Abstract
Duchenne muscular dystrophy (DMD) is a genetic disease affecting about one in every 3,500 boys. This X-linked pathology is due to the absence of dystrophin in muscle fibers. This lack of dystrophin leads to the progressive muscle degeneration that is often responsible for the death of the DMD patients during the third decade of their life. There are currently no curative treatments for this disease but different therapeutic approaches are being studied. Gene therapy consists of introducing a transgene coding for full-length or a truncated version of dystrophin complementary DNA (cDNA) in muscles, whereas pharmaceutical therapy includes the use of chemical/biochemical substances to restore dystrophin expression or alleviate the DMD phenotype. Over the past years, many potential drugs were explored. This led to several clinical trials for gentamicin and ataluren (PTC124) allowing stop codon read-through. An alternative approach is to induce the expression of an internally deleted, partially functional dystrophin protein through exon skipping. The vectors and the methods used in gene therapy have been continually improving in order to obtain greater encapsidation capacity and better transduction efficiency. The most promising experimental approaches using pharmaceutical and gene therapies are reviewed in this article.
Introduction
Muscular dystrophies are characterized by progressive degeneration and weakness of multiple muscle groups depending on the specific dystrophy. Duchenne muscular dystrophy (DMD) is an X-linked pathology due to the absence of dystrophin in muscle fibers.1,2 The first symptoms of the disease appear during early childhood, usually before 3 years of age, and death occurs in the mid to late twenties.
The dystrophin gene, called DMD gene, extends over 2.4 megabases of the X chromosome, thus ~90 times the size of most genes. It contains 79 exons that code for a 14 kb mRNA.3,4 Its translation generates a large protein of 3,685 amino acids with a molecular size of 427 kDa5 called dystrophin. This protein is localized beneath the sarcolemma of the muscle fibers.6
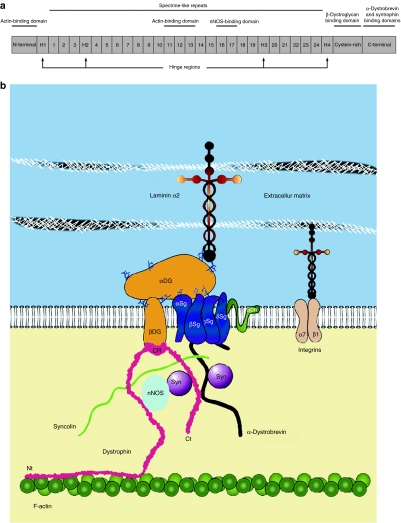
Dystrophin can be divided into four main regions (Figure 1a). The N-terminal domain interacts with actin filaments.7 The central rod domain also links to actin filaments8 and, in addition, to neuronal nitric oxide synthase (nNOS).9 This enzyme is implicated in several physiological functions of the muscle such as its regeneration and its contraction.10 The central domain also contains four hinge regions that provide flexibility.11 The third region is the cystein-rich domain that interacts with the sarcolemmal β-dystroglycan, which in turn interacts with the transmembrane α-dystroglycan.12 The dystrophin C-terminal region is associated with α-, β-, and γ-syntrophins.13,14,15 Since dystroglycans and syntrophins are also linked to other proteins, dystrophin thus interacts with many proteins in a complex called dystrophin-associated glycoprotein complex (DGC) (Figure 1b).16,17,18,19 The main function of dystrophin is to stabilize and link the muscle fiber cytoskeleton to the membrane. The lack of functional dystrophin results in the loss of the DGC, thereby rendering the muscle fibers less resistant to mechanical stress.16,20
Figure 1.
The dystrophin protein. (a) Schema representing the four main domains of dystrophin: the N-terminal part, central rod domain (containing 24 spectrin-like repeats and four hinge domains), cystein-rich region and the C-terminal part. The protein binding domains are also indicated. (b) Diagram of the dystrophin-associated glycoprotein complex (DGC). This complex includes dystrophin with its C-terminal (Ct), cysteine-rich (CR), and N-terminal (Nt) regions as well as proteins associated in this complex. DG, dystroglycan; nNOS, neuronal nitric oxide synthase; Sg, sarcoglycan; Syn, syntrophin. Modified from Odom et al.19
In DMD, the DMD gene mutations almost always result in a premature stop codon due to frameshift mutations or nonsense mutations. There are >4,700 different mutations divided into three main categories: deletion of one or more exons, duplication of one or more exons and small mutations. Depending on the cohorts studied, the proportion of these categories varies from 60 to 80% for deletions, from 7 to 11% for duplications and from 10 to 30% for more subtle DNA changes including nonsense mutations, splice-site mutations, and small insertions/deletions that disrupt the reading frame.21,22,23,24,25,26 As mentioned, most of the deletions in the DMD gene result in a frameshift.27 Those that do not produce a frameshift result in the production of an internally deleted dystrophin and give rise to a dystrophy called Becker muscular dystrophy (BMD).28 The BMD phenotype varies according to the functional loss of the missing exons but is generally less severe than DMD.29,30,31 For example, a deletion in the rod domain will often be less severe than a deletion in N-terminal. The life expectancy of BMD patients is also variable: some may suffer life threatening complications in their late twenties and have a similar life expectancy as DMD patients whereas many live a normal lifespan beyond 50 years of age.
DMD symptoms are very severe. Thus, even if there are currently no curative treatments for this disease, the medical monitoring and the care coverage of these patients contribute to prevention of some complications and to improvement in their quality of life. For that purpose, the follow-up of patients must be considered at various levels: rehabilitation, cardiac, pulmonary, orthopedic, psychosocial, and nutrition.32,33
Following the initial open-label trials of corticosteroids, the potential benefit of prednisone was clearly demonstrated >20 years ago in a double-blind randomized controlled trial for 6 months in a study of >100 boys.34 Subsequent reports showed equal benefit using deflazacort, a sodium-sparing steroid.35 These results were confirmed by other studies (see refs. 32,36,37 for an exhaustive list of these studies). Long-term follow-up of open-label administration of corticosteroids reveals prolonged ambulation for about 2 years. In addition, the lower prevalence of scoliosis through the use of long-term corticosteroid treatment represents a significant change in the natural progression of DMD.38 Prednisone prescription to DMD patients is now openly authorized in many countries but many patients are forced to stop taking the drug because of unwanted side effects that include weight gain, bone demineralization, vertebral compression fractures, hypertension, and/or behavior disorders.
Besides the DMD patient's follow-up, different therapeutic approaches are currently in development to improve the DMD phenotype. This review focuses more specially on the current status of pharmaceutical and of gene therapy approaches in DMD. We have not reviewed the different potential cell therapies for DMD; however, some ex vivo gene therapies have been included.
Pharmaceutical Approach
The great advantage of a pharmacological approach is that nearly all drugs can be delivered systemically (orally, intravenously, subcutaneously) and thus will reach and potentially treat all muscles which is critical for clinical success in DMD. However, the development and testing of new drugs for the DMD population is far from being a simple task.
Dystrophin restoration approaches
Stop codon read-through. About 10–15% of DMD patients have a mutation that converts an amino acid into a premature nonsense codon, while the rest of the mRNA is unaffected.21,22,23,24,25,26 Some drugs have been shown to enable stop codon read-through by introducing an amino acid at the premature stop codon to continue the mRNA translation. This phenomenon called “stop codon read-through” has been intensively investigated.
Gentamicin: Gentamicin is an aminoglycoside antibiotic interacting with the translational machinery (40S ribosomal subunit) when it recognizes a stop codon.39,40,41 This interaction induces the introduction of an amino acid at stop codons in the mRNA and thus allows the translational machinery to continue the mRNA translation.42,43 It specially occurs in premature stop codons since the context of nucleotide sequences surrounding nonsense mutations and regular stop codons are different.44 Gentamicin was tested as a therapeutic approach for DMD. When used in dystrophic (mdx) mice, this drug induced up to 20% dystrophin-positive fibers.45 After this positive result, two clinical trials on DMD and BMD patients were undertaken. However, the results were moderate46,47 as was also the case for some further studies in animals.48,49 Recently, a clinical trial showed that a 6 months gentamicin administration resulted in up to 15% dystrophin expression in three DMD patients, lower percentages in three other patients, and no expression in the remaining patients.50 The different results obtained in mouse and in human are probably due to the presence of different gentamicin isomers, which are not all equally potent in inducing read-through41 and since each gentamicin batch consists of a mix of different isomers, some batches may be more effective than others.
Given that gentamicin has variable effects and exhibits some toxicity, less toxic effective derivatives of this drug need to be developed for an effective DMD treatment.
