insertion of nucleotides that leads to gene disruption. Another repair mechanism yields targeted gene addition via HR of the damaged DNA with either a sister chromatin or a homologous DNA donor template. By incorporating ZFNs and a DNA donor template, we investigated the feasibility of using BVs to achieve site-specific integration of a DNA target sequence.
We demonstrated that permanent gene disruption can be achieved in established cells by BV delivery of ZFNs. We showed that by transducing CCR5-expressing cells with our engineered BV carrying ZFNs, we could generate cells composed of the CCR5low, CCR5med, and CCR5high populations and obtain gene disruption with an overall efficiency of up to 36% by the Surveyor nuclease assay (Figure 2). An increased disruption of CCR5 alleles (60%) was seen after sorting out the CCR5low population containing ~70% CCR-null cells, suggesting a correlation between the disruption efficiency induced by Bac-ZFN and its target expression. We failed to sort out the pure CCR5-null population regardless of the stringency of gating criteria we applied on the ZFN-treated cells stained with the anti-CCR5 antibody. This suggests the limitation of the Ghost-CCR5 cell line and assays we chose for this investigation. The Ghost-CCR5 cells are mixed-cell populations generated by retrovirus-mediated gene delivery.36 Although it is estimated for the Ghost-CCR5 cells to have an average of four CCR5 copies per cell, the copy number for individual cells varies. We could estimate that our average ZFN-disrupted CCR5 alleles were 1.44 copies per cell, but disruption for individual cells might differ greatly. The mixed Ghost-CCR5 cells, combined with the heterogeneous ZFN-transduction and disruption, really complicated our analysis and limited our ability to conduct further quantitative data interpretation.
We further demonstrated that targeted gene integration can be achieved by transducing cell lines with BV encoding ZFNs and a donor template (Figure 4). To further expand the baculoviral delivering system to primary cell lines of therapeutic value, we transduced hES cells and showed that the modification efficiency is comparable to that of IDLV-delivered ZFNs (5%) and ~20-fold higher than that of virus-free system (0.25%).12,17 The BV-transduced hES cells displayed neither karyotypic abnormalities nor any alteration to their pluripotency.
This study also highlighted the advantages of using BV as a gene delivery vehicle. First, the large cloning capacity of the vector allows us to generate a single construct to deliver both ZFNs and the donor template. The genome modification efficiency is comparable to that of the two-construct version (~7%) (Figure 4). Second, since mammalian promoters, such as the CMV promoter used in this study, have been shown to be ineffective in driving the expression of transgenes in insect cells, the expression of ZFNs in these cells is highly restrained, resulting in greater viability of the producing cells.37 In contrast, the CMV promoter has been shown to highly transcribe the transgene in vector-producing mammalian cells. The strong expression of both ZFNs in a single vector-producing mammalian cell could lead to cytotoxicity, resulting in a decrease in vector production. Indeed, with a lentiviral vector carrying 2A-linked ZFNs, we observed a production issue associated with this type of ZFN-mediated cytotoxicity (Figure 3).
In summary, we developed a novel BV that confers permanent gene disruption and genome modification in various mammalian cell lines as well as in hES cells by delivering ZFNs and a donor template. This system allows for safe and efficient genome modification of the target cell. Traditionally, genome is modified via integrating viral vectors such as retroviruses and lentiviruses, which tend to integrate into coding or regulatory regions of transcriptionally active genes, leading to possible gene silencing and insertional mutagenesis.38,39 Alternatively, transponsons such as Sleeping Beauty, PiggyBac, or phage integrase such as π31 could also introduce permanent genome modifications into target cells.40,41,42 However, modifications via these approaches may still impose position effects, which could still lead to insertional mutagenesis. Our approach of using site-specific HR via a ZFN-mediated double-strand break allows us to pinpoint the exact site of integration, which is less likely to cause gene silencing and/or insertional mutagenesis. In principle, by engineering the ZFN binding domain and selecting the appropriate donor sequence, we could engineer BV-ZFN to direct the modification to any desirable genome site. Moreover, by selecting the target site, it will be possible to predict the level of transgene expression and exploit endogenous promoters to drive transgene expression, thus preventing gene silencing. Modification via HR can also be applied to treat diseased cells by replacing the defective genes with functional genes. The combination of ZFNs and the BV system can enable rapid generation of new cell lines and facilitate the study of the functions of specific genes, which can advance novel developments in biotechnology and medicine.
