SUMMARY
A critical feature of the hypothesized RNA world would have been the ability to control chemical processes in response to environmental cues. Riboswitches present themselves as viable candidates for a sophisticated mechanism of regulatory control in RNA-based life. These regulatory elements in the modern world are most commonly found in the 5′-untranslated regions of bacterial mRNAs, directly interacting with metabolites as a means of regulating expression of the coding region via a secondary structural switch. In this review, we focus on recent insights into how these RNAs fold into complex architectures capable of both recognizing a specific small molecule compound and exerting regulatory control over downstream sequences, with an emphasis on transcriptional regulation.
Many bacterial mRNAs contain sequences that regulate their transcription or translation by directly binding metabolites. Similar “riboswitches” may have allowed RNA-based life to respond to its environment.
1. INTRODUCTION
Life in an RNA world would have relied on RNA as both a medium for heritable genetic information and chemical catalysis. In addition to these functions, life would have had to react to changing environmental conditions—that is, be capable of regulating biological functions. Insights into how RNA can accomplish this crucial task have been revealed through recent discoveries that this molecule accomplishes a wide variety of regulatory functions in modern biology. One of the most striking recent examples of how RNA regulates gene expression was revealed by the discovery of riboswitches, a common means of genetic regulation at the mRNA level in the bacterial kingdom (Barrick and Breaker 2007).
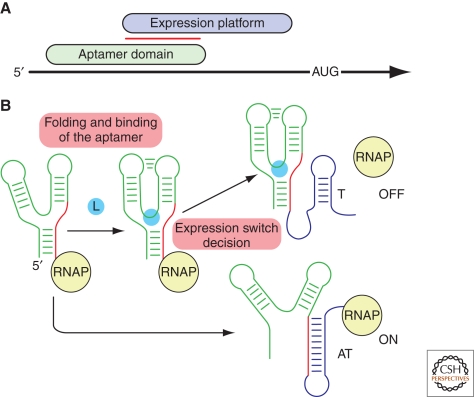
Riboswitches are elements commonly found in the 5′-untranslated region (UTR) of mRNAs that exert their regulatory control over the transcript in a cis-fashion by directly binding a small molecule ligand (McDaniel et al. 2003; Mironov et al. 2002; Nahvi et al. 2002; Winkler et al. 2002a; Winkler et al. 2002b). The typical riboswitch contains two distinct functional domains (Fig. 1A). The effector molecule is recognized by an aptamer domain, which adopts a compact three-dimensional fold to scaffold the ligand binding pocket (Winkler and Breaker 2003). As with proteins, these RNA receptors must discriminate between chemically related metabolites with high selectivity to elicit the appropriate regulatory response. A second domain, the expression platform, contains a secondary structural switch that interfaces with the transcriptional or translational machinery. Regulation is achieved by virtue of a region of overlap between these two domains, known as the switching sequence, whose pairing directs folding of the RNA into one of two mutually exclusive structures in the expression platform that represent the on and off states of the mRNA (Fig. 1B).
Figure 1.
Organization and mechanism of the typical riboswitch. (A) Arrangement of riboswitch structural elements in primary sequence of the mRNA transcript. The aptamer domain (green) and the expression platform (purple) overlap through a sequence that can base pair with either domain (red bar). (B) During transcription of the riboswitch, several events occur to elicit the appropriate regulatory outcome. Early events during transcription include the folding of the aptamer domain and potential binding of the effector (L, cyan). Depending on whether effector is bound, the RNA adopts one of two potential folds in the expression platform (an antiterminator, AT, or rho-independent terminator, T) that determines the regulatory response.
The above model of the riboswitch immediately suggests two fundamental questions regarding their regulatory mechanism. First, how does RNA create a binding pocket that achieves both high affinity and specificity for the effector molecule? This requires knowledge of the atomic-level structure of the RNA-ligand complex. The second pertinent question is: How is effector binding communicated to the expression platform to yield a regulatory outcome? This requires knowledge both of potential ligand-induced conformational changes in the aptamer and the cotranscriptional folding pathway of the mRNA. Given that transcriptional attenuation is the predominant mode of regulation for riboswitches (Barrick and Breaker 2007), they must rapidly adopt the requisite conformations to act before escape of polymerase beyond its 3′-boundary. Thus, the above questions must be considered in the context of RNA folding. although most of this article will focus on the mechanism of transcriptional regulation, it has been suggested that tight coupling of transcription and translation in bacteria may impart the same temporal constraints on translation (Link and Breaker 2009).
2. CONSTRUCTING AN RNA RECEPTOR: THE BASIC ARCHITECTURAL BUILDING BLOCKS
Sequence and structural analyses of over ten riboswitch aptamer domains in complex with their effectors (Table 1) have revealed these RNAs are organized using many of the same architectural principles observed in other large RNAs (Hendrix et al. 2005; Leontis et al. 2006). The sequence of the aptamer domain of the purine riboswitch, one of the simplest aptamers, contains three conserved helical elements (P1, P2, and P3) (Fig. 2A) that are present in all ∼500 identified sequences (Mandal et al. 2003). These helices are organized into higher order structures by two principles (Batey et al. 1999; Holbrook, 2008). First, they form sets of coaxial stacks in which two or more individual helices are arranged in a single pseudocontinuous helix, exemplified by the P1 and P3 stack (Fig. 2B) (Batey et al. 2004; Serganov et al. 2004). Second, helices and helical stacks tend to be arranged in a parallel configuration (Fig. 2B). This architectural behavior is manifest in almost all RNAs containing three or more helices (Holbrook, 2008). A notable exception to this is tRNA, in which the two coaxial stacks are arranged perpendicularly via the interaction of the D- and T-loops.