Ataluren: Ataluren (PTC124) is a new molecule recently identified by PTC Therapeutics (South Plain Field, NJ). It is presumed to work similarly to gentamicin except that PTC124 binds to the 60S ribosomal subunit.51 Its efficiency is comparable to gentamicin in mouse: between 20 and 25% dystrophin-positive fibers were observed in treated mdx mice.52 Three phase II clinical studies began on DMD and BMD patients but these studies were halted prematurely on March 2010 since the predetermined primary outcome (30 m improvement compared to placebo in the 6-minute walk test) was not reached53 while ataluren was generally well tolerated in DMD patients.54 No information is available concerning the dystrophin expression in treated muscles.
Even though gentamicin and ataluren have shown good efficiency in the mdx mouse model, the clinical studies that have been done up to date showed that these drugs still need further improvements before they can be used clinically in DMD patients.
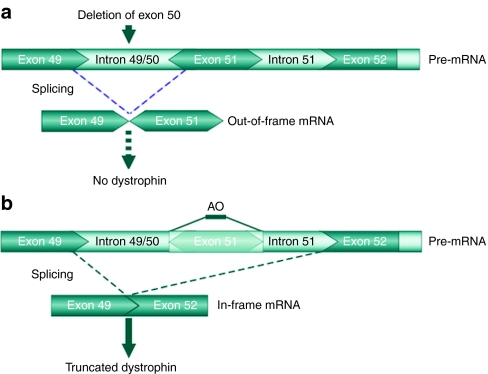
Exon skipping. In BMD patients, dystrophin is internally deleted, but still partially functional due to the presence of the essential N- and C-terminal domains. Using antisense molecules which were able to interfere with splicing signals, the skipping of the targeted specific exons in the dystrophin pre-mRNA can restore the open reading frame and allow the expression of an internally deleted but functional dystrophin in DMD patients (Figure 2). These molecules are small synthetic modified RNAs or DNAs called antisense oligonucleotides (AOs) able to bind specific intronic or exonic sites of pre-mRNA. Annealing to selected splice motifs, the AO essentially masks the targeted exon from the splicing machinery, thereby promoting specific exon exclusion from the mature mRNA. Two types of AO are mainly used: 2′-O-methyl-phosphorothioate (2OMP) and phosphorodiamidate morpholino oligomer (PMO) (Supplementary Figure S1).
Figure 2.
Example of exon skipping in Duchenne muscular dystrophy (DMD) patient who has a deletion of exon 50. (a) The absence of exon 50 in the dystrophin gene leads to an out-of-frame mRNA creating a premature stop codon in exon 51, thus aborting dystrophin synthesis during translation. (b) Using an antisense oligonucleotides (AO) targeting exon 51, this exon is skipped during splicing. This restores the open reading frame of the transcript and allows the synthesis of an internally deleted dystrophin. Modified from Van Deutekom et al.65
2′-O-methyl-phosphorothioates: 2OMPs contain around 20 nucleotides and are obtained by modifying the classic synthesis of oligonucleotides.55 The first modification is the replacement of the negatively charged oxygen by sulfur. The second one is the methylation of the hydroxyl group at the 2nd position of ribose. These modifications make the AOs more resistant to nucleases, improve their affinity for RNA, provide favorable pharmacokinetic properties and prevent RNase H to induce cleavage of RNA:RNA hybrids.56,57,58
Several 2OMPs designed to target several human DMD exons were tested with success in DMD patient-derived myotubes.59,60 In parallel, 2OMPs were designed to target the exon 23 of the mouse DMD gene since the nonsense mutation of mdx mouse is localized in this exon. Intramuscular administration of an AO targeting the exon 23 donor splice-site in these mice induced the restoration of dystrophin (without the exon 23) in the treated muscles.61 These AOs were also intravascularly injected in mdx mice. Treated mice showed dystrophin restoration in many muscles.62 However, low levels of dystrophin restoration were detectable in the heart.63 A study demonstrated that repeated 2OMP injections increased the AO efficiency without increasing its toxicity.62 A subcutaneous 2OMP injection has also been tested and this type of injection showed better pharmacokinetics and pharmacodynamics than intramuscular or intravenous injections.64
After these positive results in the mdx mouse model, a clinical trial on four DMD patients with the PRO051/GSK2402968 (2OMP targeting exon 51) was done. The muscle injected with 0.8 mg of this 2OMP showed 64–97% dystrophin-positive fibers (not corrected for positive muscle fibers in saline-injected contralateral muscle) with a level of dystrophin expression between 17 and 35%.65 No adverse effects were found in the treated muscles. A phase I/II clinical trial, in which this same AO was injected subcutaneously, was recently completed and showed that this AO was well tolerated in all patients and that novel dystrophin expression was detected in each treated patient in a dose dependent manner.66 A phase III study has started with this AO on DMD patients.67
Despite the fact that long-term toxicity studies in animal models with 2OMP are lacking, this approach seems promising.
Phosphorodiamidate morpholino oligomer: Similar to 2OMPs, PMOs (commonly referred to as morpholinos) are obtained by modifying the classic synthesis of oligonucleotides. Their ribose is replaced by a morpholine ring and the oxygen present in the phosphodiester link (the one that is not negatively charged) is replaced by a nitrogen atom. These modifications allow morpholinos to be biologically stable68 and have antisense properties.69
Exon 23 of the mouse DMD gene was the first target of morpholinos. Restoration of dystrophin was observed in the treated mdx mouse muscles when morpholinos were intramuscularly injected70 and in many muscles when intravenously71 or intraperitoneally injected.72 A partial restoration of dystrophin in the heart of mdx mice was also shown but the morpholino dose used was 50 times superior to the one used to treat skeletal muscles.72 Recent studies of long-term repeated systemic treatment of mdx mice over a year with naked PMO at doses of 5 and 50 mg/kg have shown significant improvement in pathology and complete normalization of locomotor behavior without signs of renal or hepatic toxicity.73 A morpholino designed to restore dystrophin expression in dystrophic (golden retriever muscular dystrophy) dogs was also synthesized and intravenously injected in these dogs. Five months later, treated dogs showed about 25% dystrophin-positive fibers throughout the body with a global improvement in muscle pathology in PMO-treated dogs compared to pretreated and untreated control dogs.74 No significant signs of toxicity were found.
To enhance the cellular uptake of PMOs, they can be conjugated to peptides or other conjugates. The delivery of a morpholino conjugated with a dendrimeric octaguanidine (Vivo-Morpholino) was efficient to induce dystrophin expression in mdx mouse muscles.75 Indeed, repeated injections at biweekly intervals achieved near 100% dystrophin-positive fibers in many skeletal muscles without eliciting a detectable immune response; the dystrophin restoration in the cardiac muscle reached up to 40%. PMOs conjugated with arginine-rich cell-penetrating peptides,76 called pPMOs, also produced excellent restoration of dystrophin expression in mdx mice.77,78 A pPMO targeting exon 23 was applied as well in utrophin−/− mdx mice by intraperitoneal injection. Whereas untreated animals typically died by 15 weeks of age, treated animals showed few signs of weakness, improved histopathology and appeared essentially normal at 1 year of age.79 A muscle-targeting heptapeptide (MSP) fused to an arginine-rich cell-penetrating peptide (B-peptide) and conjugated to a PMO, called B-MSP-PMO, was also shown to be efficient for restoring dystrophin in mdx muscles.80 Indeed, using an intravenous dose of 6 mg/kg of B-MSP-PMO administered biweekly over the course of 12 weeks, the dystrophin expression was found at a level of 100% in several muscles except for the heart. These pPMO seem well tolerated in mdx mice. Indeed, a pPMO targeting the exon 23 of the mouse DMD gene exhibited no toxic effects in kidneys at either 20 mg/kg weekly injection to the wild-type mice for 6 weeks or 30 mg/kg biweekly injection to mdx mice for 3 months. However, the same peptide conjugated to the PMO targeted to human exon 50 (AVI-5038) was found to cause mild tubular degeneration in the kidneys of nonhuman primates at 9 mg/kg weekly injections for 4 weeks.81
To target more dystrophin mutations occurring in DMD patients, other exons such as the exon 51 in mdx52 mice were targeted.82 In addition, it is possible to remove in-frame exons from the dystrophin pre-mRNA and induce specific internally deleted dystrophin by using AOs. This has been done for exons 19/20 and 52/53 in wild-type mice.83
After these positive results in animal models, a clinical trial in seven DMD patients was undertaken to skip exon 51 and thus to restore the reading frame of their dystrophin mRNA using unmodified morpholinos. The morpholino (AVI-4658) was intramuscularly injected and biopsies were taken 3–4 weeks later. Two patients were treated with a low dose of this morpholino (0.09 mg) and five patients with a higher dose (0.9 mg). Only the patients receiving the higher dose produced dystrophin although exon skipping was observed in all patients by reverse transcriptase PCR. In the five patients receiving the higher dose, the muscles injected with the AO showed 44–79% dystrophin-positive fibers (corrected for positive fibers in saline-injected contralateral muscle) with a level of dystrophin expression between 22 and 32%.84 No signs of toxicity were observed. After these encouraging results, a systemically delivered morpholino phase Ib/II clinical trial was undertaken. According to a press release from AVI Biopharma (Bothell, WA),85 19 DMD patients were enrolled in six dose cohorts (0.5, 1, 2, 4, 10, or 20 mg/kg) and treated during 12 weeks by weekly intravenous infusion. Some patients expressed dystrophin-positive fibers; those treated with the higher doses of morpholino had more uniform and widespread dystrophin-positive fiber distribution than patients who received lower doses. The morpholino was well tolerated in all patients. A phase II clinical trial is currently in preparation to evaluate higher weekly doses of AVI-4658 (50 and 100 mg/kg).85
Although pPMO seems to cause some toxicity in nonhuman primates, there are other ways to modify the peptide conjugate, which are hopefully less toxic, to allow clinical development for DMD patients.
