Abstract
Retinitis pigmentosa (RP) is a disease that primarily affects the peripheral retina and ultimately causes visual impairment. X-chromosomal forms of RP are frequently caused by mutations in the retinitis pigmentosa GTPase regulator (RPGR) gene. We show that the novel splice donor site (SDS) mutation c.1245+3A>T in intron 10 of RPGR cosegregates with RP in a five-generation Caucasian family. The mutation causes in-frame skipping of exon 10 from RPGR transcripts in patient-derived primary fibroblasts. To correct the splice defect, we developed a gene therapeutic approach using mutation-adapted U1 small nuclear RNA (U1). U1 is required for SDS recognition of pre-mRNAs and initiates the splice process. The mutation described herein interferes with the recognition of the SDS by U1. To overcome the deleterious effects of the mutation, we generated four U1 isoforms with increasing complementarity to the SDS. Lentiviral particles were used to transduce patient-derived fibroblasts with these U1 variants. Full complementarity of U1 corrects the splice defect partially and increases recognition of the mutant SDS. The therapeutic effect is U1-concentration dependent as we show for endogenously expressed RPGR transcripts in patient-derived cells. U1-based gene therapeutic approaches constitute promising technologies to treat SDS mutations in inherited diseases including X-linked RP.
Introduction
The term retinitis pigmentosa (RP) denotes a subgroup of retinal degenerations that is clinically characterized by night blindness, concentric constriction of the visual field, pigment deposits in the retinal periphery, and thinning of retinal vessels. The phenotype is caused by rod photoreceptor dysfunction and progressive photoreceptor cell death, which starts at the periphery of the retina. In later stages, the disease may cause complete blindness, involving the death of cone photoreceptors. The prevalence of RP is currently estimated at one in 3,500 individuals. Mutations in >40 genes have been associated with the disorder (for review see ref. (1) or RetNet http://www.sph.uth.tmc.edu/retnet/).
X-linked forms of RP often show an early disease onset and rapid progression. Up to 80% of all X-linked forms of RP cases are caused by mutations in the retinitis pigmentosa GTPase regulator (RPGR) gene.2,3,4 RPGR contains 23 exons of which several different transcript isoforms are generated.5,6,7,8 Approximately 50% of RPGR mutations locate to the alternatively spliced, tissue–specific exon ORF15.2,9,10,11 Pathogenic sequence alterations also occur in exons 1–15, whereas mutations in exons 16–19 have not been described. Interestingly, ~20% of all mutations in exon 1–15 affect splice sites (for reference see the Human Gene Mutation Database Professional 2010.3) and thus are likely to generate aberrant splice products. Although alterations in the splicing of RPGR transcripts often cause the disease, they can also act as a modifier of the phenotype.7
Natural occurring splice variants of RPGR are mostly found at the 3′ region of the transcript and are predicted to change the C-terminal part of the protein. These RPGR isoforms are not altered between exons 2 and 11, a region that encodes the RCC1 homologous domain of RPGR. It has been described that this domain mediates interactions to several binding partners including RPGRIP1, SMC1 and 3, and PDE6δ.12,13,14,15 The majority of RPGR-binding partners have been associated with retinal disorders.
Here, we show that a novel mutation in the splice donor site (SDS) of RPGR exon 10 interferes with normal splicing. We observed skipping of exon 10, which can be corrected by treatment with mutation-adapted U1 small nuclear RNA (U1). Endogenously expressed RPGR transcripts of a patient-derived cell line are successfully treated by this gene therapeutic approach.
U1 is a splice factor required for recognition of SDS in pre-mRNAs. The 5′ part of U1 binds to the SDS by Watson–Crick base pairing.16 This step initializes the splice process and leads to the recruitment of several spliceosomal components that are essential for the recognition of exons and the removal of introns (reviewed by17). Aberrant splicing is often the result of disturbed U1 binding to mutated SDS.18,19,20 Increased complementarity of U1 with the target can lead to a highly efficient correction of splice defects in eye diseases.21 This technique holds great promises for the treatment of inherited diseases caused by SDS mutations.
Results
Family pedigree and molecular genetic analysis
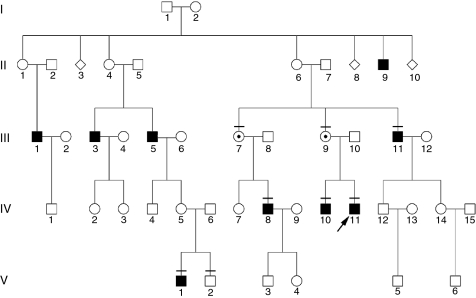
The Caucasian family described herein shows a classic X-linked inheritance pattern (Figure 1). The index patient was clinically diagnosed with RP, including pigmentary deposits in the retina, concentric constriction of the visual field and abolished scotopic electroretinogram (data not shown). The index patient described his first signs of the disease to start at childhood with night blindness, followed by clearly noticeable visual problems in his thirties. The five-generation pedigree documents that only males are affected, whereas obligate female carriers are asymptomatic. In total, nine males in four generations of the family are affected by RP.