Table 1.
Structures of riboswitches
| Riboswitch | PDB | References |
|---|---|---|
| Purine | ||
| Guanine | 1U8D, 1Y27 | (Batey et al. 2004; Serganov et al. 2004) |
| Adenine | 1Y26 | (Serganov et al. 2004) |
| 2′-deoxyguanosine | 3DS7 | (Edwards and Batey 2009) |
| S-adenosylmethionine (SAM) | ||
| SAM-I | 2GIS | (Montange and Batey 2006) |
| SAM-II | 2QWY | (Gilbert et al. 2008) |
| SAM-III | 3E5C | (Lu et al. 2008) |
| Thiamine pyrophosphate (TPP) | ||
| Bacterial (Escherichia coli) | 2GDI, 2HOJ | (Edwards and Ferre-D’Amare 2006; Serganov et al. 2006) |
| Eukaryotic (A. thaliana) | 2CKY | (Thore et al. 2006) |
| Lysine | 3DOU, 3DIL | (Garst et al. 2008; Serganov et al. 2008) |
| Flavin mononucleotide (FMN) | 3F2Q | (Serganov et al. 2009) |
| Magnesium | 2QBZ | (Dann et al. 2007) |
| glmS (ribozyme) | 2NZ4, 2Z75 | (Cochrane et al. 2007; Klein and Ferré-D’Amaré 2006) |
| Pre-Q1 | 3FU2, 2KFC, 3GCA | (Kang et al. 2009; Klein et al. 2009; Spitale et al. 2009) |
| Cyclic-di-GMP | 3IRW, 3IWN | (Kulshina et al. 2009; Smith et al. 2009) |
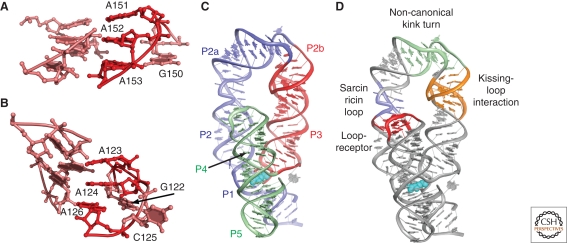
Figure 2.
Global architectures of the purine riboswitch. (A) The secondary structure of the aptamer domain of the Bacillus subtilis xpt-pbuX guanine riboswitch, drawn to reflect its tertiary architecture. Base interactions are depicted using the nomenclature of Leontis and Westhof (Leontis and Westhof 2001). Nucleotides that are >95% conserved in phylogeny are highlighted in red. (B) Crystal structure of the guanine riboswitch showing the organization of paired (P), loop (L), and joining (J) regions. The ligand is shown in cyan.
RNA helices are further organized through sequence motifs that adopt a variety of secondary structural elements: terminal loops, internal bulges and loops, and multi-helix junctions (Batey et al. 1999; Hendrix et al. 2005). One of the most common RNA motifs is the GA3 tetraloop, a terminal loop whose first nucleotide is a guanosine followed by three adenosine residues (Varani 1995). This tetraloop presents a stack of the adenosines for recognition by other RNA motifs, such as the tetraloop receptor (Cate et al. 1996; Jaeger et al. 1994; Murphy and Cech 1994) (Fig. 3A), which was recently identified in a subclass of the di-cyclic guanosine monophosphate riboswitches (Sudarsan et al. 2008). Interaction between the loop and the receptor serves to anchor the two helices together in a side-by-side arrangement, facilitating parallel packing. Variations of this terminal loop-internal loop theme are observed in the structures of the lysine (Fig. 3B) and thiamine pyrophosphate (TPP) riboswitches to achieve similar structural outcomes (Garst et al. 2008; Serganov et al. 2008; Serganov et al. 2006; Thore et al. 2006). Other common RNA building blocks that have been identified in riboswitches include kink-turns (K-turns) (Blouin and Lafontaine 2007; Winkler et al. 2001), kissing-loop interactions (Blouin and Lafontaine 2007), sarcin-ricin loops (Grundy et al. 2003; Sudarsan et al. 2003), T-loops (Barrick and Breaker 2007), and pseudoknots (McDaniel et al. 2005). Multi-helix junctions also represent an important component of many riboswitches but their sequences and structures are diverse, reflecting unique requirements for positioning the flanking helices or hosting sites of activity (Montange and Batey 2008).
Figure 3.
Organization of the lysine riboswitch. (A) The GAAA tetraloop-tetraloop receptor motif found across phylogeny in diverse RNAs, emphasizing the docking of the three adenosine residues into the minor groove of the receptor. (B) Mimicry of the tetraloop-tetraloop receptor interaction by the conserved pentaloop-helix interaction found in the lysine riboswitch. (C) The lysine riboswitch structure emphasizing the three-helix bundle formed by the P1/P2a, P2b/P3, and P4/P4 coaxial stacks (blue, red, and green, respectively). Lysine is shown in cyan. (D) Four recurrent structural motifs found in the lysine riboswitch that serve to organize the three-helix bundle.
The lysine riboswitch, one of the largest of the known riboswitches, illustrates how these architectural themes are used to establish a complex global fold. The aptamer domain of the Thermotoga maritima lysine riboswitch is organized as a bundle of three coaxially stacked sets of helices: P1/P2a, P2b/P3, and P4/P5 (Garst et al. 2008; Serganov et al. 2008) (Fig. 3C). The helical stacks are arranged in a parallel fashion by variants of the kink-turn, kissing-loop, tetraloop-tetraloop receptor and sarcin-ricin loop motifs (Fig. 3D). The P2 helix contains two internal loops: a canonical sarcin-ricin loop in the center of P2 and an unconventional kink-turn motif. The sarcin-ricin loop serves to help establish a receptor for the docking of the terminal loop L4, which forms a tetraloop-like structure (Fig. 3B), thereby arranging P4 parallel to P2. The noncanonical kink turn near the terminus of P2 bends the helix to direct the terminal loop L2 toward L3, which creates a kissing-loop interaction. This brings the P3 helix in parallel to P2 and P4, establishing the overall structure.