Modification of the DMD gene with meganucleases or zinc finger nucleases. A new alternative treatment for DMD relies on the restoration of the dystrophin reading frame by inducing a micro-deletion or a micro-insertion in the DMD gene.86 This can be done by inducing double strand breaks at the end of the exon, which precedes a deletion, or at the beginning of an exon, which follows a deletion. These double strand breaks can be induced with specially engineered meganucleases or zinc finger nucleases. They are spontaneously repaired by a process called nonhomologous end-joining, which introduces a micro-insertion or a micro-deletion. Alternatively, double strand breaks can be repaired by homologous recombination by providing a donor plasmid containing the coding sequence that is deleted in the patient's genome.
Other approaches
Myostatin. A potential therapeutic method to improve muscle strength is to block myostatin. Myostatin is a member of the transforming growth factor-β family implicated in muscle size regulation. Indeed, in the myostatin gene knockout mouse, robust muscular hypertrophy and hyperplasia are observed.87 Antibodies against myostatin were produced and intraperitoneally injected in mdx mice. The treated mice showed muscular hypertrophy, muscle strength increase, and histological improvement.88 There are also other methods to block the myostatin pathway such as the use of follistatin89 or of myostatin propeptide.90 Another approach is to directly mutate the myostatin receptor, the activin type-II receptor91 or to inject a soluble form of this receptor.92 All these approaches led to improvements of the treated mouse phenotype similar to that observed in myostatin−/− mice. Recently, the use of destructive exon skipping of the myostatin pre-mRNA induced by 2OMP and PMO has been described to induce skeletal muscle hypertrophy, which along with dystrophin exon skipping (see above) may thus provide a potential combined antisense strategy to simultaneously reactivate dystrophin expression and increase muscle bulk.93 In a recent clinical trial, the use of an antibody against myostatin (MYO-029) was undertaken. Although the antibody was well tolerated, no muscle strength improvements were detected perhaps due to a lower dose of antibody.94 Other clinical trials with myostatin inhibitors are currently undertaken by at least four biotechnology and pharmaceutical companies.95
Utrophin. Utrophin shares 80% sequence identity with dystrophin and is expressed in the muscles during embryonic development.96 However, in adult myofibers, it is located only at the neuromuscular junction and at the myotendinous junctions. Utrophin is over-expressed in muscle fibers of dystrophic mice and of DMD patients.97,98 Since it has sequence homology with dystrophin, it was suggested that its upregulation could slow down DMD development. When its expression is increased three- to fourfold in transgenic mdx mice, their phenotype is similar to wild-type mice.99 Therefore, an increase of the utrophin expression may be a potential therapy to improve DMD patients. The injection of heregulin in mdx mice increased utrophin expression by two to threefold and led to histological improvements.100 The injection of -arginine or nitric oxide also allowed utrophin upregulation in mdx mice.101 Recently, the intraperitoneal injection of a TAT-utrophin protein in mdx mice increased their muscle strength.102 A drug developed by Summit PLC (C110/BM195) to upregulate the utrophin expression was carried out by BioMarin pharmaceuticals in a phase I clinical trial with normal individuals. No adverse effects were reported but the pharmokinetics of the drug did not allow them to continue the development of this drug. Summit PLC is currently working on a new formulation, which may improve the pharmokinetics. Further investigation in increasing utrophin expression is required since the molecules tested so far in mdx mice did not increase utrophin expression sufficiently to completely suppress the symptoms due to the dystrophin deficiency in mdx mice.103,104 Moreover, utrophin does not seem to anchor nitric oxide synthase at the sarcolemma like dystrophin does, thus leading to a premature muscle ischemia.105 However, the levels of utrophin upregulation may be sufficient to alleviate most of the DMD symptoms.
Gene Therapy
Since the first clinical trial of gene therapy in 1990,106 there has been a strong interest for this therapeutic approach. However in 1999, a major setback occurred due to the death of a patient treated with an adenovirus for ornithine transcarbamylase deficiency.107 This death is believed to have been triggered by a severe innate immune response to the adenoviral vector. In 2002, another death occurred in a clinical trial for severe combined immunodeficiency with the use of a retrovirus where one of the treated patients died due to the activation of an oncogene.108 However, the fatality rate of gene therapy is still much lower than that of the standard bone marrow transplantation treatment for severe combined immunodeficiency patients.109 Moreover, >45 patients have now been treated via gene therapy, resulting in one death and >40 cures. Gene therapy is thus an appealing approach to cure many hereditary diseases such as DMD.
Gene therapy in DMD consists of the introduction of a functional copy of the DMD gene in muscle fibers with the aim of restoring muscle function including force generation and resistance to muscle contraction induced damage. The concept of dystrophin internally deleted genes that would fit the packaging capacity of small viral vectors came from clinical observations that some BMD patients with internally deleted dystrophins could maintain ambulation for many decades. This gave rise to the concept of mini-dystrophin (mDYS) or micro-dystrophin (µDys). Gene therapy is divided in two distinct categories: those using viral vectors to transfer the gene are referred to as “viral gene therapy” and those employing naked DNA as “nonviral gene therapy”.
Internally deleted dystrophin genes
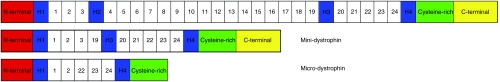
In gene therapy, the transgenes generally contain complementary DNA (cDNA) corresponding only to coding regions of a gene, i.e., exons without introns. The dystrophin cDNA size is about 11 kb and is called full-length dystrophin (FLDYS). Apart from this FLDYS, several mDYS and µDys internally deleted versions exist (Figure 3). Indeed, a BMD patient with a deletion of the exons 17–48 in the DMD gene was reported to have only a mild dystrophic phenotype.30 The missing region was located in the spectrin-like repeats of the rod domain resulting in an internally deleted dystrophin with only eight of these repeats instead of 24. The corresponding transgene was thus constructed110 and other, even smaller, truncated versions were designed subsequently.111,112 These constructions were called mDYS, or µDys when the C-terminal part is also missing.
Figure 3.
Dystrophin versions. The full-length dystrophin cDNA (11 kb) is represented at the top. The middle schema represents an example of a mini-dystrophin cDNA with an H2-R18 deletion; the approximate size of mini-dystrophins is about 6 kb. The bottom representation is a schema of a micro-dystrophin cDNA (around 4 kb) with an R3-R21 and C-terminal deletion.
Several transgenic mice expressing these internally deleted dystrophins were generated and analyzed112,113,114,115; all these mice showed the restoration of the DGC. The simple fact of restoring the DGC improves the muscle histology as well as the reduced leukocyte infiltration and the decreased number of centro-nucleated muscle fibers. The muscle strength is also increased but does not reach wild-type levels. However, the observed improvements vary depending on which exons are deleted. The use of internally deleted dystrophins is attractive but the best phenotypic restorations are still obtained with the use of FLDYS.
Viral gene therapy
Different viral vectors could be used for DMD gene therapy. Adenoviral vectors show poor efficiency in adult animal models compared to newborns. Moreover, the use of adenoviral vectors is complicated since half of the human population already has neutralizing antibodies against the adenoviral capsid and also tends to be far more immunogenic than adeno-associated viral vectors (AAV) and retroviral vectors. Due to these limitations, only AAV and lentiviral vectors are described below.
AAV. There are many different AAV, i.e., >100 different sequences are available. Some of the differences lead to different serotypes. The serotypes 1, 2, 6, 8, and 9 are more frequently used for muscle gene therapy. The AAV vector is the only efficient vector for local or systemic delivery to the skeletal muscle and heart116,117 but its packaging capacity limits the size of the dystrophin transgene.
AAV1118 and AAV2111 carrying transgenes encoding for µDYS were injected in mdx mouse muscles with success. Indeed, up to 80% dystrophin-positive fibers were found in the treated muscles. These AAV injections also restored the DGC. The results on the mdx mouse model being conclusive, experiments using AAV vectors were done in larger animal models. AAV6 and AAV8 coding for µDYS were injected in the dog model. Although dystrophin expression was observed, cytotoxic immune response against the viral capsid was detected,119,120 which has also been observed for other transgenes delivered by AAV vectors in the dog model.121 The AAV vector was also tested in nonhuman primates. Five months after the intramuscular injection of an AAV8 coding for µDYS, the transgene expression reached 80% in the treated muscle but this percentage decreased to 40% when the animal already had pre-existing antibodies against the AAV.122 In small rodent studies, AAV vectors rarely cause cellular immune responses against either the capsid proteins or the transgene products. But in large animal and human studies, variable immunological outcomes have been observed.
