Abstract
Plant growth and morphogenesis depend on the levels and distribution of the plant hormone auxin. Plants tightly regulate cellular levels of the active auxin indole-3-acetic acid (IAA) through synthesis, inactivation, and transport. Although the transporters that move IAA into and out of cells are well characterized and play important roles in development, little is known about the transport of IAA precursors. In this review, we discuss the accumulating evidence suggesting that the IAA precursor indole-3-butyric acid (IBA) is transported independently of the characterized IAA transport machinery along with the recent identification of specific IBA efflux carriers and enzymes suggested to metabolize IBA. These studies have revealed important roles for IBA in maintaining IAA levels and distribution within the plant to support normal development.
Keywords: Auxin, auxin transport, indole-3-butyric acid, IBA
INTRODUCTION
Auxin regulates many critical aspects of plant growth and development by directing cell division, elongation, and differentiation (reviewed in Perrot-Rechenmann, 2010). Levels of the active auxin indole-3-acetic acid (IAA) are tightly regulated (reviewed in Woodward and Bartel, 2005; Normanly, 2010), and the contributions of transport and de novo IAA synthesis to auxin homeostasis have been extensively studied. However, the importance of indole-3-butyric acid (IBA) transport and IBA-derived IAA in regulating auxin homeostasis has only recently been revealed.
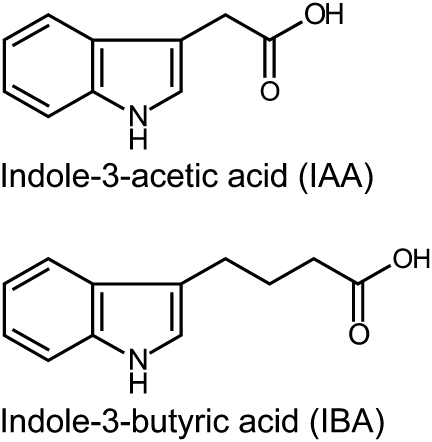
The auxin precursor indole-3-butyric acid (IBA) was originally discovered as a synthetic compound that induced root initiation in a variety of plants (Zimmerman and Wilcoxon, 1935). IBA and IAA are nearly identical, but IBA carries a four-carbon side chain whereas IAA carries a two-carbon side chain (Figure 1). IBA was found as an endogenous constituent of potato tubers by paper chromatography (Blommaert, 1954), and subsequently has been identified by gas chromatography–mass spectrometry in a variety of plants, including pea, cypress, maize, carrot, tobacco, and Arabidopsis (reviewed in Ludwig-Müller, 2000). The occurrence of IBA in phylogenetically diverse Angiosperms suggests a conserved role for IBA in auxin homeostasis.
Figure 1.
IAA and IBA.
Indole-3-acetic acid (IAA) is a naturally occurring auxin. Indole-3-butyric acid (IBA) is a naturally occurring IAA precursor.
Early bioassays demonstrated that IBA application affects not only rooting, but also other auxin-regulated processes such as leaf epinasty, cell division, and stem bending (Zimmerman and Wilcoxon, 1935). These auxin responses, which were often distant from the site of application, hinted at the existence of an IBA transport system. This review explores our current understanding of the roles of IBA transport and IBA-derived IAA in plant development.
IBA TRANSPORT
Long-Distance Transport of IBA
The active auxin IAA moves in several distinct transport streams within the plant (see review by Peer in this issue). In stems, such as the hypocotyl and inflorescence stem, IAA moves basipetally from the apex towards the root. In the root, IAA moves not only acropetally from the root–shoot junction towards the root tip in the stele, but also basipetally from the root tip back towards the shoot in the epidermis.
Early reports suggested that IBA also moves long distances within the plant. However, these early studies relied on bioassays to monitor IBA movement, such as the ability of IBA to promote root formation distant from the application site (Went and White, 1938; Leopold and Lam, 1961; Yang and Davies, 1999). More recent studies using radiolabeled IBA demonstrated both acropetal and basipetal radiolabel movement in Arabidopsis stem cuttings (Ludwig-Müller et al., 1995). Interpreting these assays is complicated by the possibility that the transported compound may be IBA or a compound derived from IBA, such as IAA.