The importance of these architectural motifs for function is highlighted by studies in which altering these sequences renders a riboswitch nonfunctional under physiological conditions. Deletion of the L2–L3 interaction in the purine riboswitch results in a ∼1000-fold decrease in affinity for guanine (Gilbert et al. 2007; Lemay et al. 2006; Stoddard et al. 2008). Siimilarly, mutation of the kink-turn or kissing-loop interaction in the B. subtilis lysC lysine riboswitch debilitates ligand-dependent transcription termination in vitro (Blouin and Lafontaine 2007). Studies of the SAM-I riboswitch show similar results (Heppell and Lafontaine 2008; McDaniel et al. 2005; Winkler et al. 2003).
3. FOLDING PATHWAYS OF THE PURINE RIBOSWITCH APTAMER
Crystal structures of riboswitch aptamer domains are static pictures that do not lend insight into the process by which these RNAs acquire their three-dimensional structures. For riboswitches to function during transcription, they must rapidly form the tertiary interactions described earlier and avoid kinetic folding traps commonly found in RNA (Pan and Sosnick 1997; Treiber and Williamson 1999). In other words, slow folding or misfolding will impede their ability to elicit a proper regulatory response during the short temporal window of transcription of the 5′-UTR. This folding process can be monitored by a range of methods including classical approaches that use temperature and chemical denaturants to measure relative stability of structural elements, and more recently developed single molecule techniques that monitor folding transitions in real time. Because of its simple architecture, the purine riboswitch has emerged as the dominant model system for understanding aptamer domain folding.
A technique that uses the reactivity of 2′-hydroxyl groups to N-methylisatoic anhydride (NMIA) as a probe of RNA folding (Wilkinson et al. 2006) was recently used to study folding of the B. subtilis xpt-pbuX guanine riboswitch (Stoddard et al. 2008). An advantageous feature of this chemical probing method—commonly called “SHAPE”—is that it can be performed over a broad temperature range (Merino et al. 2005). This allows for determination of the melting temperature (Tm, defined as the temperature at which the RNA is half folded and half unfolded) of every nucleotide in the RNA of interest (Merino et al. 2005). Applied to the purine aptamer, at high temperatures (>70°C), only the helical elements of the aptamer are structured, as expected from the large free energy associated with RNA secondary structure formation (Fig. 4A) (Brion and Westhof 1997). As the temperature drops below 60°C, the L2–L3 interaction forms along with nucleotides becoming protected in J3/1, indicating that the peripheral loop–loop interaction promotes partial preorganization of the three-way junction via coaxial stacking of P1 and P3. Notably, J1/2 and J2/3 remain conformationally flexible in the absence of ligand, which would allow the effector access to the core of the RNA (see next section).
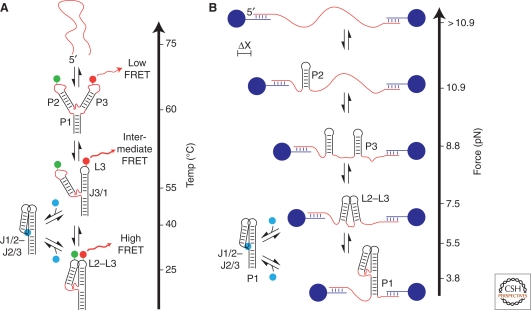
Figure 4.
Folding of the purine riboswitch aptamer domain. (A) Thermal denaturation and smFRET of the purine riboswitch reveal similar heirarchical folding landscapes. The Tm of each structural element (denoted by red strands) of the xpt-pbuX purine riboswitch reveals folding intermediates similar to those observed in smFRET experiments of the pbuE riboswitch at ambient temperature. smFRET experiments consisted of a fluorescein donor (green) and Cy3 acceptor (red) placed at L2 and L3 respectively allow observation of the loop–loop interaction in real time. (B) Unfolding the pbuE riboswitch by force measured as changes in distance (ΔX). This experiment reveals a pathway in which individual helices form sequentially from 5′ to 3′. Although denaturation studies show that ligand (cyan sphere) binding stabilizes the three way junction by ∼15°C, force spectroscopy supports a model in which P1 is also stabilized by ligand binding.
The previous analysis using SHAPE chemistry agrees well with single molecule fluorescence resonance energy transfer (smFRET) experiments with the B. subtilis pbuE adenine riboswitch (Lemay et al. 2006). In this approach, folding transitions are reported by changes in the FRET value of single molecules in which L2 and L3 were labeled with donor and acceptor fluorophores, respectively (Fig. 4A). Importantly, the smFRET data provide additional insight into the rates of folding and the influences of magnesium (Mg++) that is not possible using ensemble techniques. Addition of Mg++ above 50 µM was shown to significantly increase the folding rate of the loop-loop interaction; at physiologically relevant Mg++ concentrations, the global fold of the aptamer is adopted within ∼1 s (Lemay et al. 2006). Considering that bacterial polymerases have elongation rates of around ∼45 nucleotides per second at 25°C (Ryals et al. 1982), transcription of this 70-nucleotide aptamer would require ∼1.5 s. Thus, folding and transcription occur on similar timescales, as would be expected for a cotranscriptional process.