Recently, a clinical trial was undertaken on six DMD patients with an AAV vector coding for a functional µDYS. Of the six treated patients, two showed pre-existing T-cells recognizing the rare dystrophin-positive revertant fibers that presented peptide epitopes deemed by the host as nonself. This was detected in ELISpots of peripheral blood mononuclear cells before and after intramuscular injection of the AAV.123 Another patient had T-cells recognizing an epitope that encoded the transgene product but absent in the revertant fibers. Although the clinical trial was safe and muscle biopsies from the gene vector-treated arms and the contralateral control arms showed no difference in lymphocytes infiltration, these intriguing findings strongly suggest that additional work is required to determine how many patients have T-cells to dystrophin epitopes and whether those T-cells will prevent successful gene therapy in DMD. In addition, choices of AAV vector serotypes and promoters may also make an impact on the clinical outcome.
Exon skipping was also investigated in combination with AAV vectors. AAV1 coding for the U7 snRNA or U1 small nuclear RNA (snRNA) genes modified to target the mouse dystrophin exon 23 were injected in mdx mice. The expression of the internally deleted dystrophin was observed up to 3 months following the injection of an AAV1 coding for the U7 snRNA124 and for at least 1 year and half with an AAV1 coding for the U1 snRNA.125 These results are encouraging but this approach has to be further investigated in larger animals such as nonhuman primates or dogs.
AAV vectors were also used to interfere with the myostatin pathway. An AAV vector coding for the myostatin propeptide, a myostatin inhibitor, was designed and injected in mdx mice. Muscle hypertrophy leading to phenotypic improvements was observed in the treated mice.126 Dogs were also treated with the same vector. Unfortunately, few parameters were studied in this experiment and only the hypertrophy of some muscles was noted.127 In contrast to the other dog studies using AAVs coding for µDYS, no immune responses against the AAV capsid were observed in this study. In the mouse, a recent experiment used an AAV coding for the activin type-II receptor to block the myostatin pathway. The effects of this AAV injection were similar to those observed in the mouse following the injection of the purified activin type-II receptor alone.128
The results obtained with AAV vectors are interesting for the development of a DMD therapy. Nevertheless long-term studies of the transgene expression and the immune response against the capsid will be required before this can be considered as potential treatment for DMD.
Lentivirus. The lentivirus encapsidation size is limited to carry the mDYS. Thus, a lentiviral vector carrying this internally deleted DMD gene was intramuscularly injected in adult and newborn mdx mouse muscles. The best results were obtained in younger mice where 65% of muscle fibers expressed the transgene.129 In addition, better strength and protection against contraction induced injury were observed in the treated muscles. The lentivirus injection also transduced satellite cells.130 Despite favorable results in small animals, no studies are available for larger animal models. Moreover, the random integration of lentiviral vectors, according to the target tissues and the enhancers used in a construct, predisposes to induction of tumors (insertional mutagenesis) even though they have not been observed to date in the described experiments.
Lentivirus can also be used to genetically modify cells, which can be transplanted or injected in animal models or eventually in patients. This technique is called ex vivo gene therapy. A lentiviral vector coding for µDYS was used to integrate this gene in the genome of side population cells, which were then intravenously injected in mdx mice. Only 1% of muscle fibers expressed the transgene in the treated muscles,131 though this percentage was increased to 5% when these cells were intra-arterially injected.132 Dystrophic dog mesoangioblasts were also transduced with a lentiviral vector coding for the human µDYS and intra-arterially injected in the same dogs.133 The treated dogs showed good expression of human µDYS but two of the three treated dogs died of pneumonia during the experiment. The cause of this death was not explained by the investigators but the accumulation of the injected cells in the lungs could be involved in this mortality. Other cell types such as human and nonhuman primate myoblasts were transduced with human µDYS and transplanted with success in immunodeficient mouse and in nonhuman primate muscles respectively.134 A lentiviral vector coding for dog µDYS was also used to transduce human and dystrophic dog myoblasts. Subsequently, these cells were transplanted in mouse muscles and transgene-positive fibers were observed in the treated muscles.135
In addition to the possibility of delivering an internally deleted dystrophin, the lentiviral vector may be used to induce exon skipping as well. A lentiviral vector coding for the U7 snRNA gene modified to induce the skipping of human dystrophin exon 51 was designed. Myoblasts of DMD patients having a deletion of exons 49 and 50 were transduced with this lentivirus and transplanted in immunodeficient mouse muscles. One month later, the expression of internally deleted dystrophin (without the exons 49–51) was detected in the treated muscles.134 This approach was also used successfully with AC133+ cells.136
The use of lentiviral vector is promising for DMD but its efficacy and the risk of tumorigenicity from cells transduced by direct injection of a lentiviral vector or by ex vivo genetic modification need to be evaluated in clinical trials.
Nonviral gene therapy
Nonviral gene therapy allows the introduction of a transgene into a tissue without using a viral vector. Thus, the main advantage of this method is to avoid any immune response due to viral capsids or other viral proteins. There are also no limitations concerning the transgene size but the transfection efficiency of nonviral gene therapy is progressively reduced with the increasing plasmid size.
Naked DNA. The simplest method to deliver a plasmid into muscle is its direct injection. Plasmids coding for µDYS and for FLDYS were injected in mdx mice110; however, the transfection efficiency was very low. Nevertheless, there is a possibility for prolonged transgene expression in muscles since muscle fibers are postmitotic. A phase I clinical trial was undertaken in 2004 on nine dystrophic patients137 that were intramuscularly injected with a plasmid coding for human FLDYS. The three treated DMD patients just showed rare dystrophin-positive fibers. In the six treated BMD patients, the average level of dystrophin expression was slightly higher (about 3%). Although the application of naked DNA is appealing since this method is fast and the plasmids are easy to produce, the efficiency of direct intramuscular injection is currently too low to be clinically relevant. To improve gene delivery, chemical and physical methods can be used. However, due to the low effectiveness of chemical methods in vivo, only physical approaches are included in the present review.
Physical approach
Hydrodynamic pressure: Good expression levels were obtained following a rapid injection of a large quantity of plasmid DNA coding for luciferase or β-galactosidase.138 This intravenous injection of a large volume while using a tourniquet to occlude blood flow allows good dissemination of the naked DNA in muscles.139 Indeed, the intravascular pressure induced the formation of transient pores in the endothelium of blood vessels allowing macromolecules, such as plasmids, to leak into the surrounding muscle and thereby access the muscle fibers.140 The safety of this method was demonstrated in mice and in nonhuman primates.139,141 The hydrodynamic limb vein injection used in mdx mice with a plasmid coding for FLDYS resulted in dystrophin expression in up to 20% of muscle fibers for >1 year.142 The phenotype of the treated mice was also improved. Golden retriever muscular dystrophy dogs were also treated with this technique. The procedure appeared safe in the treated animals and enabled to obtain dystrophin expression but further work is required to determine the exact level of dystrophin expression.143 This approach seems thus promising to introduce naked DNA in muscles.
Electroporation: A second method to improve the efficiency of muscle transfection is electroporation. The electric field used in this method enhanced the uptake of a plasmid previously injected in the muscle.144,145 Indeed, the electric pulses permeabilized the cellular membrane, creating transient pores that facilitated the plasmid entry into the cell. However, these pores also increased calcium entry and activated proteases.146 Therefore, it is important to select voltage settings, which allow maximal efficiency with the least amount of damage. As with the hydrodynamic pressure method, the electroporation of naked DNA in muscles resulted in transgene expression for >1 year.147 The heart can also be treated by electroporation according to a recent research article.148 A study showed that satellite cells can be transfected with this technique.149 However, this study has not been confirmed. According to Schwann's equation, the threshold intensity of the applied electric field necessary to obtain membrane permeabilization is inversely proportional to the cell radius.150 Since the radius of satellite cells is smaller than that of muscle fibers, the satellite cells and the muscle fibers cannot be electroporated simultaneously.
Since its first use in a clinical trial in 1991,151 plasmid electroporation has proven to be safe and effective for transgene delivery to several tissues.152,153,154 In the DMD context, a plasmid coding for mouse FLDYS was electroporated in mdx mouse muscles. The electroporated muscle fibers expressed the transgene for at least 1 month and exhibited a reduced number of centro-nucleated muscle fibers as well.155,156 Dog FLDYS was also introduced with success in dystrophic dog muscle.157 In this case, a specific immune response was observed in the treated dog muscle. Further studies are thus required to determine whether this immune response was against dystrophin or against the product of another transgene also present in the plasmid.