Materials and Methods
Maintenance and differentiation of the hES cells. The hES cell line H943 was maintained as described previously.44 Briefly, hES colonies were maintained on mitotically inactivated mouse embryonic fibroblasts in media consisting of Dulbecco's modified Eagle's medium-F12 (Invitrogen, Carlsbad, CA) supplemented with 20% Knockout Serum Replacer (Invitrogen) and fibroblast growth factor-2 (4 ng/ml; Peprotech, Rocky Hill, NJ) plus antibiotics and -glutamine (1 mmol/l). Colonies were passaged weekly using mechanical dissection. To induce differentiation, hES colonies were detached using 1 mg/ml of collagenase (Invitrogen). The hES cell clumps were transferred to suspension culture for 5 days to foster the formation of EBs. The EBs were then transferred to cell culture dishes coated with matrigel to promote attachment and outgrowth of differentiated cells. Differentiated cultures were collected for reverse transcriptase-PCR analysis 5 days following transfer.
Plasmid construction. The plasmid pFasBac-Dual from Invitrogen was used as a shuttle vector to generate the recombinant BVs (pFB-ZFN, pFB-Donor, and pFB-ZFN-Donor). The complementary DNAs (cDNAs) of the reported ZFNs targeting the human CCR517,30 were synthesized by GeneArt (Regensburg, Germany). The ZFNs were cloned into the pcDNA3 vector (Invitrogen) via BamHI and EcoRI restriction sites. The resulting ZFN expression cassette including a CMV internal promoter and a BGH polyA tail was then cloned into pFastBac-Dual to yield pFB-ZFN. To generate the donor plasmid, the ZFN-targeted DNA was amplified with a primer pair (CCR5FW: TAC CGA GCT CGG ATC CTT AGA CCC TCT ATA ACA GTA ACT TCC TTT TAA AAA AGA CCT CTC CCA C; CCR5BW: TAG ATG CAT GCT CGA CTA GCG TCA ATA AAA ATG TTA AGA CTG AGT TGC AGC CG) and cloned into a TOPO plasmid (Invitrogen) via XhoI and BamHI restriction sites. A reporter expression cassette containing an human elongation factor-1α promoter, the GFP cDNA, and the polyA tail was then cloned into an EcoRV site that was inserted between the CCR5-ZFN-binding sites. To generate the pFB-Donor plasmid, the donor vector was PCR amplified with a pair of primers (CEGAFW: ATG GCT CGA GAT CCC CTT AGA CCC TCT ATA ACA GTA ACT TCC TTT TAA AAA AGA CCT CTC CCA C; CEGABW: ACT TCT CGA CAA GCT CTA GCG TCA ATA AAA ATG TTA AGA CTG AGT TGC AGC CG) and cloned into pFastBac-Dual via SmaI and HindIII restriction sites using the BD Clontech In-Fusion cloning method. Similarly, the pFB-ZFN-Donor plasmid was generated by amplifying with primers (CCR5ZFNFW: GCA ATT GTT GTT GTT GTT GAC ATT GAT TAT TGA CTA GTT ATT AAT AGT AAT CAA TTA CGG GGT CAT TAG; CCR5ZFNBW: TGC AAT AAA CAA GTT CCA TAG AGC CCA CCG CAT CCC CAG C) and cloned into pFB-Donor via the PvuII restriction site using the BD Clontech In-Fusion cloning method. To generate the lentiviral plasmid FUW-ZFN, the DNA of F2A-linked ZFNs was cloned into FUW via EcoR1 and BamH1 restriction sites.