Another method for monitoring folding processes uses force, which has gained popularity because of technological advances that allow higher spatial and temporal resolution (King et al. 2009; Li et al. 2008). Application of constant force to the ends of a macromolecule alters the energy landscape such that specific structural elements become bistable (equally occupying the folded and unfolded states) at a specific applied force, known as a F1/2 (King et al. 2009; Li et al. 2008). Like smFRET, this technique simultaneously measures thermodynamic and kinetic parameters of folding and unfolding. Observations of Vibrio vulnificus add adenine riboswitch folding by force spectroscopy revealed that as the applied force was decreased, extension changes corresponding to formation of P2 occurred, followed by P3, the loop-loop interaction, and finally P1 (Fig. 4B) (Greenleaf et al. 2008). By fully extending the RNA and allowing it to refold for predetermined time intervals before reapplying force, the authors obtained estimates for the rate of aptamer domain folding similar to those derived from smFRET measurements.
The folding model obtained by force spectroscopy is distinct from that obtained using other techniques (Greenleaf et al. 2008). Although chemical probing and smFRET propose a classic hierarchical folding pathway in which all secondary structure is acquired before tertiary structure, the folding pathway proposed using force spectroscopy proceeds from the 5′ to 3′ direction. Such a model is more consistent with expectations for folding during transcription. Nevertheless, both models illustrate the importance of tertiary structure formation before ligand binding, as this has cooperative influences on ligand binding by preorganizing the binding site.
4. RECOGNITION OF EFFECTOR MOLECULES BY RIBOSWITCH RECEPTORS
Following proper folding, the aptamer domain must accomplish two interconnected tasks: specific recognition of the effector and coupling binding to a regulatory outcome. The first objective requires that the RNA effectively discriminates between closely related compounds in the cell. For example, the purine riboswitches achieve over a 10,000-fold level of discrimination between guanine and adenine (Mandal et al. 2003; Mandal and Breaker 2004), whereas the lysine riboswitch discriminates between lysine and ornithine, amino acids that differ by a single methylene group in their side chain, by at least 5000-fold (Garst and Batey, unpublished data).
The purine riboswitch family is divided into three classes that are defined by their effector molecules: guanine/hypoxanthine, adenine, or 2′-deoxyguanosine (Kim and Breaker 2008). This family of aptamers is defined by a common conserved secondary and tertiary structure. In each class, the effector is bound using a pocket formed by the RNA three-way junction (Kim et al. 2007; Mandal et al. 2003; Mandal and Breaker 2004). Within the junction are five base triples between nucleotides that define the local architecture of the binding pocket (Fig. 2A). At the center is a base triple involving the ligand and two pyrimidine residues that serve to specifically recognize the nucleobase (Fig. 5A) (Batey et al. 2004; Noeske et al. 2005; Serganov et al. 2004). Every functional group in the ligand is directly recognized through hydrogen bonding interactions, in part explaining the ability of the RNA to achieve a high specificity for this compound. Surprisingly, the nucleobase does not completely stack on the bases above or below it, a feature common to other riboswitches that recognize nucleobase-containing effectors (Gilbert et al. 2008; Montange and Batey, 2006; Serganov et al. 2009; Serganov et al. 2006; Thore et al. 2006).
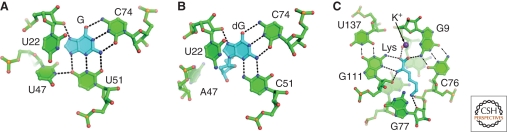
Figure 5.
Details of ligand-riboswitch interactions. (A) Recognition of guanine (cyan) by a pocket of conserved pyrimidine residues (green). The ligand interacts with the RNA primarily through hydrogen bonding interactions (black dashed lines) between pyrimidine 51 and 74. The binding pocket for adenine-binding riboswitches is nearly identical except that the ligand interacts with a uridine residue at position 74. (B) 2′-deoxyguanosine (cyan) is recognized through interactions with two cytosine residues (green). Note the shift of C51 relative to U51 to accommodate the 2′-deoxyribose moiety (C) Lysine (cyan) recognition by nucleotides in the the lysine riboswitch binding pocket (green). A potassium ion (purple sphere) partially mediates the interaction between the carboxylate group and the RNA.
Purine specificity is achieved through the identity of two pyrimidine residues that participate in a base triple with the ligand. Nucleotide 74 forms a Watson-Crick pair with the ligand, allowing the RNA to discriminate between adenine and guanine/2′-deoxyguanosine (Batey et al. 2004; Mandal et al. 2003; Mandal and Breaker 2004; Serganov et al. 2004). This position is always uridine in adenine riboswitches and always cytosine in guanine/2′-deoxyguanosine riboswitches (Fig. 5A). Similarly, the Watson-Crick face of nucleotide 51 is used to discriminate between nucleobases (adenine and guanine) and nucleosides (Edwards and Batey 2009). Residue 51 is always uridine in guanine and adenine riboswitches and cytosine in 2′-deoxyguanosine riboswitches (Fig. 5B). This cytosine shifts toward nucleotide 74 such that it opens up the binding pocket to allow for the presence of the 2′-deoxyribose sugar moiety (Edwards and Batey, 2009). Thus, small sequence changes to the aptamer can generate riboswitches that respond to chemically distinct effectors.
Like the purine riboswitch, the lysine riboswitch fully encapsulates its ligand within a binding pocket that directly interacts with all of the effector’s polar functional groups (Garst et al. 2008; Serganov et al. 2008). The main chain atoms of lysine are recognized through minor groove interactions with invariant G-C and G•U pairs, and by a potassium ion bound near the carboxyl group (Fig. 5C). The positively charged amino group of the side chain is recognized through an electrostatic interaction with a nonbridging phosphate oxygen positioned at the opposite end of the pocket. Discrimination between lysine and ornithine is likely caused by the inability of a shorter ornithine side chain to position its amino group adjacent to the nonbridging phosphate group. The ability of the effector to form this electrostatic interaction is also the basis for discrimination against other abundant amino acids such as serine and aspartate that could sterically fit in this pocket.