Discussion
DMD is a devastating pathology leading to severe muscle weakness. This disease is due to the lack of dystrophin in smooth, cardiac, and skeletal muscles. Although there are currently no curative treatments for DMD, several therapeutic approaches are undergoing clinical evaluation such as pharmaceutical approaches and gene therapy.
Pharmaceutical approaches
The stop codon read-through is one of pharmaceutical approaches. The last clinical trial with ataluren showed that it was unable to achieve its primary outcome for improved muscle function. The long-term gentamicin clinical trials gave mixed results and showed too many toxicity issues to consider this antibiotic as a feasible approach to treat DMD patients having nonsense mutation. Moreover, stop codon read-through would only be relevant to only about 10 to 15% of DMD patients.
Exon-skipping can in theory be applied to 80% of DMD patients.25 This method has shown its efficiency in mouse and dog models. Clinical trials using 2OMPs and morpholinos were also undertaken on DMD patients. In both cases, dystrophin expression was observed in the treated muscles and no significant adverse effects have been encountered. Only the results of intramuscular exon skipping trials have been published so far with results restricted to the site of delivery. However, the first results on the clinical trials using a morpholino (AVI-4658) or a 2OMP (PRO051/GSK2402968) systemically delivered showed good dystrophin expression.66,85 Even though there are no long-term toxicity studies (>6 months) available on 2OMPs and morpholinos in nonhuman primate, these two compounds are promising for DMD.
Currently, no molecules upregulate utrophin expression sufficiently to restore the phenotype of dystrophic mouse models. Therefore, utrophin upregulation must be further improved before applying it in DMD.
Gene therapy
Another method to obtain a functional dystrophin is to introduce a cDNA in muscle fibers using gene therapy. The most promising viral vector to introduce a micro-dystrophin cDNA in muscle fibers is currently the AAV vector. The results obtained with this vector in mice, dogs, and nonhuman primates are good despite the fact that antibodies against the AAV capsid were sometimes found in the treated animals (humans also have pre-existing antibodies against AAV and adenovirus). However, a recent clinical trial using an AAV coding for micro-dystrophin did not demonstrate significant transgene expression in the treated DMD patient muscles. Moreover, this study detected lymphocytes reacting with dystrophin in response to transgene expression.123
One way to eventually avoid the potential toxicity following the dissemination of viral vectors throughout the body158 is to transplant autologous cells, which have been genetically modified ex vivo. This ex vivo gene therapy has shown positive results in mice and nonhuman primates but is nevertheless limited by the same problems as myoblast transplantation, i.e., the difficulty of reaching small muscles and the high number of injection trajectories necessary to obtain a high percentage of dystrophin-positive fibers. Exon skipping can also be induced by viral vectors carrying the U7 snRNA gene modified to target a specific exon. Since no results are yet available in large animals with this gene, the AO technology currently remains the most efficient and most frequently used method to induce exon skipping in DMD.
An alternative to ex vivo gene therapy is the use of naked plasmid delivered by hydrodynamic pressure or by electroporation. These two techniques have shown good efficiency to deliver dystrophin cDNA or internally deleted versions of it in mouse model, although these physical methods are less efficient than systemic injection of viral vectors. Moreover, only a few preliminary results are available in larger animal models, such as dogs and nonhuman primates. The main limiting factor for electroporation is that at this time only a small number of muscle fibers can be treated with this technique since it requires penetration with electrodes into each muscle. The hydrodynamic method can be applied only to arm and leg muscles but not to muscles of the head and trunk.
Response to dystrophin in clinical trials
During clinical trials on DMD patients, anti-dystrophin antibodies were observed following nondystrophic myoblast transplantation159 and dystrophin-specific T-cells were detected following the injection of AAV coding for micro-dystrophin. The presence of dystrophin-specific T-cells was also detected in one patient after treatment with gentamicin.123 No anti-dystrophin antibodies were found in the DMD patients treated with AVI-4658 or with PRO051 but the presence of dystrophin-specific T-cells was not investigated. Apparently, there were no T-cell responses, or if there were, it was not effective enough to hamper dystrophin expression. This seems to indicate that if a therapeutic approach is effective to restore dystrophin in muscle fibers, some DMD patients may have to be under a sustained immunosuppression treatment.
Conclusion
Even though the DMD gene was discovered 23 years ago, there are still no curative treatments for DMD although the use of steroids and assisted ventilation have greatly improved the quality of life and extended life span by nearly 50%.33
When a therapeutic approach is found to restore dystrophin in the DMD patient's muscles, the problems of fat infiltration or fibrosis in the muscles will still need to be resolved, as well as the existing muscle weakness or bone deformation. An approach to improve muscle strength is to block the myostatin pathway. Indeed, myostatin inhibition leads to muscle hypertrophy and muscle strength increases in animals. The process of fat infiltration and fibrosis in DMD patient's muscles is not well understood and needs to be further investigated. The best approach will thus be to treat DMD patients when they are still young to avoid most of the consequences due to the absence of dystrophin. Moreover, all muscles (or a large proportion of them) will need to be treated to obtain a curative treatment.
SUPPLEMENTARY MATERIAL Figure S1. AOs used in DMD. Schematic representation of 2OMP (left) and PMO (right). R and R′ indicate respectively the continuation of the oligomer chain in the 5′ and 3′ direction.
Acknowledgments
We thank Jerry R Mendell for his helpful suggestions. J.P.T. is the president and hold shares in CellGene, a biotech company involved in the development of cell and gene therapies.
Supplementary Material
AOs used in DMD. Schematic representation of 2OMP (left) and PMO (right). R and R′ indicate respectively the continuation of the oligomer chain in the 5′ and 3′ direction.
REFERENCES
- Zatz M, Vianna-Morgante AM, Campos P., and, Diament AJ. Translocation (X;6) in a female with Duchenne muscular dystrophy: implications for the localisation of the DMD locus. J Med Genet. 1981;18:442–447. doi: 10.1136/jmg.18.6.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KE, Pearson PL, Harper PS, Murray JM, O'Brien T, Sarfarazi M.et al. (1983Linkage analysis of two cloned DNA sequences flanking the Duchenne muscular dystrophy locus on the short arm of the human X chromosome Nucleic Acids Res 112303–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM., and, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C., and, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Koenig M, Monaco AP., and, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Zubrzycka-Gaarn EE, Bulman DE, Karpati G, Burghes AH, Belfall B, Klamut HJ.et al. (1988The Duchenne muscular dystrophy gene product is localized in sarcolemma of human skeletal muscle Nature 333466–469. [DOI] [PubMed] [Google Scholar]
- Levine BA, Moir AJ, Patchell VB., and, Perry SV. The interaction of actin with dystrophin. FEBS Lett. 1990;263:159–162. doi: 10.1016/0014-5793(90)80728-2. [DOI] [PubMed] [Google Scholar]
- Amann KJ, Renley BA., and, Ervasti JM. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J Biol Chem. 1998;273:28419–28423. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- Lai Y, Thomas GD, Yue Y, Yang HT, Li D, Long C.et al. (2009Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy J Clin Invest 119624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS., and, Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- Koenig M., and, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265:4560–4566. [PubMed] [Google Scholar]
- Ishikawa-Sakurai M, Yoshida M, Imamura M, Davies KE., and, Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to β-dystroglycan. Hum Mol Genet. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA., and, Froehner SC. Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-m. [DOI] [PubMed] [Google Scholar]
- Ahn AH., and, Kunkel LM. Syntrophin binds to an alternatively spliced exon of dystrophin. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams ME, Mueller HA., and, Froehner SC. In vivo requirement of the alpha-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J Cell Biol. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM., and, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Pilgram GS, Potikanond S, Baines RA, Fradkin LG., and, Noordermeer JN. The roles of the dystrophin-associated glycoprotein complex at the synapse. Mol Neurobiol. 2010;41:1–21. doi: 10.1007/s12035-009-8089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom GL, Gregorevic P., and, Chamberlain JS. Viral-mediated gene therapy for the muscular dystrophies: successes, limitations and recent advances. Biochim Biophys Acta. 2007;1772:243–262. doi: 10.1016/j.bbadis.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KP., and, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ., and, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- Scheuerbrandt G. Approaching therapies for boys with Duchenne muscular dystrophy. Parent Project Muscular Dystrophy. Annual conference in Cincinnati/Ohio, 13-16 July 2006. Acta Myol. 2006;25:77–97. [PubMed] [Google Scholar]
- Cunniff C, Andrews J, Meaney FJ, Mathews KD, Matthews D, Ciafaloni E.et al. (2009Mutation analysis in a population-based cohort of boys with Duchenne or Becker muscular dystrophy J Child Neurol 24425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, United Dystrophinopathy Project Consortium et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30:1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ.et al. (2009Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations Hum Mutat 30293–299. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Buzin CH, Feng J, Yan J, Serrano C, Sangani DS.et al. (2001Diagnosis of Duchenne dystrophy by enhanced detection of small mutations Neurology 57645–650. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H., and, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Becker PE., and, Kiener F. [A new x-chromosomal muscular dystrophy] Arch Psychiatr Nervenkr Z Gesamte Neurol Psychiatr. 1955;193:427–448. doi: 10.1007/BF00343141. [DOI] [PubMed] [Google Scholar]