Figure 1.
Pedigree of index patient 70423 (indicated by an arrow) showing a X-chromosomal inheritance pattern. Retinitis pigmentosa segregates in four generations of the family. Squares represent males, circles represent females. Filled symbols show affected males. Individuals included in cosegregation analysis of the mutation and phenotype are indicated by a horizontal bar. Only females who are confirmed carriers of the mutation are shown with a dot in the circle. Of note, additional carriers are likely to exist in this family.
Two genes, RPGR and RP2, have been associated with X-linked RP.22,23,24 Routine diagnostic testing of the complete coding region of RPGR and RP2 identified the sequence alteration c.1245+3A>T in RPGR (reference sequence NM_000328). This sequence change affects the SDS of exon 10 at the third position of the intron 10 (Figure 2a,b). Analysis of eight family members in three generations revealed cosegregation of c.1245+3A>T with the phenotype of RP. Together, these data strongly suggest that the detected sequence alteration in RPGR is disease causing.
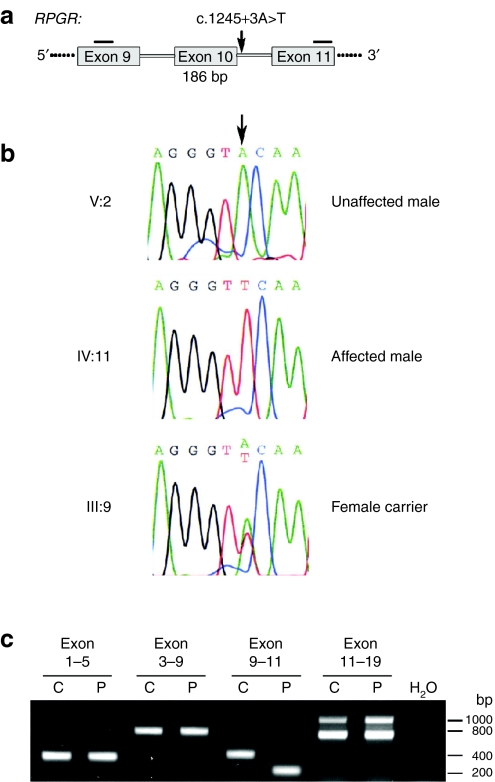
Figure 2.
DNA and RNA analyses of RPGR mutation c.1245+3A>T. (a) Parts of the gene structure of RPGR are shown in a schematic drawing. The position of the mutation is indicated by an arrow. Horizontal bars represent reverse transcription-PCR (RT-PCR) primers. Skipping of RPGR exon 10 during splicing leads to a 186 base pair deletion in the transcript. (b) Sequencing electropherograms of family members V:2, IV:11, III:9. The arrow indicates the nucleotide position which is mutated. Sequencing profiles are shown for the splice donor site of RPGR exon 10: the first three nucleotides are of exonic origin, whereas the last six nucleotides are part of intron 10. (c) RT-PCR products of RPGR exons 1–19. RNA from control (C) or patient (P) fibroblasts were analyzed for splice alterations. The only difference in splicing was detected by amplification of exons 9–11. Sequencing confirmed that exon 10 skipping occurred in the RPGR transcript of the patient cell line. RPGR, retinitis pigmentosa GTPase regulator gene.
Analysis of RPGR splicing
Primary skin fibroblasts were cultured from the index patient (IV:11). To analyze the effect of the mutation c.1245+3A>T on RPGR splicing, we performed reverse transcription-PCR (RT-PCR) using RNA from these patient-derived cells. We analyzed splicing of RPGR exons 1 through 19 and detected skipping of exon 10 exclusively in the patient sample (Figure 2c). In contrast, the control cell line showed normal splicing including exon 10 into the transcript (Figure 2c). These results document that the mutation c.1245+3A>T interferes with splicing of RPGR and causes exon skipping from its pre-mRNA. Exon 10 skipping leads to an in-frame deletion of 186 bp and is predicted to remove 62 amino acid residues from the RCC1 homologous region of RPGR, a domain that is required for interaction with several binding partners.