A distinct challenge faced by RNA is recognition of negatively charged moieties. Instead of directly contacting negative charges presented by ligands, aptamers tend to use metal cations as a bridge. For example, the phosphate groups of TPP and flavin mononucleotide (FMN) are coordinated by Mg++ ions to mediate RNA contacts (Serganov et al. 2009; Serganov et al. 2006; Thore et al. 2006). Similarly, the carboxylic acid moiety of lysine interacts with the RNA via a potassium cation (Fig. 5C) (Serganov et al. 2008). The SAM-I and SAM-II riboswitches represent an exception in which the carboxylic acid moiety interacts directly with the Watson-Crick face of a guanine and adenine residue, respectively (Gilbert et al. 2008; Montange and Batey 2006).
These structures highlight what appears to be a general feature of the majority of riboswitch aptamer-ligand interactions: The ligand is highly solvent inaccessible, buried within the core of the RNA. On average, 90% of the metabolite surface area is solvent inaccessible, which is greater than that of ligands bound to artificial aptamers (71%) (Edwards et al. 2007). At first glance, this might be attributed to the need to discriminate between closely related metabolites in the cell (Gilbert and Batey 2005). However, in vitro selected RNAs can easily achieve comparable specificities for their ligands without the need for extensive burial. The theophylline aptamer is capable of binding its cognate purine with an affinity that rivals the adenine riboswitch, capable of discriminating against caffeine by >10,000-fold (Jenison et al. 1994). Like many riboswitches, the theophylline aptamer undergoes a significant ligand-dependent conformational change (Zimmermann et al. 1997; Zimmermann et al. 2000), indicating that folding and a high degree of ligand burial are not necessarily correlated phenomena.
5. LIGAND BINDING INDUCES CONFORMATIONAL CHANGES IN THE RECEPTOR
A more likely reason for extensive ligand burial is related to interdomain communication in the riboswitch. Solvent inaccessibility of the effector molecule implies that the aptamer must adopt a flexible “open” state that allows the ligand entry into the pocket, followed by a conformational change that encapsulates the ligand. This phenomenon, referred to as an induced-fit binding mechanism, is a common feature of RNA binding reactions, such as in RNP assembly (Williamson 2000). Conformational changes in the riboswitch aptamer domain are used to couple ligand binding to folding of downstream structural switch in the expression platform (see next section) that in turn instructs the transcriptional or translational machinery.
The inherent flexibility of riboswitches in the absence of a ligand typically precludes crystallographic examination of these states. Instead, they are studied in solution using techniques such as chemical probing and NMR spectroscopy, which provide information on conformational dynamics at the global and local levels. NMR studies of both the pbuE and xpt-pbuX purine aptamers show that in the absence of ligand, nucleotides in the three-way junction are completely disordered even at ambient temperatures as indicated by the absence of peaks for corresponding nucleotides (Noeske et al. 2007a; Noeske et al. 2007b). This is consistent with the high degree of reactivity in these regions identified by in-line probing (Mandal et al. 2003) and SHAPE chemistry (Stoddard et al. 2008). Both NMR and chemical probing data indicate the J1/2 and J2/3 regions of the aptamer domain are disordered in the absence of the effector (Fig. 4A) (Noeske et al. 2007b; Ottink et al. 2007; Stoddard et al. 2008), suggesting that these elements form a flexible lid that closes around the docked ligand.
Addition of the appropriate purine nucleobase induces structure in the three-way junction, as evidenced both by the appearance of new peaks in NMR spectra (Noeske et al. 2005) and a >15°C increase in the Tm of these nucleotides by NMIA probing (Fig. 4A) (Stoddard et al. 2008). Along with formation of intermolecular interactions between the ligand and RNA, these data reveal formation of RNA–RNA interactions in the junction. Importantly, a subset of these ligand-dependent interactions involves formation of two base triples between J2/3 and the 3′-side of the P1 helix (Stoddard et al. 2008). Formation of these interactions was proposed to stabilize incorporation of the switching sequence into the receptor domain, serving as the basis for the communication of binding to the signal transduction domain (Batey et al. 2004). This was further supported by force spectroscopy of the pbuE aptamer, in which an additional ∼2.2 pN of force was required to make the P1 helix bistable in the presence of adenine, providing direct evidence for adenine induced stabilization of this helix (Fig. 4B) (Greenleaf et al. 2008).
Ligand-induced folding of the lysine riboswitch is similar to that of the purine riboswitches in many respects. Small angle X-ray scattering (SAXS), which provides a measure of the global conformation in solution (Lipfert and Doniach 2007; Putnam et al. 2007), demonstrates that the lysine aptamer is largely preorganized by Mg++, whereas NMIA chemical probing data reveal that a subset of nucleotides in the five-way junction remains disordered under these conditions (Garst et al. 2008). In the presence of saturating lysine concentrations, these nucleotides become nonreactive to NMIA, indicating that lysine induces local order to the junction. Interestingly, this RNA crystallizes well in the absence of lysine, allowing the free state of this aptamer to be characterized (Garst et al. 2008; Serganov et al. 2008). In contrast to the solution studies, the crystal structures of the free and bound state were nearly identical. This suggests that in solution the aptamer exists as an ensemble of interconverting structures and that lysine recognizes a subpopulation of binding-competent RNAs. Although the aptamer can adopt a “bound-like” state in the absence of ligand, it requires lysine to fully stabilize the crystallographically observed structure.