More recent studies include demonstrations that most of the IBA remains intact during the timeframe of the transport experiment. For example, in a detailed comparison of [3H]IAA and [3H]IBA transport in Arabidopsis, Rashotte et al. (2003) found that roots transport both IBA and IAA both basipetally and acropetally. Also, similarly to IAA, IBA moves basipetally (towards the root) but not acropetally (towards the apex) in seedling hypocotyls (Rashotte et al., 2003). In inflorescence stems, however, IBA is not transported, whereas IAA moves basipetally (Rashotte et al., 2003). The directional transport of IBA in roots, hypocotyls, and other tissues suggested the existence of active IBA transporters, and the differences between IBA and IAA transport suggested that IBA might use transporters distinct from those used to move IAA.
Many IAA Transporters Do Not Transport IBA
Specialized influx and efflux carriers mediate IAA transport (reviewed in Vieten et al., 2007). Despite the chemical similarity of IAA and IBA (Figure 1), examination of many of these IAA carriers has revealed that they do not transport IBA.
Like IAA uptake, IBA uptake is saturable in Arabidopsis (Ludwig-Müller et al., 1995; Rashotte et al., 2003), suggesting the existence of protein carriers rather than passive diffusion. IAA influx into cells is mediated by the AUXIN RESISTANT1 (AUX1) family of proteins (reviewed in Vieten et al., 2007). The four members of the AUX1 family—AUX1, LIKE AUX1 (LAX1), LAX2, and LAX3—are similar to amino acid permeases and contain 11 transmembrane domains (Figure 2A; Swarup et al., 2004). Because both IAA and the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) are brought into cells by the AUX1 transporter, aux1 loss-of-function mutants are resistant to IAA and 2,4-D (Maher and Martindale, 1980; Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Yang et al., 2006). In contrast, aux1 responds normally to NAA, which is not transported by AUX1 (Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Yang et al., 2006). Although aux1 is moderately resistant to IBA-mediated inhibition of root elongation in long-term (multiple day) assays (Zolman et al., 2000), this resistance is likely due to aux1 resistance to IAA derived from IBA after import by a different carrier, as several lines of evidence suggest that IBA is not an AUX1 substrate. For example, root acropetal and basipetal IBA transport (Rashotte et al., 2003) and IBA accumulation in excised root tips (Strader and Bartel, 2009) are unaltered in aux1 mutants. Moreover, IBA does not competitively inhibit [3H]IAA uptake when AUX1 is heterologously expressed in Xenopus oocytes (Yang et al., 2006). Although it is unlikely that AUX1 is an IBA uptake carrier, other AUX1 family members are potential candidates for this role. For example, when LAX3 is heterologously expressed in Xenopus oocytes, IBA competes with [3H]IAA uptake, although not as well as IAA (Swarup et al., 2008). Whether LAX proteins or some other carriers mediate IBA uptake, the inability of AUX1 to transport IBA indicates that IAA and IBA influx are at least partially distinct.
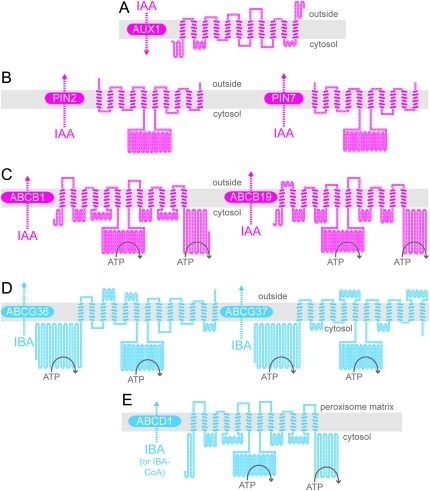
Figure 2.
Predicted Topologies of IAA Carrier Proteins that Do Not Transport IBA (A–C) and Proteins Demonstrated (D) or Suggested (E) to Transport IBA.
Schematic diagrams illustrating the predicted topologies of AUX1 (A), PIN2 and PIN7 (B), ABCB1 and ABCB19 (C), ABCG36 and ABCG37 (D), and ABCD1 (E) based on the outputs of ARAMEMNON (http://aramemnon.botanik.uni-koeln.de; Schwacke et al., 2003) and TOPO2 (www.sacs.ucsf.edu/TOPO2). Each amino acid residue is represented by a circle; filled circles represent residues predicted to span the membrane (gray rectangle). Positions of nucleotide-binding domains in the ABC proteins are schematized by ATP hydrolysis. For AUX1, the ARAMEMNON TmConsens prediction of transmembrane domains predicted 10 transmembrane domains with strong scores and one additional transmembrane domain with a weak score. Because AUX1 was experimentally shown to have 11 transmembrane domains (Swarup et al., 2004), all 11 transmembrane domains are depicted in the diagram. We used the ARAMEMNON TmConsens predictions for PIN2, PIN7, ABCB1, ABCB19, and ABCG37 to create the corresponding models, and the ARAMEMNON TmHMM_v2 prediction (Sonnhammer et al., 1998) to create the ABCG36 diagram. The ARAMEMNON MemSat_v3 (Jones et al., 1994) predicted 13 transmembrane domains for ABCD1. However, because the ATPase domains of ABCD1 are cytosolic (Nyathi et al., 2010), we did not include the first predicted transmembrane domain in the ABCD1 diagram.
