Abstract
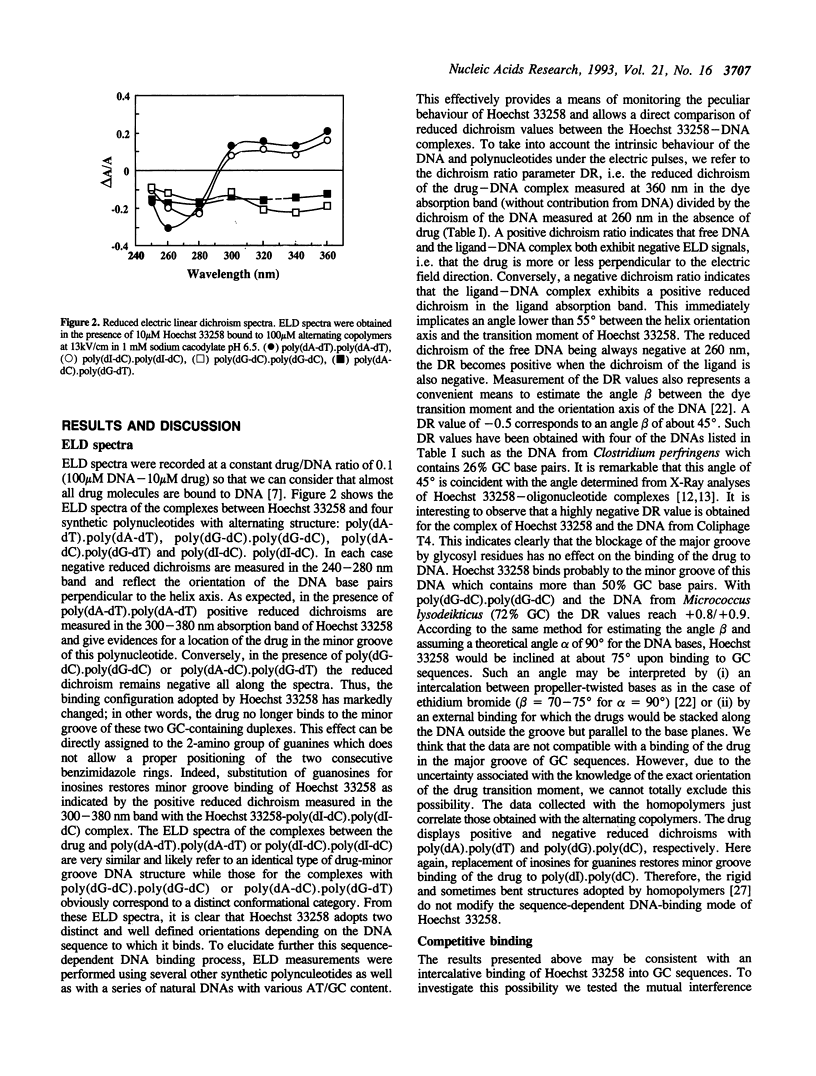
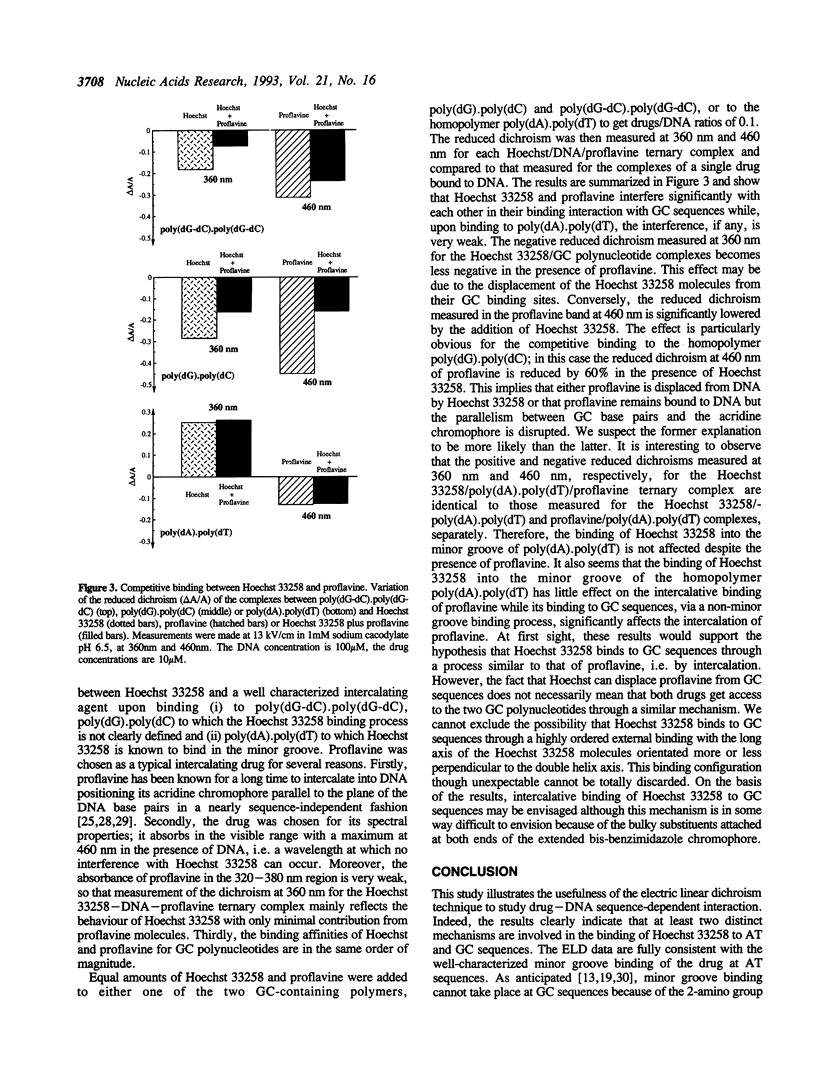
The binding mode of the bisbenzimidazole derivative Hoechst 33258 to a series of DNAs and polynucleotides has been investigated by electric linear dichroism. Positive reduced dichroisms were measured for the poly(dA-dT).poly(dA-dT)- and poly(dA).poly(dT)-Hoechst complexes in agreement with a deep penetration of the drug into the minor groove. Similarly, the drug displays positive reduced dichroism in the presence of the DNAs from calf thymus, Clostridium perfringens and Coliphage T4. Conversely, negative reduced dichroisms were obtained when Hoechst 33258 was bound to poly(dG-dC).poly(dG-dC), poly(dA-dC).poly(dG-dT) and poly(dG).poly(dC) as well as with the GC-rich DNA from Micrococcus lysodeikticus indicating that in this case minor groove binding cannot occur. Substitution of guanosines for inosines induces a reversal of the reduced dichroism from negative to positive. Therefore, as anticipated it is the 2-amino group of guanines protruding in this groove which prevents Hoechst 33258 from getting access to the minor groove of GC sequences. The ELD data obtained with the GC-rich biopolymers are consistent with an intercalative binding. Competition experiments performed with the intercalating drug proflavine lend credence to the involvement of an intercalative binding rather than to an external or major groove binding of Hoechst 33258 at GC sequences.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bontemps J., Houssier C., Fredericq E. Physico-chemical study of the complexes of "33258 Hoechst" with DNA and nucleohistone. Nucleic Acids Res. 1975 Jun;2(6):971–984. doi: 10.1093/nar/2.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrondo M. A., Coll M., Aymami J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Binding of a Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry. 1989 Sep 19;28(19):7849–7859. doi: 10.1021/bi00445a047. [DOI] [PubMed] [Google Scholar]
- Harshman K. D., Dervan P. B. Molecular recognition of B-DNA by Hoechst 33258. Nucleic Acids Res. 1985 Jul 11;13(13):4825–4835. doi: 10.1093/nar/13.13.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssier C., Bontemps J., Emonds-Alt X., Fredericq E. Electric dichroism and birefringence of DNA, chromatin, and their complexes with cationic dyes. The structure of chromatin. Ann N Y Acad Sci. 1977 Dec 30;303:170–189. doi: 10.1111/j.1749-6632.1977.tb55930.x. [DOI] [PubMed] [Google Scholar]