Although the purine and lysine aptamers appear to undergo localized conformational changes on binding ligand, others show more dramatic structural organizations. For example, studies of the TPP riboswitch using site-specific labeling with the fluorescent reporter 2-aminopurine (2AP) revealed both the ligand binding pocket and a terminal loop-helix interaction fold with comparable rates on addition of ligand (Lang et al. 2007). However, a sevenfold disparity between the folding rate of the loop-helix interaction and the top of the P1 helix was measured, indicating that the free state of the TPP aptamer is largely disordered and ligand-induced organization of the RNA propagates from the periphery to the central junction. Similarly, fluorescence and NMR spectroscopic analyses of the preQ1 (a biosynthetic precursor of the queuosine nucleoside found in the anticodon of many tRNAs) riboswitch revealed that the free state of this RNA folds into a hairpin structure with a highly disordered single stranded 3'-tail (Kang et al. 2009; Rieder et al. 2009). Addition of preQ1 is required for association of the hairpin loop with the 3'-tail to complete the pseudoknot structure.
6. TEMPORAL EFFECTS OF TRANSCRIPTION
To this point, we have focused on studies of isolated riboswitch aptamers, which typically bind their effector ligands with nanomolar equilibrium dissociation constants (KD). However, a number of studies have shown higher ligand concentrations are required to effectively regulate gene expression. The FMN, SAM-I, and lysine riboswitches require ligand concentrations ∼100–1000-fold above the KD of the aptamer to achieve half maximal transcription termination (T50) (Blouin and Lafontaine 2007; Tomsic et al. 2008; Wickiser et al. 2005b). This is a hallmark of a kinetically controlled process in which the aptamer domain has insufficient time to equilibrate with the cellular environment before the expression platform commits the RNA to an alternative folding route that may be largely irreversible. Studies of a number of riboswitches have found that transcripts encompassing the entire riboswitch are deficient in effector binding, strongly arguing for irreversible folding (Lemay et al. 2006; Mironov et al. 2002; Winkler et al. 2002a).
The time sensitivity of riboswitch regulation is explained in part by the slow kinetics of ligand binding, an inherent feature of induced fit processes. Measurement of the association kinetics of 2-aminopurine to the xpt and pbuE purine riboswitches yielded rate constants on the order of 105 M−1s−1 for both aptamers (Gilbert et al. 2006; Wickiser et al. 2005a). Similar values were obtained for the FMN and TPP riboswitches (Wickiser et al. 2005b). These rates suggest that at 1 µM intracellular ligand concentration, at least 10 s would be required for effector to fully saturate the aptamer. If transcription occurred at a constant rate for this duration, a bacterial polymerase would transcribe nearly 500–1000 nucleotides, well beyond the boundaries of even the largest riboswitches, making regulation by the riboswitch impossible.
One means by which riboswitches deal with this time limitation is by manipulating the rate of transcription using programmed pause sites embedded within the expression platform. Two such sites were identified by synchronized transcription assays of the FMN aptamer that have pause lifetimes of 1 and 10 s respectively (Wickiser et al. 2005b). Mutations that ablate pausing in these sites cause significant elevation of the T50 values in the context of the same aptamer, as expected when transcription proceeds through the riboswitch more rapidly. Uridine-rich tracts in the expression platform of the pbuE riboswitch have also been suggested to serve as transcriptional pause sites, though they remain to be experimentally validated (Wickiser et al. 2005a). Although the general importance of pausing remains to be addressed for the majority of aptamers, this phenomenon may provide an important mechanism for tuning the response range of a riboswitch to ligand concentrations relevant for the cell.
Pausing may also provide time at important points during transcription to allow structural rearrangements that guide more efficient folding of the downstream sequence. For example, RNAs such as RNase P, tmRNA, and SRP RNAs all form labile, nonnative structural intermediates at the point of transcriptional pauses (Wong et al. 2007). These intermediates sequester upstream portions of long-range helices in which the 5′ and 3′ sides are separated in primary sequence by more than 50 nucleotides. It has been proposed that these intermediates provide the RNA with a mechanism for preventing misfolding and enhancing the rate of the overall folding reaction in vivo. By analogy, the long range P1 helix in riboswitch aptamers may be formed as part of a labile folding intermediate that is trapped by ligand binding.
7. MODELS FOR STRUCTURAL SWITCHING
Riboswitches, as well as a number of other types of RNA regulatory elements in bacteria and eukarya, use mutually exclusive secondary structures to direct expression machinery. As these processes are often cotranscriptional, it is important to consider the rate at which RNA transcribes and folds, as well as the relative thermodynamic stability of competing secondary structures. As opposed to the majority of studies that observe folding of a fully synthesized RNA (see previous discussion), the directionality of transcription imprints order that biases the final conformation because upstream sequences can fold before transcription of downstream sequences. For riboswitches, this means that folding of the aptamer can occur in the absence of the expression platform, and the antiterminator can form before synthesis of the terminator stem-loop.