Bioinformatic splice donor site analysis in RPGR
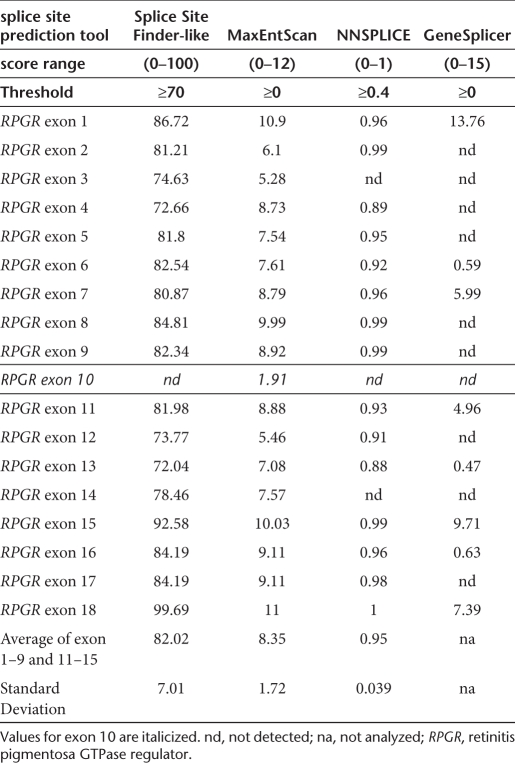
We analyzed the wild-type SDS of exon 10 using splice site prediction tools and found that its conservation is low compared to other RPGR exons (Table 1). Only one of four prediction tools was able to identify the SDS using the recommended threshold. Moreover, the calculated MaxEntScan splicing score of 1.91 for RPGR exon 10 was the lowest among 18 RPGR SDSs. On average, the splicing score of exons 1–18 was 8.35 with a standard deviation of 1.73. These data suggest that the SDS of exon 10 is weak.
Table 1. Bioinformatic prediction of splice donor sites in retinitis pigmentosa GTPase regulator.
Gene therapeutic correction of the splice defect
It has been described that U1 recognizes a SDS by complementary base pairing16 with nine nucleotides at the exon-intron border. The last three base pairs of the exon and the first six base pairs of the intron are predominately recognized by this mechanism. In the case of RPGR exon 10, only four of nine nucleotides match the U1 wild type sequence (Figure 3a). Such low complementarity with the U1 consensus sequence is in agreement with the findings of the bioinformatic splice site prediction and further supports that this SDS is weak. It also explains why mutations in this SDS are not tolerated by the splicing machinery and lead to splice defects.
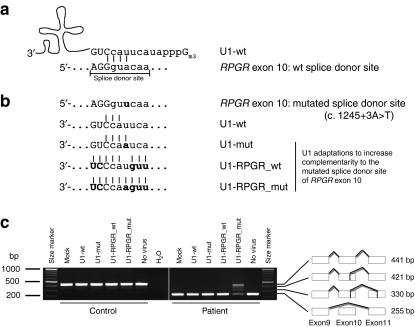
Figure 3.
Gene therapeutic approach to correct splice defects in patient-derived cells. (a) U1 small nuclear RNA (U1) annealing to the splice donor site of RPGR exon 10, predominantly including nine base pairs. Upper case letters show exonic nucleotides, whereas the lower case letters denote intronic nucleotides. Only four nucleotides of the splice donor site of RPGR exon 10 build Watson–Crick base pairs with the U1 as indicated by vertical lines. (b) Strategy to increase complementarity of the U1 with the mutated splice donor site of RPGR. In addition to wild-type U1 (U1-wt), three U1 adaptations were tested. U1 complementarity was increased stepwise by adaptation to the mutation (U1-mut), or to the wild-type splice donor site of RPGR exon 10 (U1-RPGR_wt), or to the mutant splice donor site of RPGR exon 10 (U1-RPGR_mut). Bold letters either mark the mutation or adapted nucleotides in U1. (c) Correction of the mutation-induced splice defect in RPGR. Reverse transcription-PCR (RT-PCR) was performed from patient-derived fibroblasts that have been transduced with lentiviral particles containing the different U1 isoforms. Fully adapted U1 resulted in a correction of RPGR exon 10 skipping exclusively in the patient cell line. Neither the control fibroblast cell line, nor less than completely adapted U1 showed an effect on splicing of exon 10. Sequencing results of all RT-PCR products are illustrated by schematic drawings. RPGR, retinitis pigmentosa GTPase regulator gene.
In order to overcome the deleterious effect of the mutation, we generated U1 variants with increased complementarity to the mutated SDS of exon 10. These mutation-specific adaptations resulted in four different U1 variants: (i) the wild-type U1; (ii) U1 adapted only to the mutation; (iii) U1 matching all nucleotides of the wild-type exon 10 SDS; (iv) U1 matching all nucleotides of the patient exon 10 SDS including the mutation (Figure 3a,b). We used lentiviral particles expressing the U1 variants to transduce primary fibroblasts of the patient and of a control.
RT-PCR analysis of RPGR transcripts showed a correction of the mutation-induced splice defect in the patient-derived cell line. Treatment with the fully adapted U1 isoform resulted in a strong reduction in exon skipping (255 bp fragment) and a significant increase in correctly spliced RPGR (441 bp fragment) (Figure 3c). We also observed two additional splice products (421 bp and 330 bp fragments) where cryptic splice sites in exon 10 or 11 were used. Treatment with U1 isoforms showing less than full sequence-complementarity to the mutant SDS did not reveal therapeutic effects on RPGR splicing.