Vector production. Recombinant BVs with the described expression cassettes were produced and propagated in SF9 cells according to the manual of the Bac-to-Bac Baculovirus Expression system from Invitrogen. Budded viruses were filtered through a 0.45-µm pore size filter (Nalgene, Rochester, NY) to remove any cell debris, and concentrated via centrifugation at 10,000 r.p.m. for 5 hours and 20 °C. Viral pellets were resuspended in 1 ml of HBSS (Invitrogen). To determine the titers of the recombinant BVs, we adapted a qPCR assay to first quantify the physical number of the viral particles.45 The BacPAK Baculovirus Rapid Titer Kit (Clontech, Mountain View, CA) was then used to determine the infectious units. To generate the lentiviral vector FUW-ZFN, 293T cells were seeded in a 6 cm culture dish in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (Sigma, St Louis, MO, 10%), -glutamine (10 ml/l), penicillin, and streptomycin (100 units/ml) the night before transfection. 293T cells were transfected at a confluence of 80–90% with 5 µg of the lentiviral backbone vector (FUW-ZFN or FUW), and 2.5 µg each of pMDLg/pRRE, pRSV-Rev, and pVSVG via the standard calcium phosphate precipitation technique.46 Cells were replenished with prewarmed media 4 hours post-transfection. Vectors were harvested 2 days post-transfection and filtered through a 0.45-µm pore size filter (Nalgene).
Surveyor nuclease assay. Genomic DNA was extracted from modified and control cells using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). A 292-bp fragment of the CCR5 locus containing the ZFNs-binding site was amplified with the primer sets ZFNFW: AAG ATG GAT TAT CAA GTG TCA AGT CC and ZFNBW: CAA AGT CCC ACT GGG CG using Taq DNA polymerase (Fermentas, Glen Burnie, MD) and supplemented with 5 µCi α-P32 dATP and 5 µCi α-P32 dCTP. The PCR product was then purified with a G-50 column (GE Healthcare, Piscataway, NJ), denatured, reannealed and digested with Surveyor Nuclease (Transgenomic, Omaha, NE). The resulting products were resolved on a nondenaturing 10% TBE polyacrylamide gel (Bio-Rad, Hercules, CA). The gel was dried and exposed and the resulting bands were quantified by phosphorimager (Bio-Rad). The proportion of ZFN-disrupted CCR5 alleles in the original sample is calculated using the formula: [1-SQRT (parental fraction)] × 100.
Viral transduction. For viral transduction to the Ghost-CCR5 cell line, 0.05 million Ghost-CCR5 cells were seeded on a 24-well culture plate and spin-transduced with the BVs (Bac-ZFN, MOI = 500) or lentiviral vectors (FUW-ZFN/VSVG or FUW/VSVG) at 25 °C and 2,500 r.p.m. for 90 minutes using a RT Legend Centrifuge. After replacing with fresh media, the treated cells were cultured for an additional 5 days at 37 °C and 5% CO2. To analyze the CCR5 disruption efficiency, the treated cells were stained by biotin-conjugated anti-CCR5 antibody (Invitrogen), followed by a secondary staining with phycoerythrin-conjugated streptavidin (BD Biosciences, San Jose, CA). Flow cytometry (BD FACSort; BD Biosciences) was then used to analyze the disruption efficiency.
For viral transduction to the 293T or U87 cell line, 0.5 million cells were seeded on a 24-well cell culture plate and spin-transduced with the indicated BVs (Bac-ZFN, Bac-Donor, or Bac-ZFN-Donor) at 25 °C and 2,500 r.p.m. for 90 minutes using a RT Legend Centrifuge. After replacing with fresh media, the treated cells were cultured for additional 30 days at 37 °C and 5% CO2. Flow cytometry (BD FACSort) was then used to analyze gene addition efficiency.