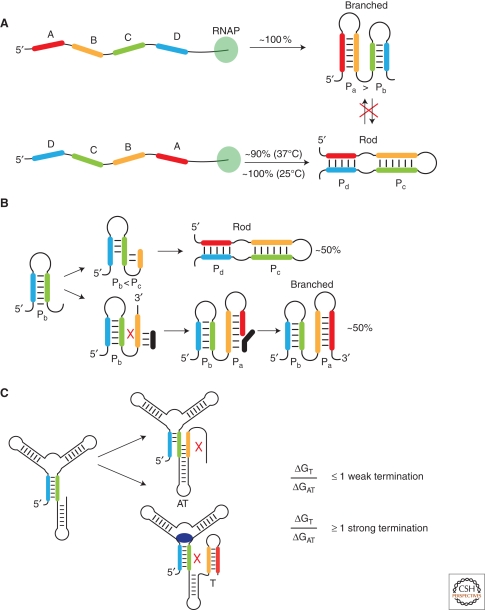
The effects of transcription order have been studied for many RNAs, but perhaps most clearly so using a simple bistable switch that mimics many of the features of naturally occurring riboswitches (Fig. 6) (Xayaphoummine et al. 2007). This RNA was engineered to fold into mutually exclusive branched or rodlike structures that can be differentiated using native gel electrophoresis. Transcription of the “forward sequence” (e.g., abc … xyz) of the switch (Fig. 6A) yields exclusively the branched structure, whereas transcription of the reversed sequence (e.g., zyx … cba) leads to the alternative rodlike structure with ∼90% frequency. In contrast, heat renaturation of the RNA leads to an equal distribution of the two states, which do not significantly interchange at room temperature. In other words, RNA can readily adopt nonequilibrium structures during cotranscriptional folding because of high activation energies associated with their interconversion (Crothers et al. 1974). This result emphasizes that traditional methods for studying RNA folding might fail to capture some of the most crucial features of cotranscriptional folding which are inherently tied to the mechanism of riboswitch regulation.
Figure 6.
Cotranscriptional folding of synthetic RNA switches. (A) A synthetically designed RNA sequence transcribed in forward and reverse directions illustrating the influence of transcription order on the folding outcome. The free energy of each sequential helix dictates the outcome of folding. (B) A model of the reverse synthetic sequence reveals that structural transitions can be redirected by addition of a competitor oligonucleotide (black) to the transcription reaction. (C) Regulatory efficiency of purine riboswitches correlates with the relative free energy of the terminator (T) and anti terminator (AT) stems, pointing to similarities between riboswitches and their synthetic counterparts.
A second key conclusion gained from studies of the engineered switch is that the relative stability of helices (Pa > Pc, and Pc > Pb) is sufficient to encode a precise folding pathway (Xayaphoummine et al. 2007). Similar models have been devised to explain the variable regulatory activities of purine riboswitches within the B. subtilis genome (Mulhbacher and Lafontaine 2007). In cases where the relative stability of the antiterminator hairpin (ΔGAT) is greater than that of the terminator hairpin (ΔGT) (Fig. 6C), reporter gene assays demonstrate that there is a significant amount of transcription read through in vivo, even at saturating ligand concentrations in the growth media. In contrast, increasing the stability of the terminator hairpin is correlated with the enhanced ability of the riboswitch to promote transcription termination under high ligand concentrations (Mulhbacher and Lafontaine 2007). Similar factors may account for the large range of behaviors observed for the 12 SAM-I riboswitches in the B. subtilis genome (Tomsic et al. 2008).
The analogy between the engineered and natural riboswitches can be extended by considering self-induced folding transitions occurring during folding of the reverse sequence (Fig. 6B). In the absence of a competitor oligonucleotide, the reverse switch sequence destabilizes the Pb stem in favor of the Pc stem; lower temperatures bias the structure further toward Pc because of slower transcription rates. This refolding process can be disrupted by introduction of a competitor oligonucleotide (Fig. 6B, black line) that pairs with the Pc stem early in transcription. The oligonucleotide efficiently redirects folding into the branched structure, in the same fashion as ligand binding prevents self-induced transitions during riboswitch folding that would lead to aberrant antitermination (Fig. 6C). Elements of the unbound aptamer can be thought of as behaving like the antisense oligonucleotide by disrupting terminator stem formation while simultaneously nucleating the antiterminator stem. This comparison warrants further investigation of the simple secondary structures in both domains of the RNA that must interchange during transcription to elicit regulatory control.
8. RIBOSWITCHES IN THE RNA WORLD
Although the exact nature of the RNA world cannot be reconstructed, riboswitches present themselves as an elegant solution to the problem of biological regulation. These RNAs are capable of recognizing and responding to compounds that were likely to have been metabolites in the RNA world, such as the purine nucleobases. In addition, the secondary structural switch that interfaces with the expression machinery can function by directing RNA–RNA interactions. In modern riboswitches that act at the translational level, effector binding to the aptamer domain occludes the Shine-Dalgarno sequence that recruits the 30S subunit by pairing with a site near the 3′-end of the 16S ribosomal RNA. Thus, riboswitches contain the essential features to efficiently function in the hypothesized RNA world.
If riboswitches are truly biological fossils of an ancient world, it is even more impressive that they have survived billions of years of evolutionary pressure to serve in modern organisms. Some riboswitches that are widely distributed in phylogeny, such as the thiamine pyrophosphate and adenosylcobalamin classes, might have survived because they occupy a niche in metabolic regulation that perhaps is better suited to RNA than protein. They are an energetically cost-effective means for genetic regulation compared with protein synthesis, as only mRNA transcription needs to occur. Furthermore, riboswitches offer an immediate feedback response as riboswitch regulation involves fewer steps compared with that of regulatory proteins. Thus, modern organisms continue to capitalize on the unique abilities of RNA to fine-tune gene expression levels using a mechanism that could have easily found a home in the RNA world.