- Jin R., Breslauer K. J. Characterization of the minor groove environment in a drug-DNA complex: bisbenzimide bound to the poly[d(AT)].poly[d(AT)]duplex. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8939–8942. doi: 10.1073/pnas.85.23.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson K. F., Varshney U., van de Sande J. H. Interaction of Hoechst 33258 with repeating synthetic DNA polymers and natural DNA. J Biomol Struct Dyn. 1988 Apr;5(5):1005–1023. doi: 10.1080/07391102.1988.10506446. [DOI] [PubMed] [Google Scholar]
- Kapuscinski J. Interactions of nucleic acids with fluorescent dyes: spectral properties of condensed complexes. J Histochem Cytochem. 1990 Sep;38(9):1323–1329. doi: 10.1177/38.9.1696951. [DOI] [PubMed] [Google Scholar]
- Kumar S., Joseph T., Singh M. P., Bathini Y., Lown J. W. Structure and dynamics of ligand-template interactions of topoisomerase inhibitory analogs of Hoechst 33258: high field 1H-NMR and restrained molecular mechanics studies. J Biomol Struct Dyn. 1992 Apr;9(5):853–880. doi: 10.1080/07391102.1992.10507963. [DOI] [PubMed] [Google Scholar]
- Kumar S., Yadagiri B., Zimmermann J., Pon R. T., Lown J. W. Sequence specific molecular recognition and binding by a GC recognizing Hoechst 33258 analogue to the decadeoxyribonucleotide d-[CATGGCCATG]2: structural and dynamic aspects deduced from high field 1H-NMR studies. J Biomol Struct Dyn. 1990 Oct;8(2):331–357. doi: 10.1080/07391102.1990.10507809. [DOI] [PubMed] [Google Scholar]
- LERMAN L. S. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961 Feb;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- Loontiens F. G., McLaughlin L. W., Diekmann S., Clegg R. M. Binding of Hoechst 33258 and 4',6'-diamidino-2-phenylindole to self-complementary decadeoxynucleotides with modified exocyclic base substituents. Biochemistry. 1991 Jan 8;30(1):182–189. doi: 10.1021/bi00215a027. [DOI] [PubMed] [Google Scholar]
- Loontiens F. G., Regenfuss P., Zechel A., Dumortier L., Clegg R. M. Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A-T)], and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry. 1990 Sep 25;29(38):9029–9039. doi: 10.1021/bi00490a021. [DOI] [PubMed] [Google Scholar]
- Lown J. W. Lexitropsins: rational design of DNA sequence reading agents as novel anti-cancer agents and potential cellular probes. Anticancer Drug Des. 1988 Jun;3(1):25–40. [PubMed] [Google Scholar]
- Norden B., Kubista M., Kurucsev T. Linear dichroism spectroscopy of nucleic acids. Q Rev Biophys. 1992 Feb;25(1):51–170. doi: 10.1017/s0033583500004728. [DOI] [PubMed] [Google Scholar]
- Parkinson J. A., Barber J., Douglas K. T., Rosamond J., Sharples D. Minor-groove recognition of the self-complementary duplex d(CGCGAATTCGCG)2 by Hoechst 33258: a high-field NMR study. Biochemistry. 1990 Nov 6;29(44):10181–10190. doi: 10.1021/bi00496a005. [DOI] [PubMed] [Google Scholar]