As with influx, IBA efflux seems to use carriers distinct from those that efflux IAA. Two types of proteins facilitate IAA efflux: PIN-FORMED (PIN) proteins (Figure 2B) and members of the ABCB class of ATP-Binding-Cassette (ABC) transporters (Figure 2C). The eight members of the PIN family localize to either the plasma membrane or the endoplasmic reticulum membrane (reviewed in Grunewald and Friml, 2010). The plasma-membrane PIN proteins often display specific polar localization to a particular cellular face, suggesting that they determine the direction of IAA flow through cells and tissues (reviewed in Grunewald and Friml, 2010). Members of the ABCB/MULTIDRUG RESISTANCE/P-GLYCOPROTEIN family, such as ABCB1, ABCB4, and ABCB19, also localize to the plasma membrane but are more symmetrically localized than PIN proteins and may facilitate non-polar IAA efflux to contribute to long-distance IAA transport, potentially delivering IAA to the PIN proteins (Bandyopadhyay et al., 2007; Blakeslee et al., 2007; Bailly et al., 2008; Mravec et al., 2008).
Neither the PIN family nor the ABCB family appears to facilitate IBA efflux. The polar auxin transport inhibitors naphthylphthalamic acid (NPA) and 2,3,5-triiodobenzoic acid (TIBA) decrease auxin transport by blocking auxin efflux carrier complexes (Thomson et al., 1973; Cande and Ray, 1976). Including NPA or TIBA in transport assays effectively blocks polar transport of [3H]IAA, but not [3H]IBA, in roots and hypocotyls (Rashotte et al., 2003), suggesting that NPA- and TIBA-sensitive IAA efflux carriers are not required for IBA efflux. Moreover, auxin efflux deficiency causes pin2 roots to bend into agar containing substrates of the PIN2 effluxer, such as IAA and NAA (Utsuno et al., 1998). However, pin2 roots do not bend to enter IBA-containing agar (Poupart and Waddell, 2000; Zolman et al., 2000), suggesting that IBA is not a PIN2 substrate. In addition, root basipetal IBA transport is unaffected in a pin2 mutant (Rashotte et al., 2003) and PIN2, PIN7, ABCB1, and ABCB19 heterologously expressed in mammalian cells do not transport [3H]IBA (Růžička et al., 2010). Because IBA undergoes long-distance movement, and because none of the tested IAA efflux carriers appears to transport IBA, distinct carrier proteins likely facilitate IBA and IAA movement.
ABCG/PDR IBA EFFLUX CARRIERS
ABCG36 and ABCG37 Promote IBA Efflux
Genetic approaches in Arabidopsis have begun to identify molecular components facilitating IBA transport (Table 1). IBA efflux from root cells is promoted by at least two members of the PLEIOTROPIC DRUG RESISTANCE (PDR) subclade of the ABCG family of ABC transporters. Loss of PDR8/ABCG36 function increases sensitivity to IBA, but not IAA (Strader and Bartel, 2009). Similarly, loss of PDR9/ABCG37 function results in IBA hypersensitivity and wild-type IAA sensitivity (Strader et al., 2008; Růžička et al., 2010). Unlike abcg36, however, abcg37 mutants also are hypersensitive to 2,4-D (Ito and Gray, 2006), and several auxin transport inhibitors (Fujita and Syōno, 1997; Ito and Gray, 2006; Růžička et al., 2010). The IBA hypersensitivity phenotypes of mutants defective in these transporters suggest that IBA is a common substrate effluxed by both ABCG36 and ABCG37 (Figure 2D).
Table 1.