Acknowledgments
A.D.G. and A.L.E. contributed equally to this work. We thank members of the Batey laboratory and Jennifer Pfingsten for careful reading of this manuscript. This work was supported by grants from the National Institutes of Health.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Barrick JE, Breaker RR 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 [DOI] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA 1999. Tertiary Motifs in RNA Structure and Folding. Angew Chem Int Ed Engl 38: 2326–2343 [DOI] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA 2007. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion P, Westhof E 1997. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct 26: 113–137 [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685 [DOI] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA 2007. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol 14: 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers DM, Cole PE, Hilbers CW, Shulman RG 1974. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol 87: 63–88 [DOI] [PubMed] [Google Scholar]
- Dann CE 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130: 878–892 [DOI] [PubMed] [Google Scholar]
- Edwards AL, Batey RT 2009. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol 385: 938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Ferre-D’Amare AR 2006. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 14: 1459–1468 [DOI] [PubMed] [Google Scholar]
- Edwards TE, Klein DJ, Ferre-D’Amare AR 2007. Riboswitches: Small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol 17: 273–279 [DOI] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Batey RT 2005. Riboswitches: natural SELEXion. Cell Mol Life Sci 62: 2401–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Love CE, Edwards AL, Batey RT 2007. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry 46: 13297–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182 [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT 2006. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol 359: 754–768 [DOI] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Lehman SC, Henkin TM 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A 100: 12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DK, Brenner SE, Holbrook SR 2005. RNA structural motifs: building blocks of a modular biomolecule. Q Rev Biophys 38: 221–243 [DOI] [PubMed] [Google Scholar]
- Heppell B, Lafontaine DA 2008. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47: 1490–1499 [DOI] [PubMed] [Google Scholar]
- Holbrook SR 2008. Structural principles from large RNAs. Annu Rev Biophys 37: 445–464 [DOI] [PubMed] [Google Scholar]
- Jaeger L, Michel F, Westhof E 1994. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol 236: 1271–1276 [DOI] [PubMed] [Google Scholar]
- Jenison RD, Gill SC, Pardi A, Polisky B 1994. High-resolution molecular discrimination by RNA. Science 263: 1425–1429 [DOI] [PubMed] [Google Scholar]
- Kang M, Peterson R, Feigon J 2009. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell 33: 784–790 [DOI] [PubMed] [Google Scholar]
- Kim JN, Breaker RR 2008. Purine sensing by riboswitches. Biol Cell 100: 1–11 [DOI] [PubMed] [Google Scholar]
- Kim JN, Roth A, Breaker RR 2007. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci 104: 16092–16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM, Carter AR, Churnside AB, Eberle LS, Perkins TT 2009. Ultrastable atomic force microscopy: Atomic-scale stability and registration in ambient conditions. Nano Lett 9: 1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D’Amaré AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Edwards TE, Ferre-D’Amare AR 2009. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol 16: 343–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferre-D’Amare AR 2009. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol 16: 1212–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K, Rieder R, Micura R 2007. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res 35: 5370–5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA 2006. Folding of the adenine riboswitch. Chem Biol 13: 857–868 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E 2006. The building blocks and motifs of RNA architecture. Current Opinion in Structural Biology 16: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PT, Vieregg J, Tinoco I Jr, 2008. How RNA unfolds and refolds. Annu Rev Biochem 77: 77–100 [DOI] [PubMed] [Google Scholar]
- Link KH, Breaker RR 2009. Engineering ligand-responsive gene-control elements: Lessons learned from natural riboswitches. Gene Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Doniach S 2007. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct 36: 307–327 [DOI] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A 2008. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol 15: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR 2004. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11: 29–35 [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577–586 [DOI] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Henkin TM 2005. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol 57: 1008–1021 [DOI] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM 2003. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci 100: 3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM 2005. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231 [DOI] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111: 747–756 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2008. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37: 117–133 [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, Lafontaine DA 2007. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res 35: 5568–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FL, Cech TR 1994. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol 236: 49–63 [DOI] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043. [DOI] [PubMed] [Google Scholar]
- Noeske J, Schwalbe H, Wohnert J 2007b. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res 35: 5262–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J 2007a. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res 35: 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J 2005. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci 102: 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS 2007. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA 13: 2202–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sosnick TR 1997. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat Struct Biol 4: 931–938 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA 2007. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Rieder U, Lang K, Kreutz C, Polacek N, Micura R 2009. Evidence for pseudoknot formation of class I preQ1 riboswitch aptamers. Chembiochem 10: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Ryals J, Little R, Bremer H 1982. Temperature dependence of RNA synthesis parameters in Escherichia coli. J Bacteriol 151: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2008. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ 2006. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441: 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ 2004. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol 11: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE 2009. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem 284: 11012–11016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT 2008. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev 17: 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N 2006. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 312: 1208–1211 [DOI] [PubMed] [Google Scholar]
- Tomsic J, McDaniel BA, Grundy FJ, Henkin TM 2008. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol 190: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR 1999. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol 9: 339–345 [DOI] [PubMed] [Google Scholar]
- Varani G 1995. Exceptionally stable nucleic acid hairpins. Annu Rev Biophys Biomol Struct 24: 379–404 [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM 2005a. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44: 13404–13414 [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM 2005b. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]
- Williamson JR 2000. Induced fit in RNA-protein recognition. Nat Struct Biol 7: 834–837 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker R 2003. Genetic control by metabolite-binding riboswitches. Chembiochem 4: 1024–1032 [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR 2002a. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR 2002b. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A 99: 15908–15913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Grundy FJ, Murphy BA, Henkin TM 2001. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707 [DOI] [PubMed] [Google Scholar]
- Wong TN, Sosnick TR, Pan T 2007. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci 104: 17995–18000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xayaphoummine A, Viasnoff V, Harlepp S, Isambert H 2007. Encoding folding paths of RNA switches. Nucleic Acids Res 35: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A 1997. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol 4: 644–649 [DOI] [PubMed] [Google Scholar]
- Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A 2000. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA 6: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]