In parallel to the patient cell line, we treated control fibroblast cells. Only correctly spliced transcripts were detected (441 bp fragment) (Figure 3c). None of the U1 variants induced alterations in RPGR splicing in the control, suggesting a highly specific therapeutic effect of the U1 adaptation on the mutated SDS. Furthermore, to test the effect of the U1 treatment on nontarget transcripts, we analyzed splicing of five RP-associated genes that contain potential binding sites to the fully adapted U1. We did not find events of mis-splicing, supporting our above described observation that the virus-mediated U1 treatment does not lead to massive alteration of splice patterns of nontarget transcripts (Supplementary Figure S1).
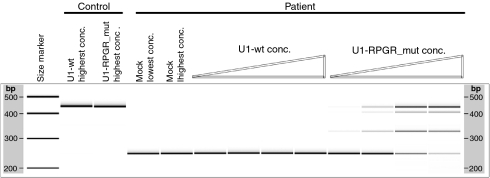
Concentration dependence of the U1 treatment
To evaluate whether the therapeutic effect is dose dependent, we treated the patient and control cell lines with increasing amounts of viral particles containing the different U1 isoforms. Specifically for fully adapted U1, an increasing concentration of viral particles resulted in higher levels of corrected RPGR transcripts (Figure 4). Similar concentration gradients with mock and wild-type U1 isoforms did not influence the splicing of RPGR in the patient cell line (Figure 4). Comparably, no effect on the control cell line was detected, thus confirming that the U1 adaptations, independent of the concentration, did not interfere with normal splicing of RPGR.
Figure 4.
The therapeutic efficiency of the U1 approach is dose dependent. Increasing concentrations of the fully adapted U1 (U1-RPGR_mut) result in increased levels of correctly spliced RPGR transcripts in the patient cell line. In contrast, similar treatments of the patient and control fibroblasts with wild type U1 (U1-wt) and mock (lentiviral particles without U1 isoforms) have no detectable influence on RPGR exon 10 splicing. RPGR, retinitis pigmentosa GTPase regulator gene.
Discussion
The eye is an ideal target organ for gene therapeutic interventions. Its easy accessibility and transparency constitute advantages for the application of therapeutics and the follow up of therapeutic effects in vivo. Especially the retina shows an interconnected system of spatially well organized cell types, which facilitates the application of therapeutics to the target cells. Moreover, the eye has an efficient blood–retina barrier that prevents exchange of most therapeutics with other organs and thus reduces side effects and therapy-provoked immune responses. This is best illustrated by the finding that systemic virus particles have not been detected following gene replacement therapy in the human eye.25
Previous studies have demonstrated that retinal diseases in human can be successfully treated with gene therapeutic approaches.25,26,27 These clinical trials have studied gene replacement for RPE65, a gene that is associated with autosomal recessive, early-onset RP or Leber congenital amaurosis. In addition, several other genes causing retinal diseases have been replaced successfully in animal models.28,29
Despite these encouraging results, the heterogeneity of retinal diseases is an unsolved problem. To develop and evaluate therapeutic approaches for each of the at least 170 genes associated with retinal diseases1 will require enormous resources and time. Furthermore, the treatment of autosomal dominant retinal disorders is more challenging than recessively inherited diseases, because gain of function effects may complicate treatment approaches.30
Genetic diseases are caused by alterations of the nucleotide composition within the genome of affected patients and their relatives. Most frequently, point mutations are causative of the disease and the majority of them (~85%) are reported as missense or nonsense mutations. Several possibilities exist for how these mutations interfere with the function of the disease gene. Often it is speculated that the mutation affects the protein composition as a consequence of mRNA translation. In recent years, the impact of point mutations on splicing received more attention. It has been shown that about 20% of the mutations cause mis-splicing of the pre-mRNA rather than only affecting the protein composition.31,32,33,34 Of note, even disease-associated repeat expansions were shown to be involved in pathogenic splice alterations.35 In summary, mis-splicing is among the most frequent pathogenic mechanisms underlying genetic diseases.
Therapeutic approaches to correct mutation-induced mis-splicing will be independent from the disease gene and applicable to ~20% of the patients with inherited diseases. These strategies aim to modify the splice pattern of a particular gene in order to overcome the pathogenic process leading to the disease. Different treatment approaches for various diseases have been investigated: small-molecule drugs, antisense oligonucleotides, trans-splicing approaches, and RNA interference.34,36,37,38,39 In addition, isoform-specific antibodies can elicit similar effects.