- Pjura P. E., Grzeskowiak K., Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol. 1987 Sep 20;197(2):257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Assignment of DNA binding sites for 4',6-diamidine-2-phenylindole and bisbenzimide (Hoechst 33258). A comparative footprinting study. Biochim Biophys Acta. 1988 Feb 28;949(2):158–168. doi: 10.1016/0167-4781(88)90079-6. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Lipanov A. A., Dickerson R. E. Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G. Biochemistry. 1991 Oct 22;30(42):10294–10306. doi: 10.1021/bi00106a030. [DOI] [PubMed] [Google Scholar]
- Rao K. E., Lown J. W. Molecular recognition between ligands and nucleic acids: DNA binding characteristics of analogues of Hoechst 33258 designed to exhibit altered base and sequence recognition. Chem Res Toxicol. 1991 Nov-Dec;4(6):661–669. doi: 10.1021/tx00024a011. [DOI] [PubMed] [Google Scholar]
- Searle M. S., Embrey K. J. Sequence-specific interaction of Hoechst 33258 with the minor groove of an adenine-tract DNA duplex studied in solution by 1H NMR spectroscopy. Nucleic Acids Res. 1990 Jul 11;18(13):3753–3762. doi: 10.1093/nar/18.13.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram M., van der Marel G. A., Roelen H. L., van Boom J. H., Wang A. H. Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342. EMBO J. 1992 Jan;11(1):225–232. doi: 10.1002/j.1460-2075.1992.tb05045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke T., Steen H. B. Multiple binding modes for Hoechst 33258 to DNA. J Histochem Cytochem. 1985 Apr;33(4):333–338. doi: 10.1177/33.4.2579998. [DOI] [PubMed] [Google Scholar]
- Strekowski L., Wilson W. D., Mokrosz J. L., Mokrosz M. J., Harden D. B., Tanious F. A., Wydra R. L., Crow S. A., Jr Quantitative structure-activity relationship analysis of cation-substituted polyaromatic compounds as potentiators (amplifiers) of bleomycin-mediated degradation of DNA. J Med Chem. 1991 Feb;34(2):580–588. doi: 10.1021/jm00106a017. [DOI] [PubMed] [Google Scholar]
- Tang P., Juang C. L., Harbison G. S. Intercalation complex of proflavine with DNA: structure and dynamics by solid-state NMR. Science. 1990 Jul 6;249(4964):70–72. doi: 10.1126/science.2367853. [DOI] [PubMed] [Google Scholar]
- Tanious F. A., Veal J. M., Buczak H., Ratmeyer L. S., Wilson W. D. DAPI (4',6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry. 1992 Mar 31;31(12):3103–3112. doi: 10.1021/bi00127a010. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W. D., Barton H. J., Tanious F. A., Kong S. B., Strekowski L. The interaction with DNA of unfused aromatic systems containing terminal piperazino substituents. Intercalation and groove-binding. Biophys Chem. 1990 Apr;35(2-3):227–243. doi: 10.1016/0301-4622(90)80011-u. [DOI] [PubMed] [Google Scholar]