Arabidopsis Mutants Implicated in IBA Transport.
| Mutant | Gene product | Mutant phenotype | References |
| pdr8 / pen3 / abcg36 | At1g59870 plasma membrane ABC transporter; IBA effluxer | IBA and 2,4-dichlorophenoxybutyric acid (2,4-DB) hypersensitivity; reduced IBA efflux from root tips; long root hairs | Strader and Bartel, 2009 |
| pdr9 / eta4 / pis1 / abcg37 | At3g53480 plasma membrane ABC transporter; IBA effluxer | IBA, 2,4-D, 2,4-DB, and TIBA hypersensitivity; reduced IBA efflux from root tips | Ito and Gray, 2006; Strader et al., 2008; Růžička et al., 2010 |
| pxa1 / cts1 / acn2 / ped3 / abcd1 | At4g39850 peroxisomal ABC transporter; moves IBA (and other substrates) into peroxisome | IBA resistance; reduced lateral root formation; decreased filament elongation; fatty acid β-oxidation defects | Hayashi et al., 1998; Zolman et al., 2001; Footitt et al., 2002; Hooks et al., 2004; Footitt et al., 2007 |
| rib1 | Unidentified | IBA and 2,4-D resistance; altered IBA transport; gravity response defects | Poupart and Waddell, 2000; Poupart et al., 2005 |
Root tips from abcg36 and abcg37 display wild-type [3H]IAA accumulation but hyperaccumulate [3H]IBA in a simplified auxin transport assay (Strader et al., 2008; Strader and Bartel, 2009), suggesting that ABCG36 and ABCG37 promote IBA efflux from root tips. Indeed, abcg36 root tips loaded with [3H]IBA efflux IBA slowly (Strader and Bartel, 2009), and root tips from the abcg36 abcg37 double mutant accumulate even more [3H]IBA than either parent (Růžička et al., 2010), consistent with partially redundant roles for ABCG36 and ABCG37 in promoting IBA efflux.
When expressed in heterologous systems, ABCG37 directly effluxes IBA. Schizosaccharomyces pombe cells expressing ABCG37 accumulate less [3H]IBA than control cells but accumulate [3H]IAA like control cells (Růžička et al., 2010), consistent with abcg37 mutant phenotypes of hyper-responsiveness to IBA and wild-type IAA responsiveness (Ito and Gray, 2006; Strader et al., 2008; Růžička et al., 2010). Additionally, mammalian cells expressing ABCG37 export both [3H]2,4-D and [3H]IBA (Růžička et al., 2010). Although direct demonstration of IBA efflux by heterologously expressed ABCG36 has not been reported, the IBA-hypersensitivity and reduced [3H]IBA efflux from root tips of the abcg36 mutant are consistent with ABCG36 acting as an IBA efflux carrier. Thus, at least two members of the PDR subclade of the ABCG transporters, ABCG36 and ABCG37, are likely to efflux the auxin precursor IBA from root cells.
ABCG36 and ABCG37 Localize to the Outward Face of Root Epidermal Cells
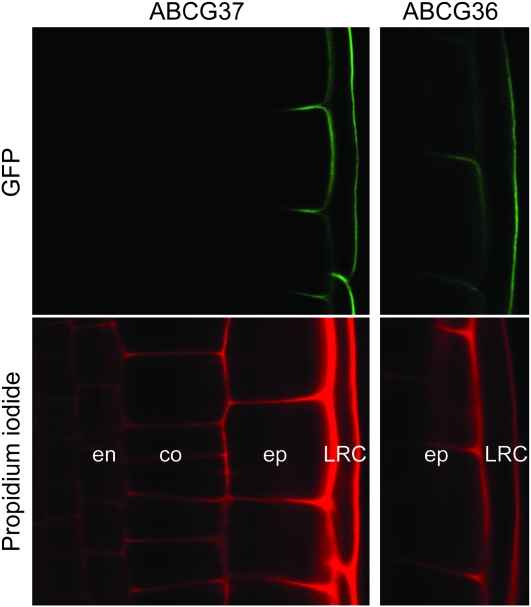
Consistent with increased [3H]IBA retention in root tips from abcg36 and abcg37 mutants (Strader et al., 2008; Strader and Bartel, 2009), PDR8/ABCG36 and PDR9/ABCG37 localize to the outer face of root epidermal cells (Figure 3) (Strader and Bartel, 2009; Łangowski et al., 2010; Růžička et al., 2010). Because these proteins localize to the outward face of root cells regardless of the position or the apical–basal axis of a cell, they define a new polar region of the cell that has been termed the ‘outer polar domain’ (Łangowski et al., 2010). Intriguingly, maintaining these transporters in the outer polar domain does not require the same molecular components (i.e. the actin cytoskeleton, BFA-sensitive endocytosis, the ARF GEF GNOM, AXR4, the protein kinase PINOID, and the protein phosphatase PP2A) that are required to target IAA transporters to the apical and basal domains of the cell (Łangowski et al., 2010), suggesting the possibility of a novel mechanism that targets ABCG36 and ABCG37 to the proper cellular face. The outward-facing localization of ABCG36 and ABCG37 implies that plants efflux IBA into the rhizosphere. This efflux is a facet of IAA homeostasis (discussed below); whether it also aids in plant communication with soil microbes and/or contributes to long-distance IBA transport remains to be determined. In any case, the outward-facing localization of ABCG36 and ABCG37 (Figure 3) is distinct from the apical/basal localization of the plasma membrane members of the PIN family of IAA effluxers, suggesting that additional components facilitating long-distance IBA movement may remain to be discovered.