U1-based strategies provide an alternative technique to correct splice defects which cause either recessive or dominant diseases. An advantage of the U1 technique compared to gene replacement strategies is that it corrects the endogenously expressed transcript and thus is independent from the selection of appropriate promoters needed to drive the expression of a transgene. Furthermore, it has the potential to rescue the normal splice pattern and consequently reduce the amount of mutated proteins, a process particularly relevant for the treatment of dominant diseases with gain of function mutations. Using minigene assays, we previously showed that over 90% of the mis-spliced transcripts can be corrected for rhodopsin, one of the most frequently mutated genes in autosomal dominant RP.21 Nevertheless, it will be important to establish the technique for several cases and systematically compare its efficiency to correct individual SDS mutations. Furthermore, side effects of the treatment need to be evaluated in detail, including the possibility that nontarget transcripts are influenced by the U1 adaptations. Although we have not observed such side effects in patient-derived cell lines, in vivo studies using animal models are required to assess the balance between benefits of the treatment and severity of possible side effects.
In this report, we analyzed adapted U1 isoforms to correct a splice defect in RPGR, a gene frequently mutated in X-chromosomal RP patients. We show that the treatment with mutation-adapted U1 can be applied successfully to correct splice defects of RPGR transcripts that are endogenously expressed in patient-derived cell lines. This treatment resulted in an increase of correctly spliced RPGR and a significant reduction of exon skipping, thus confirming the therapeutic potential of the technique. We speculate that a comparable gene therapeutic intervention in retinal cells might result in a delayed onset and/or decreased progression rate of the disease. Of note, the lentivirus shows limited ability to infect photoreceptors. Pseudotyped lenti- or adeno-associated virus particles might be used to transduce retinal cells.40
The U1-induced correction of RPGR transcripts also generated aberrant splice variants using cryptic splice sites in exons 10 and 11. Although increased complementarity of U1 leads to clearly improved recognition of the mutated exon 10, it seems that nearby cryptic SDS complicate an unambiguous selection of the correct exon-intron border. This is in agreement with the finding that the conservation of the SDS of RPGR exon 10 is low, making alternative splice variants more likely. Interestingly, a different mutation (c.1245+3A>G) at the same position in RPGR has also been reported to result in the recognition of cryptic splice sites. In contrast to the mutation described herein (c.1245+3A>T), the c.1245+3A>G nucleotide exchange41 leads to a reduced amount of normal splice products in addition to a single aberrant splice variant. This aberrant splice product resulted from usage of the same cryptic splice sites in exons 10 and 11 as found in this study. Since the A to T exchange resulted exclusively in exon skipping without normal splice products, it can be assumed that this mutation more strongly disturbs the splice site selection compared to the A to G transition. This observation is in agreement with the conservation of SDSs across the genome, where position +3 is most frequently an A or G, whereas T and C are clearly underrepresented.42 Consequently, an A to G mutation is more likely tolerated by the splicing machinery than an A to T. It can be speculated that the U1 approach is more effective to correct an A to G exchange at position +3 of RPGR exon 10 compared to an A to T exchange.
RPGR shows several splice variants, some of which are tissue-specifically expressed. ORF15 isoforms are mostly found in neuronal tissues, but additional variants have been described.5,7,8 Even sub-populations of neuronal cells may generate specific RPGR variants, e.g., exon 9a isoforms, which were detected predominantly in cone photoreceptors of the retina.6 Although this complex expression pattern of RPGR complicates conclusions on the functional relevance of the identified mutation, it seems likely that the RCC1 homologous domain in the N-terminal part of RPGR is disturbed by the exon 10 skipping event. The RCC1 homologous domain includes amino acids encoded by exons 2 through 11. It builds a seven bladed propeller-like structure,6,43 with which several binding partners, also implicated in retinal diseases, interact.13,14,15,44 The deletion of exon 10 encoded amino acids is likely to interfere with the generation or maintenance of this “RPGR interactome.” Furthermore, RPGR has been located to the connecting cilium of photoreceptors, where protein transport between inner and outer photoreceptor segments takes place. It is plausible that altered transport properties through the connecting cilium can cause photoreceptor degeneration as found in RP.
In conclusion, skipping of RPGR exon 10, a pathogenic process that causes X-chromosomal RP, might be treatable with adapted U1. It will be important to study animal models of retinal diseases and test this promising gene therapeutic technique in vivo.
Materials and Methods
Patients and mutation screening. Prior to interview and blood sampling, the nature and consequences of this study were explained to the patient and healthy controls. Informed consent for clinical diagnostic testing and research applications were obtained before samples were collected. The study followed the Declaration of Helsinki protocols. Genomic DNA was extracted from EDTA blood samples as previously described6 and subjected to molecular genetic analysis. To verify the mutation in intron 10 of RPGR in different family members, exon 10 and flanking intronic sequences were amplified by PCR using HotFire Taq Polymerase (Solis Biodyne, Tartu, Estonia) and sequenced. PCR conditions are given in the Supplementary Table S1. Sequencing profiles were compared with the RPGR reference sequence NM_000328 (SeqScape software, ver 2.0, Applied Biosystems, Rotkreuz, Switzerland). Sequence alterations are annotated as recommended by the Human Genome Variation Society (http://www.hgvs.org/mutnomen).