- Malhotra SB, Hart KA, Klamut HJ, Thomas NS, Bodrug SE, Burghes AH.et al. (1988Frame-shift deletions in patients with Duchenne and Becker muscular dystrophy Science 242755–759. [DOI] [PubMed] [Google Scholar]
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE.et al. (1990Very mild muscular dystrophy associated with the deletion of 46% of dystrophin Nature 343180–182. [DOI] [PubMed] [Google Scholar]
- Beggs AH, Hoffman EP, Snyder JR, Arahata K, Specht L, Shapiro F.et al. (1991Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies Am J Hum Genet 4954–67. [PMC free article] [PubMed] [Google Scholar]
- Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, DMD Care Considerations Working Group et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, DMD Care Considerations Working Group et al. Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care. Lancet Neurol. 2010;9:177–189. doi: 10.1016/S1474-4422(09)70272-8. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Moxley RT, Griggs RC, Brooke MH, Fenichel GM, Miller JP.et al. (1989Randomized, double-blind six-month trial of prednisone in Duchenne's muscular dystrophy N Engl J Med 3201592–1597. [DOI] [PubMed] [Google Scholar]
- Biggar WD, Gingras M, Fehlings DL, Harris VA., and, Steele CA. Deflazacort treatment of Duchenne muscular dystrophy. J Pediatr. 2001;138:45–50. doi: 10.1067/mpd.2001.109601. [DOI] [PubMed] [Google Scholar]
- Angelini C. The role of corticosteroids in muscular dystrophy: a critical appraisal. Muscle Nerve. 2007;36:424–435. doi: 10.1002/mus.20812. [DOI] [PubMed] [Google Scholar]
- Manzur AY, Kuntzer T, Pike M., and, Swan A. Glucocorticoid corticosteroids for Duchenne muscular dystrophy. Cochrane Database Syst Rev. 2008;1:CD003725. doi: 10.1002/14651858.CD003725.pub3. [DOI] [PubMed] [Google Scholar]
- King WM, Ruttencutter R, Nagaraja HN, Matkovic V, Landoll J, Hoyle C.et al. (2007Orthopedic outcomes of long-term daily corticosteroid treatment in Duchenne muscular dystrophy Neurology 681607–1613. [DOI] [PubMed] [Google Scholar]
- Singh A, Ursic D., and, Davies J. Phenotypic suppression and misreading Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- Palmer E, Wilhelm JM., and, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D., and, Puglisi JD. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ. Correction of genetic disease by making sense from nonsense. J Clin Invest. 1999;104:367–368. doi: 10.1172/JCI8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurino S., and, Nigro V. Readthrough strategies for stop codons in Duchenne muscular dystrophy. Acta Myol. 2006;25:5–12. [PubMed] [Google Scholar]
- Manuvakhova M, Keeling K., and, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Davis ER, Cordier L, Shoturma DI, Leland SE., and, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KR, Hamed S, Hadley DW, Gropman AL, Burstein AH, Escolar DM.et al. (2001Gentamicin treatment of Duchenne and Becker muscular dystrophy due to nonsense mutations Ann Neurol 49706–711. [PubMed] [Google Scholar]
- Politano L, Nigro G, Nigro V, Piluso G, Papparella S, Paciello O.et al. (2003Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results Acta Myol 2215–21. [PubMed] [Google Scholar]
- Dunant P, Walter MC, Karpati G., and, Lochmüller H. Gentamicin fails to increase dystrophin expression in dystrophin-deficient muscle. Muscle Nerve. 2003;27:624–627. doi: 10.1002/mus.10341. [DOI] [PubMed] [Google Scholar]
- Arakawa M, Shiozuka M, Nakayama Y, Hara T, Hamada M, Kondo S.et al. (2003Negamycin restores dystrophin expression in skeletal and cardiac muscles of mdx mice J Biochem 134751–758. [DOI] [PubMed] [Google Scholar]
- Malik V, Rodino-Klapac LR, Viollet L, Wall C, King W, Al-Dahhak R.et al. (2010Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy Ann Neurol 67771–780. [DOI] [PubMed] [Google Scholar]
- PTC Therapeutics Ataluren for genetic disorders . < http://www.ptcbio.com/3.1.1_genetic_disorders.aspx >.
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P.et al. (2007PTC124 targets genetic disorders caused by nonsense mutations Nature 44787–91. [DOI] [PubMed] [Google Scholar]
- Clinical Trials. < http://www.clinicaltrials.gov/ct2/results?term=ptc124 >.
- PTC Therapeutics 2010Pivotal data presented at the world muscle society congress suggest ataluren slows the loss of walking ability in patients with nonsense mutation Duchenne/Becker muscular dystrophy . < http://ptct.client.shareholder.com/releasedetail.cfm?ReleaseID=518941 >.
- Dias N., and, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- Stein CA., and, Cheng YC. Antisense oligonucleotides as therapeutic agents–is the bullet really magical. Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ.et al. (1993Evaluation of 2'-modified oligonucleotides containing 2'-deoxy gaps as antisense inhibitors of gene expression J Biol Chem 26814514–14522. [PubMed] [Google Scholar]
- Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM.et al. (19972′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells J Biol Chem 27211994–12000. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT.et al. (2001Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells Hum Mol Genet 101547–1554. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F.et al. (2003Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients Hum Mol Genet 12907–914. [DOI] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA.et al. (2003Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse Nat Med 91009–1014. [DOI] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J.et al. (2005Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles Proc Natl Acad Sci USA 102198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk HA, de Winter CL, de Kimpe SJ, van Kuik-Romeijn P, Heuvelmans N, Platenburg GJ.et al. (2009In vivo comparison of 2′-O-methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping J Gene Med 11257–266. [DOI] [PubMed] [Google Scholar]
- Heemskerk H, de Winter C, van Kuik P, Heuvelmans N, Sabatelli P, Rimessi P.et al. (2010Preclinical PK and PD studies on 2′-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model Mol Ther 181210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M.et al. (2007Local dystrophin restoration with antisense oligonucleotide PRO051 N Engl J Med 3572677–2686. [DOI] [PubMed] [Google Scholar]
- Prosensa Developing therapies for neuromuscular diseases . < http://prosensa.eu/technology-and-products/pipeline/pro051gsk2402968 >.
- Prosensa GSK and Prosensa announce start of phase III study of investigational Duchenne muscular dystrophy medication 2011 ) < http://www.prosensa.eu/press-release/gsk-and-prosensa-announce-start-phase-iii-study-investigational-dmd-medication >.
- Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB., and, Weller DD. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 1996;6:267–272. doi: 10.1089/oli.1.1996.6.267. [DOI] [PubMed] [Google Scholar]
- Partridge M, Vincent A, Matthews P, Puma J, Stein D., and, Summerton J. A simple method for delivering morpholino antisense oligos into the cytoplasm of cells. Antisense Nucleic Acid Drug Dev. 1996;6:169–175. doi: 10.1089/oli.1.1996.6.169. [DOI] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S., and, Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD.et al. (2006Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology Nat Med 12175–177. [DOI] [PubMed] [Google Scholar]
- Wu B, Lu P, Benrashid E, Malik S, Ashar J, Doran TJ.et al. (2010Dose-dependent restoration of dystrophin expression in cardiac muscle of dystrophic mice by systemically delivered morpholino Gene Ther 17132–140. [DOI] [PubMed] [Google Scholar]
- Malerba A, Sharp PS, Graham IR, Arechavala-Gomeza V, Foster K, Muntoni F.et al. (2011Chronic systemic therapy with low-dose morpholino oligomers ameliorates the pathology and normalizes locomotor behavior in mdx mice Mol Ther 19345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S.et al. (2009Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs Ann Neurol 65667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Li Y, Morcos PA, Doran TJ, Lu P., and, Lu QL. Octa-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx mice. Mol Ther. 2009;17:864–871. doi: 10.1038/mt.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton HM, Nelson MH, Hatlevig SA, Reddy MT., and, Iversen PL. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Buckley B, Roberts J, Sazani P, Fucharoen S.et al. (2008Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice Mol Ther 161624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Moulton HM, Iversen PL, Jiang J, Li J, Li J.et al. (2008Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer Proc Natl Acad Sci USA 10514814–14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Babbs A, Powell D, Kole R, Fletcher S, Wilton SD.et al. (2010Prevention of dystrophic pathology in severely affected dystrophin/utrophin-deficient mice by morpholino-oligomer-mediated exon-skipping Mol Ther 18198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Moulton HM, Betts C, Merritt T, Seow Y, Ashraf S.et al. (2010Functional rescue of dystrophin-deficient mdx mice by a chimeric peptide-PMO Mol Ther 181822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton HM., and, Moulton JD. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim Biophys Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Nakamura A, Yokota T, Saito T, Okazawa H, Nagata T.et al. (2010In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse Mol Ther 181995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S, Adams AM, Johnsen RD, Greer K, Moulton HM., and, Wilton SD. Dystrophin isoform induction in vivo by antisense-mediated alternative splicing. Mol Ther. 2010;18:1218–1223. doi: 10.1038/mt.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C.et al. (2009Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study Lancet Neurol 8918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVI-4658 Data From Phase 1b/2 Study Presented at 15th International Congress of the World Muscle Society Supports Potential as Disease Modifying Therapy. AVI BioPharma's investigational drug candidate avi-4658 demonstrates broadly favorable profile of safety and tolerability, new dystrophin expression, stable clinical performance and inflammatory modulation in the treatment of Duchenne muscular dystrophy . < http://investorrelations.avibio.com/phoenix.zhtml?c=64231&p=irol-newsArticle&ID=1483148&highlight= >.