For viral transduction to hES cells, single hES cells in suspension were used to avoid the effect of virus absorption by the mouse embryonic fibroblast. On the day of the transduction, single cell suspensions were generated by treating hES colonies with 0.05% trypsin solution containing 0.2 g/l EDTA (Invitrogen) for 3 minutes. Dissociated cells were collected by centrifugation at 1,000 r.p.m. for 1 minute. 2.5 × 104 hES cells were resuspended in 100 µl of knockout Dulbecco's modified Eagle's medium containing 10 µmol/l of p160-Rho-associated coiled-coil kinase inhibitor (Y-27632; Sigma-Aldrich, St Louis, MO) to prevent apoptosis during culture in suspension and enable the replating of single cells. BVs (Bac-ZFN or Bac-Donor) were added at the indicated MOI. Transduction was carried out at 37 °C for 2 hours, after which the cells were replated onto fresh mouse embryonic fibroblast, in 12 well plates. The treated cells were cultured for additional 21 days at 37 °C and 5% CO2. Flow cytometry (BD LSR II; BD Biosciences) was used to analyze gene addition efficiency.
p24 Analysis of lentiviral vectors. Various lentiviral vectors (10 µl, fresh supernatant) were lysed with 90 µl of 10% Triton-X 100 in phosphate-buffered saline and the p24 levels were measured by a p24 antigen capture enzyme-linked immunosorbent assay kit (ImmunoDiagnostics, Woburn, MA)
Detection of double-strand breaks in ZFN-treated cells. SF9 or 293T cells were transfected by Cellfectin (Invitrogen) using the protocol recommended by the manufacturer. Fluorescent images were acquired on a Yokogawa spinning-disk confocal scanner system (Solamere Technology Group, Salt Lake City, UT) using a Nikon eclipse Ti-E microscope equipped with a 60×/1.49 Apo TIRF oil objective and a Cascade II: 512 EMCCD camera (Photometrics, Tucson, AZ). An acousto-optical tunable filter controlled laser-merge system (Solamere Technology Group) was used to provide illumination power at each of the following laser lines: 491, 561, and 640 nm solid state lasers (50 mW for each laser). To image 53BP1, both treated and nontreated cells were seeded into a 35-mm glass-bottom culture dish and grown at 27 °C or 37 °C overnight. The seeded cells were rinsed with cold phosphate-buffered saline and fixed with 4% formaldehyde on ice for 10 minutes, and then immunostained with anti-53BP1 rabbit polyclonal antibodies (Bethyl Laboratories, Montgomery, TX) followed by incubation with Texas-Red anti-rabbit immunoglobulin G (Invitrogen). TO-PRO-3 iodide (Invitrogen) was also used for nuclear staining.
Targeted integration analysis. Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). One hundred nanogram of genomic DNA was subjected to PCR using a primer pair of 5′CCR5FW (5′-CTT AGA ACA GTG ATT GGC ATC CAG TAT GTG CCC TC-3′) and BGHPABW (5′-GCT GGG GAT GCG GTG GGC TCT ATG G-3′) to confirm 5′ targeted integration, and a primer pair of EF-1αFW (5′-GCC GAC CCC TCC CCC CAA CTT CTC-3′) and 3′CCR5FW (5′-GGC TTA AAA GAT CTA ATC TAC TTT AAA CAG ATG CCA AAT AAA TGG ATG-3′) to confirm 3′ targeted integration. A primer pair of CCR5NTFW (5′-CTC TCC CTT CAC TCC GAA AGT TCC TTA TGT ATA TTT AAA AGA AAG C-3′) and CCR5NTBW (5′-CTT GCA GTG AGG CTT CTG TCT TTG CCA GCA ATA G-3′) was used to amplify nontargeted locus DNA. The PCR amplified products were resolved on 1% agarose gel and visualized by ethidium bromide staining.