Bioinformatic splice donor site evaluation. Splice site prediction tools were used as included in the Alamut mutation interpretation software (ver 1.53, Interactive Biosoftware, Rouen, France). Recommended thresholds were applied to calculate the splice score of four different prediction tools: Splice Site Finder-like (as included in the Alamut software), MaxEntScan (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html), NNSplice (http://www.fruitfly.org/seq_tools/splice.html), and GeneSplicer (http://www.cbcb.umd.edu/software/GeneSplicer/gene_spl.shtml).
RT-PCR analysis. Total RNA was extracted from cultured fibroblasts using NucleoSpin RNA kits (Macherey-Nagel, Oensingen, Switzerland). Reverse transcription to cDNA with random primers was performed according to the manufacturer's instructions (Superscript III; Invitrogen, Basel, Switzerland). RT-PCR reactions were performed using HotFire Taq Polymerase. RT-PCR conditions are summarized in Supplementary Table S1. PCR products were analyzed on either an agarose gel or the 2100 Bioanalyser (Agilent Technologies AG, Basel, Switzerland). All RT-PCR products were verified by sequencing.
Cell culture and lentiviral transduction. Patient-derived primary skin fibroblasts of the index patient and control were prepared and cultured as previously described.45 In brief, a standard skin biopsy was cut in small pieces and transferred to a sterile culture flask. After the explants were attached to the flask bottom by air drying, fibroblast culture medium (minimal essential medium with 20% fetal bovine serum, 1.3% -glutamine, 0.8% antibiotic, and antimycotic solution) was added and incubated at 37 °C and 5% CO2 for 5–10 days. Fibroblasts were harvested and transferred to a 75 cm2 flask for propagation and analysis.
The human U1 small nuclear RNA expression cassette46 was cloned into the lentivirus plasmid p.RRLSIN.cPPT.SFFV/GFP.WPRE47,48 using HpaI restriction sites. Mutagenesis of U1 was performed as previously described.21 HEK293T cells (6 × 106 cells were seeded the day before transfection in a 75 cm2 flask) were cotransfected with the two packaging plasmids pSPAX2 (6.5 µg) and pMD2.G (2 µg) and the U1 containing lentiviral plasmid (12 µg). For each transfection, 37.5 µg branched polyethyleneimine (Sigma-Aldrich, Steinheim, Germany) was used. The secreted virus was harvested in 20 ml culture medium and added to 1 × 105 fibroblasts for transduction in a six-well plate format. All virus particles were produced in parallel using identical conditions. Patient and control fibroblasts were treated with either 1, 3, 6, or 9 ml of the virus-containing culture medium to analyze the dose dependency of the therapeutic effect. Transduced fibroblasts were harvested after 2–4 days in culture.
SUPPLEMENTARY MATERIAL Table S1. PCR conditions included standard puffer composition. Figure S1. Analysis of side effects in patient-derived fibroblasts after U1 treatment. U1 binding to non-target transcripts was analyzed. Potential binding sites for U1-RPGR_mut were found in 5 genes which are associated with RP and expressed in fibroblasts. U1-RPGR_mut showed three potential binding sites in the intron 1 of PRPH2 as well as a single binding sites in exons 1 (5'UTR) of RP2, exon 5 (3'UTR) of RP2, intron 17 of ABCA4, and intron 4 of MERTK. We performed RT-PCR from patient-derived cells transduced with the highest virus concentration tested and compared mock, U1-wt, and U1-RPGR_mut treatments. No side effects of mis-spliced non-target transcripts were detected.
Acknowledgments
We are grateful to patients and family members for their contribution to this study. We thank Deborah Bartholdi for the skin biopsy, Barbara Kloeckener-Gruissem for comments on the manuscript, and Matteo Bertelli for gene diagnostics and DNA aliquots. The authors state that there is no conflict of interest to disclose. The study was financially supported by the Velux Foundation, the Forschungskredit of the University of Zurich, Schweizerischer Fonds zur Verhütung und Bekämpfung der Blindheit, and Hartmann Müller Foundation (to J.N.).
Supplementary Material
PCR conditions included standard puffer composition.
Analysis of side effects in patient-derived fibroblasts after U1 treatment. U1 binding to non-target transcripts was analyzed. Potential binding sites for U1-RPGR_mut were found in 5 genes which are associated with RP and expressed in fibroblasts. U1-RPGR_mut showed three potential binding sites in the intron 1 of PRPH2 as well as a single binding sites in exons 1 (5'UTR) of RP2, exon 5 (3'UTR) of RP2, intron 17 of ABCA4, and intron 4 of MERTK. We performed RT-PCR from patient-derived cells transduced with the highest virus concentration tested and compared mock, U1-wt, and U1-RPGR_mut treatments. No side effects of mis-spliced non-target transcripts were detected.