Figure 3.
ABCG37 and ABCG36 Localize to the Outer Polar Domain of Root Epidermal and Lateral Root Cap Cells.
Confocal images of root tips of 5-day-old seedlings carrying GFP–ABCG37 (pis1-1 carrying 35S:GFP–ABCG37) (Łangowski et al., 2010) and ABCG36–GFP (pen3-1 carrying PEN3:PEN3–GFP) (Stein et al., 2006). The upper panels show GFP signal and the lower panels show propidium iodide staining of endodermal (en), cortex (co), epidermal (ep), and lateral root cap (LRC) cell walls. Both GFP–ABCG37 (Łangowski et al., 2010; Růžička et al., 2010) and ABCG36–GFP (Strader and Bartel, 2009) accumulate on the outward face of epidermal and lateral root cap cells, a polar domain termed the ‘outer polar domain’ (Łangowski et al., 2010), implying that these transporters move IBA from the root into the rhizosphere.
Are Additional IBA Effluxers Found in the PDR Clade within the ABCG Family?
Within the 43-member ABCG subfamily of ABC transporters in Arabidopsis, the 15-member PDR group consists of full-sized transporters with two apparent nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs), each consisting of six membrane-spanning sequences (reviewed in van den Brule and Smart, 2002; Crouzet et al., 2006; Verrier et al., 2008). Interestingly, both the ABCB and ABCG subfamilies have full-sized and half-sized members, but only full-sized members of these subfamilies have demonstrated roles in IAA or IBA transport (Figure 2C and 2D). ABCG36 and ABCG37 are only 53% identical at the amino acid level, and yet each is implicated in IBA efflux. Because ABCG36 and ABCG37 are not closely related within the PDR group, and each promotes IBA efflux, additional PDR family members also may efflux IBA. ABCG36 is expressed in many plant organs and ABCG37 is highly expressed in roots (van den Brule and Smart, 2002); other PDR family members have diverse expression patterns (Crouzet et al., 2006), suggesting that if they function in IBA efflux, their roles may be distinct from PDR8 and PDR9 functions in moving IBA out of the root. It will be interesting to learn whether additional ABCG family members are polarly localized, efflux IBA or related molecules, or function in either long- or short-distance IBA transport.
Additional Substrates Transported by the PDR Clade of ABCG Family Members
Although ABCG36 and ABCG37 are implicated in IBA but not IAA efflux, these transporters do appear to be somewhat promiscuous. In addition to IBA hypersensitivity (Strader et al., 2008; Růžička et al., 2010), abcg37 loss-of-function alleles are hypersensitive to 2,4-D, other members of the phenoxyalkanoic acid family of herbicides (Ito and Gray, 2006), and auxin transport inhibitors (Fujita and Syōno, 1997; Ito and Gray, 2006; Růžička et al., 2010). Root tips from the abcg37 mutant hyperaccumulate not only [3H]IBA (Strader et al., 2008), but also [14C]2,4-D (Ito and Gray, 2006) and [3H]NPA (Ito and Gray, 2006) in simplified auxin transport assays, suggesting that these compounds—a naturally occurring auxin precursor, a synthetic auxin, and an auxin transport inhibitor, respectively—all may be ABCG37 substrates. Indeed, ABCG37 has been directly demonstrated to transport IBA and 2,4-D (Růžička et al., 2010). ABCG36, on the other hand, has been implicated in IBA efflux (Strader and Bartel, 2009), cadmium efflux (Kim et al., 2007), and pathogen response (Kobae et al., 2006; Stein et al., 2006). Additionally, abcg36 mutant shoots display altered callose (Clay et al., 2009), camalexin (Bednarek et al., 2009), and glucosinolate (Bednarek et al., 2009; Clay et al., 2009) accumulation in response to pathogens and their elicitors. Thus, both ABCG36 and ABCG37 are likely to efflux a range of substrates, a trait shared by many members of the PDR family (reviewed in Crouzet et al., 2006).