Cell sorting and reverse transcription-PCR analysis. For sorting the GFP+ cells, single cell suspensions were generated by treating hES colonies with 0.05% trypsin solution containing 0.2 g/l EDTA for 3 minutes. Dissociated cells were collected by centrifugation at 1,000 r.p.m. for 1 minute and resuspended in 2 ml of knockout Dulbecco's modified Eagle's medium containing 10 µmol/l of p160-Rho-associated coiled-coil kinase inhibitor. Cells were sorted with fluorescence-activated cell sorting Aria SORP equipped with 100 µm nozzle (BD Biosciences).
Total RNA was extracted from hES cells or EBs using the RNeasy kit (Qiagen). First-strand cDNA was synthesized by the PowerScript Reverse Transcriptase system (BD Biosciences). Fifty nanogram of cDNA reaction mix was subjected to PCR amplification with the following primer pairs and the resulting products were electrophoresed on a 1% agarose gel. MAP2: 5′-GAA GCA AAG GCA CCT CAC TG-3′ and 5′-TCT GAG GCA GGT GAT GGG-3′ NEUROD1: 5′-GTC CTT CGA TAG CCA TTC AC-3′ and 5′-CTT TGA TCC CCT GTT TCT TCC-3′ AFP: 5′-AGA ACC TGT CAC AAG CTG TG-3′ and 5′-GAC AGC AAG CTG AGG ATG TC-3′ DCN: 5′-CAC AAC ACC AAA AAG GCT TC-3′ and 5′-TTG CAG TTA GGT TTC CAG TAT C-3′ HAND1: 5′-TGC CTG AGA AAG AGA ACC AG-3′ and 5′-ATG GCA GGA TGA ACA AAC AC-3′ IGF2: 5′-TCC TCC CTG GAC AAT CAG AC-3′ and 5′-AGA AGC ACC AGC ATC GAC TT-3′.
Immunohistochemistry and karyotypic analysis. For immunostaining hES cell colonies were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde for 10 minutes. Cells were permeabilized with 0.1% Triton X-100 for 5 minutes at room temperature and then treated for 1 hour with the following primary antibodies: OCT4 (C-10, 1: 10 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), NANOG (H-155, 1:10 dilution; Santa Cruz Biotechnology), GCTM2 (A kind gift from Dr Martin Pera), and SSEA4 (Millipore,1 µg/ml). Cells were washed with staining buffer (phosphate-buffered saline supplemented with 2.5% fetal bovine serum) and then incubated for 1 hour with secondary antibodies: Texas-Red anti-mouse immunoglobulin G (T862, 1:400 dilution; Invitrogen) or AlexaFluor 594 anti-rabbit immunoglobulin G (A11012, 1:1000 dilution; Invitrogen). All antibodies were diluted in staining buffer. Nuclear staining was carried out using Hoescht (33258; Sigma-Aldrich). Images were acquired on an inverted fluorescent microscope (Axioimager, by Carl Zeiss). The karyotyping of the transduced hES cells was conducted by Cell Line Genetics (Madison, WI).
SUPPLEMENTARY MATERIAL Figure S1. Detection of integrated green fluorescent protein (GFP) reporter gene or baculoviral vector. (a) Copies of GFP per cellular genome of 293T clones were detected by a quantitative TaqMan PCR protocol. The copy number was determined by using a primer-probe set that specifically amplifies the GFP reporter gene, which is normalized to another probe set specific for the β-actin gene that allows for the quantification of genomes. (b–c) Detection of randomly integrated copies of either baculoviral VP39 gene (b) or baculoviral GP64 gene (c) in 293T clones using the similar protocol described in (a). (d) Copies of integrated GFP, VP39 and GP64 genes in BV-treated hES cells were detected by a similar protocol described in (a). Figure S2. Entry efficiency of lentiviral vectors to Bac-ZFN-treated Ghost cells. The sorted CCR5-high or CCR5-negative/low cells were transduced by a lentiviral vector enveloped with a CCR5-tropism HIV-1 glycoprotein (FUGW/GP160). GFP expression as the indication of successful entry was measured by flow cytometry. We consistently observed that it was difficult to sort out a pure CCR5-positive and CCR5-negative populations, presumably due to the dynamic expression of CCR5 and limitation of antibody labeling. Therefore, we further gated on the sorted cells to identify CCR5+ and CCR5 populations for the analysis of GFP expression.