REFERENCES
- Berger W, Kloeckener-Gruissem B., and, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Neidhardt J, Glaus E, Lorenz B, Netzer C, Li Y, Schambeck M.et al. (2008Identification of novel mutations in X-linked retinitis pigmentosa families and implications for diagnostic testing Mol Vis 141081–1093. [PMC free article] [PubMed] [Google Scholar]
- Bader I, Brandau O, Achatz H, Apfelstedt-Sylla E, Hergersberg M, Lorenz B.et al. (2003X-linked retinitis pigmentosa: RPGR mutations in most families with definite X linkage and clustering of mutations in a short sequence stretch of exon ORF15 Invest Ophthalmol Vis Sci 441458–1463. [DOI] [PubMed] [Google Scholar]
- Sharon D, Sandberg MA, Rabe VW, Stillberger M, Dryja TP., and, Berson EL. RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am J Hum Genet. 2003;73:1131–1146. doi: 10.1086/379379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner R, Rosenberg T, Schultz-Heienbrok R, Lenzner S, Feil S, Roepman R.et al. (1999RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa Hum Mol Genet 81571–1578. [DOI] [PubMed] [Google Scholar]
- Neidhardt J, Glaus E, Barthelmes D, Zeitz C, Fleischhauer J., and, Berger W. Identification and characterization of a novel RPGR isoform in human retina. Hum Mutat. 2007;28:797–807. doi: 10.1002/humu.20521. [DOI] [PubMed] [Google Scholar]
- Schmid F, Glaus E, Cremers FP, Kloeckener-Gruissem B, Berger W., and, Neidhardt J. Mutation- and tissue-specific alterations of RPGR transcripts. Invest Ophthalmol Vis Sci. 2010;51:1628–1635. doi: 10.1167/iovs.09-4031. [DOI] [PubMed] [Google Scholar]
- Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG.et al. (2000Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa Nat Genet 25462–466. [DOI] [PubMed] [Google Scholar]
- Breuer DK, Yashar BM, Filippova E, Hiriyanna S, Lyons RH, Mears AJ.et al. (2002A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa Am J Hum Genet 701545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Hoyos M, Garcia-Sandoval B, Cantalapiedra D, Riveiro R, Lorda-Sánchez I, Trujillo-Tiebas MJ.et al. (2006Mutational screening of the RP2 and RPGR genes in Spanish families with X-linked retinitis pigmentosa Invest Ophthalmol Vis Sci 473777–3782. [DOI] [PubMed] [Google Scholar]
- Pelletier V, Jambou M, Delphin N, Zinovieva E, Stum M, Gigarel N.et al. (2007Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: genotype-phenotype correlations and impact on genetic counseling Hum Mutat 2881–91. [DOI] [PubMed] [Google Scholar]
- Linari M, Ueffing M, Manson F, Wright A, Meitinger T., and, Becker J. The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proc Natl Acad Sci USA. 1999;96:1315–1320. doi: 10.1073/pnas.96.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JP., and, Wright AF. Identification of a novel protein interacting with RPGR. Hum Mol Genet. 2000;9:2085–2093. doi: 10.1093/hmg/9.14.2085. [DOI] [PubMed] [Google Scholar]
- Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, Ropers HH.et al. (2000The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors Hum Mol Genet 92095–2105. [DOI] [PubMed] [Google Scholar]
- Khanna H, Hurd TW, Lillo C, Shu X, Parapuram SK, He S.et al. (2005RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins J Biol Chem 28033580–33587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., and, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL., and, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Pagani F, Buratti E, Stuani C, Bendix R, Dörk T., and, Baralle FE. A new type of mutation causes a splicing defect in ATM. Nat Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- Crehalet H, Latour P, Bonnet V, Attarian S, Labauge P, Bonello N.et al. (2010U1 snRNA mis-binding: a new cause of CMT1B Neurogenetics 1113–19. [DOI] [PubMed] [Google Scholar]
- Freund M, Hicks MJ, Konermann C, Otte M, Hertel KJ., and, Schaal H. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005;33:5112–5119. doi: 10.1093/nar/gki824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner G, Glaus E, Barthelmes D, Ader M, Fleischhauer J, Pagani F.et al. (2009Therapeutic strategy to rescue mutation-induced exon skipping in rhodopsin by adaptation of U1 snRNA Hum Mutat 30255–263. [DOI] [PubMed] [Google Scholar]
- Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A.et al. (1996A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3) Nat Genet 1335–42. [DOI] [PubMed] [Google Scholar]
- Roepman R, van Duijnhoven G, Rosenberg T, Pinckers AJ, Bleeker-Wagemakers LM, Bergen AA.et al. (1996Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1 Hum Mol Genet 51035–1041. [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G.et al. (1998Positional cloning of the gene for X-linked retinitis pigmentosa 2 Nat Genet 19327–332. [DOI] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K.et al. (2008Effect of gene therapy on visual function in Leber's congenital amaurosis N Engl J Med 3582231–2239. [DOI] [PubMed] [Google Scholar]
- Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L.et al. (2008Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial Hum Gene Ther 19979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J.et al. (2008Safety and efficacy of gene transfer for Leber's congenital amaurosis N Engl J Med 3582240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Kim SR, Binley K, Pata I, Doi K, Mannik J.et al. (2008Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy Gene Ther 151311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Smith AJ, Pawlyk B, Xu X, Liu X, Bainbridge JB.et al. (2009Gene therapy for retinitis pigmentosa and Leber congenital amaurosis caused by defects in AIPL1: effective rescue of mouse models of partial and complete Aipl1 deficiency using AAV2/2 and AAV2/8 vectors Hum Mol Genet 182099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadderton N, Millington-Ward S, Palfi A, O'Reilly M, Tuohy G, Humphries MM.et al. (2009Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy Mol Ther 17593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle D., and, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA., and, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- López-Bigas N, Audit B, Ouzounis C, Parra G., and, Guigó R. Are splicing mutations the most frequent cause of hereditary disease. FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Wang GS., and, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Baralle M, Pastor T, Bussani E., and, Pagani F. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet. 2008;83:77–88. doi: 10.1016/j.ajhg.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Baker BF, Bennett CF., and, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC., and, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Gabut M., and, Tazi J. SR proteins as potential targets for therapy. Prog Mol Subcell Biol. 2006;44:65–87. doi: 10.1007/978-3-540-34449-0_4. [DOI] [PubMed] [Google Scholar]
- Sumanasekera C, Watt DS., and, Stamm S. Substances that can change alternative splice-site selection. Biochem Soc Trans. 2008;36 Pt 3:483–490. doi: 10.1042/BST0360483. [DOI] [PubMed] [Google Scholar]
- Auricchio A, Kobinger G, Anand V, Hildinger M, O'Connor E, Maguire AM.et al. (2001Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model Hum Mol Genet 103075–3081. [DOI] [PubMed] [Google Scholar]
- Fujita R, Buraczynska M, Gieser L, Wu W, Forsythe P, Abrahamson M.et al. (1997Analysis of the RPGR gene in 11 pedigrees with the retinitis pigmentosa type 3 genotype: paucity of mutations in the coding region but splice defects in two families Am J Hum Genet 61571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast G. How did alternative splicing evolve. Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M.et al. (1998The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller Nature 39297–101. [DOI] [PubMed] [Google Scholar]
- Linari M, Hanzal-Bayer M., and, Becker J. The delta subunit of rod specific cyclic GMP phosphodiesterase, PDE delta, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS Lett. 1999;458:55–59. doi: 10.1016/s0014-5793(99)01117-5. [DOI] [PubMed] [Google Scholar]
- Villegas J., and, McPhaul M. Establishment and culture of human skin fibroblasts. Curr Protoc Mol Biol. 2005;Chapter 28:Unit 28.3. doi: 10.1002/0471142727.mb2803s71. [DOI] [PubMed] [Google Scholar]
- Lund E., and, Dahlberg JE. True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J Biol Chem. 1984;259:2013–2021. [PubMed] [Google Scholar]
- Brenner S., and, Malech HL. Current developments in the design of onco-retrovirus and lentivirus vector systems for hematopoietic cell gene therapy. Biochim Biophys Acta. 2003;1640:1–24. doi: 10.1016/s0167-4889(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Werner M, Kraunus J, Baum C., and, Brocker T. B-cell-specific transgene expression using a self-inactivating retroviral vector with human CD19 promoter and viral post-transcriptional regulatory element. Gene Ther. 2004;11:992–1000. doi: 10.1038/sj.gt.3302255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR conditions included standard puffer composition.
Analysis of side effects in patient-derived fibroblasts after U1 treatment. U1 binding to non-target transcripts was analyzed. Potential binding sites for U1-RPGR_mut were found in 5 genes which are associated with RP and expressed in fibroblasts. U1-RPGR_mut showed three potential binding sites in the intron 1 of PRPH2 as well as a single binding sites in exons 1 (5'UTR) of RP2, exon 5 (3'UTR) of RP2, intron 17 of ABCA4, and intron 4 of MERTK. We performed RT-PCR from patient-derived cells transduced with the highest virus concentration tested and compared mock, U1-wt, and U1-RPGR_mut treatments. No side effects of mis-spliced non-target transcripts were detected.