- Chapdelaine P, Pichavant C, Rousseau J, Pâques F., and, Tremblay JP. Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 2010;17:846–858. doi: 10.1038/gt.2010.26. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM., and, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS.et al. (2002Functional improvement of dystrophic muscle by myostatin blockade Nature 420418–421. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Wolfman NM, Celeste AJ, Hattersley G, Hewick R., and, Rosen V. A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos. Dev Biol. 1999;208:222–232. doi: 10.1006/dbio.1998.9191. [DOI] [PubMed] [Google Scholar]
- Böttinger EP, Factor VM, Tsang ML, Weatherbee JA, Kopp JB, Qian SW.et al. (1996The recombinant proregion of transforming growth factor β1 (latency-associated peptide) inhibits active transforming growth factor β1 in transgenic mice Proc Natl Acad Sci USA 935877–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ., and, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Reed LA, Davies MV, Girgenrath S, Goad ME, Tomkinson KN.et al. (2005Regulation of muscle growth by multiple ligands signaling through activin type II receptors Proc Natl Acad Sci USA 10218117–18122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JK, Malerba A, Popplewell L, Foster K., and, Dickson G. Antisense-induced myostatin exon skipping leads to muscle hypertrophy in mice following octa-guanidine morpholino oligomer treatment. Mol Ther. 2011;19:159–164. doi: 10.1038/mt.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KR, Fleckenstein JL, Amato AA, Barohn RJ, Bushby K, Escolar DM.et al. (2008A phase I/IItrial of MYO-029 in adult subjects with muscular dystrophy Ann Neurol 63561–571. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Speed and endurance: you can have it all. J Appl Physiol. 2010;109:621–622. doi: 10.1152/japplphysiol.00618.2010. [DOI] [PubMed] [Google Scholar]
- Tinsley JM, Blake DJ, Roche A, Fairbrother U, Riss J, Byth BC.et al. (1992Primary structure of dystrophin-related protein Nature 360591–593. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishiguro T, Eguchi C, Saito K., and, Ozawa E. Expression of a dystrophin-related protein associated with the skeletal muscle cell membrane. Histochemistry. 1991;96:1–5. doi: 10.1007/BF00266753. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Nonaka I, Hirai S., and, Ozawa E. Reciprocal expression of dystrophin and utrophin in muscles of Duchenne muscular dystrophy patients, female DMD-carriers and control subjects. J Neurol Sci. 1993;119:43–52. doi: 10.1016/0022-510x(93)90190-a. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM.et al. (1998Expression of full-length utrophin prevents muscular dystrophy in mdx mice Nat Med 41441–1444. [DOI] [PubMed] [Google Scholar]
- Krag TO, Bogdanovich S, Jensen CJ, Fischer MD, Hansen-Schwartz J, Javazon EH.et al. (2004Heregulin ameliorates the dystrophic phenotype in mdx mice Proc Natl Acad Sci USA 10113856–13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubourt E, Fossier P, Baux G, Leprince C, Israël M., and, De La Porte S. Nitric oxide and l-arginine cause an accumulation of utrophin at the sarcolemma: a possible compensation for dystrophin loss in Duchenne muscular dystrophy. Neurobiol Dis. 1999;6:499–507. doi: 10.1006/nbdi.1999.0256. [DOI] [PubMed] [Google Scholar]
- Sonnemann KJ, Heun-Johnson H, Turner AJ, Baltgalvis KA, Lowe DA., and, Ervasti JM. Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med. 2009;6:e1000083. doi: 10.1371/journal.pmed.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire S, Raymackers JM, Vandebrouck C, Potter A, Tinsley J, Fisher R.et al. (2002Prevention of pathology in mdx mice by expression of utrophin: analysis using an inducible transgenic expression system Hum Mol Genet 113333–3344. [DOI] [PubMed] [Google Scholar]
- Miura P., and, Jasmin BJ. Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: how close are we. Trends Mol Med. 2006;12:122–129. doi: 10.1016/j.molmed.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Li D, Bareja A, Judge L, Yue Y, Lai Y, Fairclough R.et al. (2010Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin J Cell Sci 123Pt 122008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WF. September 14, 1990: the beginning. Hum Gene Ther. 1990;1:371–372. doi: 10.1089/hum.1990.1.4-371. [DOI] [PubMed] [Google Scholar]
- Lehrman S. Virus treatment questioned after gene therapy death. Nature. 1999;401:517–518. doi: 10.1038/43977. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E.et al. (2003A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency N Engl J Med 348255–256. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L.et al. (2009Gene therapy for immunodeficiency due to adenosine deaminase deficiency N Engl J Med 360447–458. [DOI] [PubMed] [Google Scholar]
- Acsadi G, Dickson G, Love DR, Jani A, Walsh FS, Gurusinghe A.et al. (1991Human dystrophin expression in mdx mice after intramuscular injection of DNA constructs Nature 352815–818. [DOI] [PubMed] [Google Scholar]
- Wang B, Li J., and, Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc Natl Acad Sci USA. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF.et al. (2002Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy Nat Med 8253–261. [DOI] [PubMed] [Google Scholar]
- Phelps SF, Hauser MA, Cole NM, Rafael JA, Hinkle RT, Faulkner JA.et al. (1995Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice Hum Mol Genet 41251–1258. [DOI] [PubMed] [Google Scholar]
- Rafael JA, Cox GA, Corrado K, Jung D, Campbell KP., and, Chamberlain JS. Forced expression of dystrophin deletion constructs reveals structure-function correlations. J Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DJ, Wells KE, Asante EA, Turner G, Sunada Y, Campbell KP.et al. (1995Expression of human full-length and minidystrophin in transgenic mdx mice: implications for gene therapy of Duchenne muscular dystrophy Hum Mol Genet 41245–1250. [DOI] [PubMed] [Google Scholar]
- Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, Miller DG.et al. (2004Systemic delivery of genes to striated muscles using adeno-associated viral vectors Nat Med 10828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J.et al. (2005Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart Nat Biotechnol 23321–328. [DOI] [PubMed] [Google Scholar]
- Wang B, Li J, Qiao C, Chen C, Hu P, Zhu X.et al. (2008A canine minidystrophin is functional and therapeutic in mdx mice Gene Ther 151099–1106. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS.et al. (2007Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression Mol Ther 151160–1166. [DOI] [PubMed] [Google Scholar]
- Ohshima S, Shin JH, Yuasa K, Nishiyama A, Kira J, Okada T.et al. (2009Transduction efficiency and immune response associated with the administration of AAV8 vector into dog skeletal muscle Mol Ther 1773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Storb R, Lee D, Kushmerick MJ, Chu B, Berger C.et al. (2010Immune responses to AAV in canine muscle monitored by cellular assays and noninvasive imaging Mol Ther 18617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Klapac LR, Montgomery CL, Bremer WG, Shontz KM, Malik V, Davis N.et al. (2010Persistent expression of FLAG-tagged micro dystrophin in nonhuman primates following intramuscular and vascular delivery Mol Ther 18109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Campbell K, Rodino-Klapac L, Sahenk Z, Shilling C, Lewis S.et al. (2010Dystrophin immunity in Duchenne's muscular dystrophy N Engl J Med 3631429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L.et al. (2004Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping Science 3061796–1799. [DOI] [PubMed] [Google Scholar]
- Denti MA, Incitti T, Sthandier O, Nicoletti C, De Angelis FG, Rizzuto E.et al. (2008Long-term benefit of adeno-associated virus/antisense-mediated exon skipping in dystrophic mice Hum Gene Ther 19601–608. [DOI] [PubMed] [Google Scholar]