Beyond the roles in IBA transport demonstrated for ABCG36 and ABCG37, other members of the PDR clade of ABCG family members are implicated in the transport of terpenoids, including the phytohormone abscisic acid (ABA). The first plant PDR family members to be characterized were SpTUR2 from Spirodela polyrrhiza (Smart and Fleming, 1996) and NpPDR1/NpABC1 from Nicotiana plumbaginifolia (Jasinski et al., 2001). SpTUR2 expression is up-regulated by ABA application (Smart and Fleming, 1996) and ectopic SpTUR2 expression in Arabidopsis confers resistance to sclareol, a diterpenoid antifungal agent (van den Brule et al., 2002). NpABC1 protein levels are increased by sclareolide and sclareol, and NpABC1 may promote secretion of these compounds to function in plant defense (Jasinski et al., 2001). AtPDR12/ABCG40, the closest Arabidopsis homolog of SpTUR2 and NpABC1, also is implicated in sclareol extrusion; abcg40 mutants are sclareol-hypersensitive (Campbell et al., 2003). More recently, PDR12/ABCG40 has been identified as a plasma membrane ABA influx carrier; heterologous expression of ABCG40 in yeast and BY2 cells promotes ABA uptake (Kang et al., 2010). abcg40 mutant plants display delayed ABA-responsive gene expression and impaired stress tolerance (Kang et al., 2010). In addition to its roles in transport of the terpenoids sclareol and ABA, ABCG40 is implicated in lead resistance (Lee et al., 2005). Future research will be necessary to disentangle the proposed roles of ABCG40 as both an effluxer of sclareol and lead and an influxer of ABA. The variety of phenotypes displayed by pdr/abcg mutants suggests that members of this family may act as carriers for a range of substrates, including IBA. It remains to be determined whether the apparent promiscuity of these ABCG transporters is regulated in different tissues, during development, or in response to specific environmental conditions.
IBA-TO-IAA METABOLISM
Identification of Candidate IBA β-oxidation enzymes
Numerous plants shorten the auxin precursor IBA into active IAA (Fawcett et al., 1960; reviewed in Epstein and Ludwig-Müller, 1993) and, at least in Arabidopsis, this metabolism is peroxisome-dependent (Strader et al., 2010). Genetic evidence suggests that auxin activity of IBA in Arabidopsis is completely dependent on its conversion to IAA through a multi-step process similar to fatty acid β-oxidation (Zolman et al., 2000). Indeed, some peroxisomal enzymes, such as the 3-ketoacyl–CoA thiolase encoded by PED1, may act not only in fatty acid β-oxidation (Hayashi et al., 1998), but also in IBA β-oxidation (Zolman et al., 2000). In contrast, several peroxisomal enzymes appear to be dedicated to IBA β-oxidation; mutations in genes encoding these enzymes confer IBA resistance without altering IAA response or conferring dependence on exogenous fixed carbon sources to fuel growth after germination (Table 2). Candidates for dedicated IBA β-oxidation include the predicted short-chain dehydrogenase/reductase INDOLE-3-BUTYRIC ACID RESPONSE1 (IBR1) (Zolman et al., 2008), the acyl–CoA dehydrogenase/oxidase-like IBR3 (Zolman et al., 2007), the predicted enoyl–CoA hydratase IBR10 (Zolman et al., 2008), and ENOYL–COA HYDRATASE2 (ECH2) (Strader et al., 2011). The ibr1 ibr3 ibr10 triple mutant displays additive IBA resistance (Zolman et al., 2008) and converts IBA to IAA inefficiently (Strader et al., 2010). Examination of ibr1, ibr3, ibr10, ech2, and higher-order combinations of these mutants has illuminated diverse roles for IBA-derived IAA in seedling development.
Table 2.