Acknowledgments
This work was supported by grant from the California Institute for Regenerative Medicine (RT1-01028-1). We thank Steven Froelich and April Tai for critical reading of this manuscript.
Supplementary Material
Detection of integrated green fluorescent protein (GFP) reporter gene or baculoviral vector. (a) Copies of GFP per cellular genome of 293T clones were detected by a quantitative TaqMan PCR protocol. The copy number was determined by using a primer-probe set that specifically amplifies the GFP reporter gene, which is normalized to another probe set specific for the β-actin gene that allows for the quantification of genomes. (b–c) Detection of randomly integrated copies of either baculoviral VP39 gene (b) or baculoviral GP64 gene (c) in 293T clones using the similar protocol described in (a). (d) Copies of integrated GFP, VP39 and GP64 genes in BV-treated hES cells were detected by a similar protocol described in (a).
Entry efficiency of lentiviral vectors to Bac-ZFN-treated Ghost cells. The sorted CCR5-high or CCR5-negative/low cells were transduced by a lentiviral vector enveloped with a CCR5-tropism HIV-1 glycoprotein (FUGW/GP160). GFP expression as the indication of successful entry was measured by flow cytometry. We consistently observed that it was difficult to sort out a pure CCR5-positive and CCR5-negative populations, presumably due to the dynamic expression of CCR5 and limitation of antibody labeling. Therefore, we further gated on the sorted cells to identify CCR5+ and CCR5 populations for the analysis of GFP expression.
REFERENCES
- Yates F., and, Daley GQ. Progress and prospects: gene transfer into embryonic stem cells. Gene Ther. 2006;13:1431–1439. doi: 10.1038/sj.gt.3302854. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Rivas-Carrillo JD, Soto-Gutierrez A, Fukazawa T, Chen Y, Navarro-Alvarez N.et al. (2005Gene delivery to embryonic stem cells Birth Defects Res C Embryo Today 7510–18. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]
- Boyce FM., and, Bucher NL. Baculovirus-mediated gene transfer into mammalian cells. Proc Natl Acad Sci USA. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P., and, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci USA. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I.et al. (1997Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors J Gen Virol 78 (Pt 10)2657–2664. [DOI] [PubMed] [Google Scholar]
- Kost TA., and, Condreay JP. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends Biotechnol. 2002;20:173–180. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- Sandig V, Hofmann C, Steinert S, Jennings G, Schlag P., and, Strauss M. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum Gene Ther. 1996;7:1937–1945. doi: 10.1089/hum.1996.7.16-1937. [DOI] [PubMed] [Google Scholar]
- Cheshenko N, Krougliak N, Eisensmith RC., and, Krougliak VA. A novel system for the production of fully deleted adenovirus vectors that does not require helper adenovirus. Gene Ther. 2001;8:846–854. doi: 10.1038/sj.gt.3301459. [DOI] [PubMed] [Google Scholar]
- Ho YC, Chung YC, Hwang SM, Wang KC., and, Hu YC. Transgene expression and differentiation of baculovirus-transduced human mesenchymal stem cells. J Gene Med. 2005;7:860–868. doi: 10.1002/jgm.729. [DOI] [PubMed] [Google Scholar]
- Zeng J, Du J, Zhao Y, Palanisamy N., and, Wang S. Baculoviral vector-mediated transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1055–1061. doi: 10.1634/stemcells.2006-0616. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC.et al. (2009Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases Nat Biotechnol 27851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK.et al. (2009Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells Cell Stem Cell 597–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH., and, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Porteus MH., and, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG.et al. (2001Stimulation of homologous recombination through targeted cleavage by chimeric nucleases Mol Cell Biol 21289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA.et al. (2007Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery Nat Biotechnol 251298–1306. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S.et al. (2005Highly efficient endogenous human gene correction using designed zinc-finger nucleases Nature 435646–651. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M., and, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND., and, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE.et al. (2009Precise genome modification in the crop species Zea mays using zinc-finger nucleases Nature 459437–441. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK.et al. (2009High-frequency modification of plant genes using engineered zinc-finger nucleases Nature 459442–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE.et al. (2008Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases Nature 26702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumera KJ, Trautman JK, Bozas A, Liu J-L, Rutter J, Gall JG, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G.et al. (2008Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts Stem Cells 261998–2005. [DOI] [PubMed] [Google Scholar]
- Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C.et al. (2006Transient gene expression by nonintegrating lentiviral vectors Mol Ther 131121–1132. [DOI] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM.et al. (1996Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene Nature 382722–725. [DOI] [PubMed] [Google Scholar]
- Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K.et al. (2009Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation N Engl J Med 360692–698. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O.et al. (2008Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases Nat Biotechnol 26808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I.et al. (2007An improved zinc-finger nuclease architecture for highly specific genome editing Nat Biotechnol 25778–785. [DOI] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ., and, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ., and, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Kost TA, Condreay JP., and, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo F, Monciotti A, Recchia A, Cortese R, Ciliberto G., and, La Monica N. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J Virol. 1998;72:5025–5034. doi: 10.1128/jvi.72.6.5025-5034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörner A, Björndal A, Albert J, Kewalramani VN, Littman DR, Inoue R.et al. (1999Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage J Virol 732343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Xu CY, Wang YB, Chen M, Wu T, Zhang J.et al. (2004A rapid and efficient method to express target genes in mammalian cells by baculovirus World J Gastroenterol 101612–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S.et al. (2005Genome-wide analysis of retroviral DNA integration Nat Rev Microbiol 3848–858. [DOI] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S., and, Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ., and, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Yant SR, Giering JC, Xu H, Engler JA., and, Kay MA. Somatic integration from an adenoviral hybrid vector into a hot spot in mouse liver results in persistent transgene expression levels in vivo. Mol Ther. 2007;15:146–156. doi: 10.1038/sj.mt.6300011. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS.et al. (1998Embryonic stem cell lines derived from human blastocysts Science 2821145–1147. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A., and, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Hitchman RB, Siaterli EA, Nixon CP., and, King LA. Quantitative real-time PCR for rapid and accurate titration of recombinant baculovirus particles. Biotechnol Bioeng. 2007;96:810–814. doi: 10.1002/bit.21177. [DOI] [PubMed] [Google Scholar]
- Klages N, Zufferey R., and, Trono D. A stable system for the high-titer production of multiply attenuated lentiviral vectors. Mol Ther. 2000;2:170–176. doi: 10.1006/mthe.2000.0103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of integrated green fluorescent protein (GFP) reporter gene or baculoviral vector. (a) Copies of GFP per cellular genome of 293T clones were detected by a quantitative TaqMan PCR protocol. The copy number was determined by using a primer-probe set that specifically amplifies the GFP reporter gene, which is normalized to another probe set specific for the β-actin gene that allows for the quantification of genomes. (b–c) Detection of randomly integrated copies of either baculoviral VP39 gene (b) or baculoviral GP64 gene (c) in 293T clones using the similar protocol described in (a). (d) Copies of integrated GFP, VP39 and GP64 genes in BV-treated hES cells were detected by a similar protocol described in (a).
Entry efficiency of lentiviral vectors to Bac-ZFN-treated Ghost cells. The sorted CCR5-high or CCR5-negative/low cells were transduced by a lentiviral vector enveloped with a CCR5-tropism HIV-1 glycoprotein (FUGW/GP160). GFP expression as the indication of successful entry was measured by flow cytometry. We consistently observed that it was difficult to sort out a pure CCR5-positive and CCR5-negative populations, presumably due to the dynamic expression of CCR5 and limitation of antibody labeling. Therefore, we further gated on the sorted cells to identify CCR5+ and CCR5 populations for the analysis of GFP expression.