- Qiao C, Li J, Jiang J, Zhu X, Wang B, Li J.et al. (2008Myostatin propeptide gene delivery by adeno-associated virus serotype 8 vectors enhances muscle growth and ameliorates dystrophic phenotypes in mdx mice Hum Gene Ther 19241–254. [DOI] [PubMed] [Google Scholar]
- Qiao C, Li J, Zheng H, Bogan J, Li J, Yuan Z.et al. (2009Hydrodynamic limb vein injection of adeno-associated virus serotype 8 vector carrying canine myostatin propeptide gene into normal dogs enhances muscle growth Hum Gene Ther 201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morine KJ, Bish LT, Selsby JT, Gazzara JA, Pendrak K, Sleeper MM.et al. (2010Activin IIB receptor blockade attenuates dystrophic pathology in a mouse model of Duchenne muscular dystrophy Muscle Nerve 42722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobinger GP, Louboutin JP, Barton ER, Sweeney HL., and, Wilson JM. Correction of the dystrophic phenotype by in vivo targeting of muscle progenitor cells. Hum Gene Ther. 2003;14:1441–1449. doi: 10.1089/104303403769211655. [DOI] [PubMed] [Google Scholar]
- Kimura E, Li S, Gregorevic P, Fall BM., and, Chamberlain JS. Dystrophin delivery to muscles of mdx mice using lentiviral vectors leads to myogenic progenitor targeting and stable gene expression. Mol Ther. 2010;18:206–213. doi: 10.1038/mt.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach E, Li S, Perez AL, Schienda J, Liadaki K, Volinski J.et al. (2004Systemic delivery of human microdystrophin to regenerating mouse dystrophic muscle by muscle progenitor cells Proc Natl Acad Sci USA 1013581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach E, Perez AL, Choi YH, Illigens BM, Jun SJ, del Nido P.et al. (2006Muscle engraftment of myogenic progenitor cells following intraarterial transplantation Muscle Nerve 3444–52. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Blot S, D'Antona G, Granger N, Tonlorenzi R, Innocenzi A.et al. (2006Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs Nature 444574–579. [DOI] [PubMed] [Google Scholar]
- Quenneville SP, Chapdelaine P, Skuk D, Paradis M, Goulet M, Rousseau J.et al. (2007Autologous transplantation of muscle precursor cells modified with a lentivirus for muscular dystrophy: human cells and primate models Mol Ther 15431–438. [DOI] [PubMed] [Google Scholar]
- Pichavant C, Chapdelaine P, Cerri DG, Dominique JC, Quenneville SP, Skuk D.et al. (2010Expression of dog microdystrophin in mouse and dog muscles by gene therapy Mol Ther 181002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchaouir R, Meregalli M, Farini A, D'Antona G, Belicchi M, Goyenvalle A.et al. (2007Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice Cell Stem Cell 1646–657. [DOI] [PubMed] [Google Scholar]
- Romero NB, Braun S, Benveniste O, Leturcq F, Hogrel JY, Morris GE.et al. (2004Phase I study of dystrophin plasmid-based gene therapy in Duchenne/Becker muscular dystrophy Hum Gene Ther 151065–1076. [DOI] [PubMed] [Google Scholar]
- Budker V, Zhang G, Danko I, Williams P., and, Wolff J. The efficient expression of intravascularly delivered DNA in rat muscle. Gene Ther. 1998;5:272–276. doi: 10.1038/sj.gt.3300572. [DOI] [PubMed] [Google Scholar]
- Hagstrom JE, Hegge J, Zhang G, Noble M, Budker V, Lewis DL.et al. (2004A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs Mol Ther 10386–398. [DOI] [PubMed] [Google Scholar]
- Wolff J, Lewis DL, Herweijer H, Hegge J., and, Hagstrom J. Non-viral approaches for gene transfer. Acta Myol. 2005;24:202–208. [PubMed] [Google Scholar]
- Hegge JO, Wooddell CI, Zhang G, Hagstrom JE, Braun S, Huss T.et al. (2010Evaluation of hydrodynamic limb vein injections in nonhuman primates Hum Gene Ther 21829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wooddell CI, Hegge JO, Griffin JB, Huss T, Braun S.et al. (2010Functional efficacy of dystrophin expression from plasmids delivered to mdx mice by hydrodynamic limb vein injection Hum Gene Ther 21221–237. [DOI] [PubMed] [Google Scholar]
- Thibaud JL, Huss, T, Hegge, J, Thioudellet, C, Granger, N, Gnirs, K.et al. (2006Intravascular administration of dystrophin plasmid DNA in a canine model of Duchenne muscular dystrophy: early side effects and long-term biological efficacy J Vet Intern Med 201270 [Google Scholar]
- Aihara H., and, Miyazaki J. Gene transfer into muscle by electroporation in vivo. Nat Biotechnol. 1998;16:867–870. doi: 10.1038/nbt0998-867. [DOI] [PubMed] [Google Scholar]
- Mir LM, Bureau MF, Gehl J, Rangara R, Rouy D, Caillaud JM.et al. (1999High-efficiency gene transfer into skeletal muscle mediated by electric pulses Proc Natl Acad Sci USA 964262–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissel H., and, Clausen T. Excitation-induced Ca2+ influx and skeletal muscle cell damage. Acta Physiol Scand. 2001;171:327–334. doi: 10.1046/j.1365-201x.2001.00835.x. [DOI] [PubMed] [Google Scholar]
- Trollet C, Scherman D., and, Bigey P. Delivery of DNA into muscle for treating systemic diseases: advantages and challenges. Methods Mol Biol. 2008;423:199–214. doi: 10.1007/978-1-59745-194-9_14. [DOI] [PubMed] [Google Scholar]
- Marshall WG, Jr, Boone BA, Burgos JD, Gografe SI, Baldwin MK, Danielson ML.et al. (2010Electroporation-mediated delivery of a naked DNA plasmid expressing VEGF to the porcine heart enhances protein expression Gene Ther 17419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Zhao Y, Lu H, Pang W., and, Xu Y. In vivo plasmid DNA electroporation resulted in transfection of satellite cells and lasting transgene expression in regenerated muscle fibers. Biochem Biophys Res Commun. 2005;338:1490–1498. doi: 10.1016/j.bbrc.2005.10.111. [DOI] [PubMed] [Google Scholar]
- Trollet C, Bloquel C, Scherman D., and, Bigey P. Electrotransfer into skeletal muscle for protein expression. Curr Gene Ther. 2006;6:561–578. doi: 10.2174/156652306778520656. [DOI] [PubMed] [Google Scholar]
- Mir LM, Belehradek M, Domenge C, Orlowski S, Poddevin B, Belehradek J., Jret al. (1991[Electrochemotherapy, a new antitumor treatment: first clinical trial] C R Acad Sci III, Sci Vie 313613–618. [PubMed] [Google Scholar]
- Favard C, Dean, DS., and, Rols MP. Electrotransfer as a non viral method of gene delivery. Curr Gene Ther. 2007;7:67–77. doi: 10.2174/156652307779940207. [DOI] [PubMed] [Google Scholar]
- Bodles-Brakhop AM, Heller R., and, Draghia-Akli R. Electroporation for the delivery of DNA-based vaccines and immunotherapeutics: current clinical developments. Mol Ther. 2009;17:585–592. doi: 10.1038/mt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir LM. Application of electroporation gene therapy: past, current, and future. Methods Mol Biol. 2008;423:3–17. doi: 10.1007/978-1-59745-194-9_1. [DOI] [PubMed] [Google Scholar]
- Chapdelaine P, Moisset PA, Campeau P, Asselin I, Vilquin JT., and, Tremblay JP. Functional EGFP-dystrophin fusion proteins for gene therapy vector development. Protein Eng. 2000;13:611–615. doi: 10.1093/protein/13.9.611. [DOI] [PubMed] [Google Scholar]
- Vilquin JT, Kennel PF, Paturneau-Jouas M, Chapdelaine P, Boissel N, Delaère P.et al. (2001Electrotransfer of naked DNA in the skeletal muscles of animal models of muscular dystrophies Gene Ther 81097–1107. [DOI] [PubMed] [Google Scholar]
- Pichavant C, Chapdelaine P, Cerri DG, Bizario JC., and, Tremblay JP. Electrotransfer of the full-length dog dystrophin into mouse and dystrophic dog muscles. Hum Gene Ther. 2010;21:1591–1601. doi: 10.1089/hum.2010.024. [DOI] [PubMed] [Google Scholar]
- Zaiss AK., and, Muruve DA. Immunity to adeno-associated virus vectors in animals and humans: a continued challenge. Gene Ther. 2008;15:808–816. doi: 10.1038/gt.2008.54. [DOI] [PubMed] [Google Scholar]
- Roy R, Tremblay JP, Huard J, Richards C, Malouin F., and, Bouchard JP. Antibody formation after myoblast transplantation in Duchenne-dystrophic patients, donor HLA compatible. Transplant Proc. 1993;25 1 Pt 2:995–997. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AOs used in DMD. Schematic representation of 2OMP (left) and PMO (right). R and R′ indicate respectively the continuation of the oligomer chain in the 5′ and 3′ direction.