Arabidopsis Mutants Implicated in IBA Metabolism.
| Mutant | Gene product | Mutant phenotype | References |
| ech2 | At1g76150 enoyl–CoA hydratase | IBA resistance | Strader et al., 2011 |
| ibr1 | At4g05530 putative short-chain dehydrogenase/ reductase | IBA resistance | Zolman et al., 2000, 2008 |
| ibr3 | At3g06810 putative acyl–CoA dehydrogenase | IBA resistance | Zolman et al., 2000, 2007 |
| ibr10 | At4g14430 putative enoyl–CoA hydratase | IBA resistance | Zolman et al., 2000, 2008 |
| ped1 | At2g33150 3-ketoacyl–CoA thiolase | IBA resistance; fatty acid β-oxidation defects | Hayashi et al., 1998; Zolman et al., 2000 |
| UGT74E2OE | At1g05680 IBA glucosyltransferase | Overexpression leads to increased IBA glucosylation; increased shoot branching; abiotic stress resistance | Tognetti et al., 2010 |
Roles for IBA-Derived IAA in Plant Development
Consistent with a role for IBA efflux, mutants defective in ABCG36 and ABCG37 display developmental phenotypes suggestive of high auxin levels in certain cell types. Root hairs, the tubular outgrowths from specific root cell files, provide a single-cell expansion model that is highly sensitive to auxin response and levels (reviewed in Grierson and Schiefelbein, 2002). abcg36 and abcg37 loss-of-function mutants have lengthened root hairs (Strader and Bartel, 2009; Růžička et al., 2010), suggesting that root hair IAA levels increase when IBA efflux decreases. abcg36 seedlings also display enlarged cotyledons several days after germination (Strader and Bartel, 2009), a second phenotype suggesting increased cell expansion, because post-germinative Arabidopsis cotyledons grow by cell expansion without division (Mansfield and Briarty, 1996). The abcg36 root hair and cotyledon developmental phenotypes both are suppressed when combined with the ibr1, ibr3, and ibr10 mutations (Strader et al., 2010), suggesting that these abcg36 hyper-expansion phenotypes result from increased IBA-derived IAA in these cell types. In contrast to the expanded root hairs and cotyledons of IBA efflux mutants (Strader and Bartel, 2009; Růžička et al., 2010), mutants defective in IBA β-oxidation display reduced root hair and cotyledon cell expansion (Strader et al., 2010, 2011), confirming that IBA-derived IAA, rather than IBA itself, drives cell expansion in these cell types.
Developmental roles for IBA-derived IAA are not limited to cell expansion. ech2 ibr1 ibr3 ibr10 quadruple mutant seedlings display decreased free IAA levels and wide-ranging auxin-related developmental defects (Strader et al., 2011). In addition to defects in cotyledon and root hair cell expansion, ech2 ibr1 ibr3 ibr10 displays delayed development, reduced apical hook curvature, reduced high-temperature-induced hypocotyl elongation, decreased lateral root production, and smaller root meristems, defining multiple seedling developmental processes that depend on IBA-derived IAA. The high-auxin phenotypes displayed by mutants blocked in IBA efflux combined with the low-auxin phenotypes of mutants blocked in IBA-to-IAA conversion suggest that IBA-derived IAA is a significant auxin source during seedling development. In addition, increasing IBA glucosylation by overexpressing the UDP-glucosyltransferase UGT74E2 results in increased shoot branching and improved drought and salt tolerance (Tognetti et al., 2010), implying that roles for IBA are not confined to the seedling stage.
PXA1/ABCD1 MAY TRANSPORT IBA INTO THE PEROXISOME TO BE METABOLIZED
Because IBA-to-IAA conversion is peroxisomal, a transporter is required to move IBA into the peroxisome. This transporter is likely the peroxisomal ABC transporter PXA1/ABCD1 (Figure 2E), because abcd1 loss-of-function mutants are IBA-resistant (Zolman et al., 2001) and do not efficiently convert IBA to IAA (Strader et al., 2010). In addition to functioning in IBA import into the peroxisome, ABCD1 also likely transports fatty acids (reviewed in Linka and Weber, 2010). As a result, abcd1 mutants β-oxidize seed storage lipids slowly and require exogenous fixed carbon to fuel seedling growth prior to establishment of photosynthesis (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002). Additionally, ABCD1 is thought to transport jasmonic acid (JA) precursors into the peroxisome; JA levels are reduced in abcd1 mutants (Theodoulou et al., 2005). Although ABCD1 is necessary for IBA response, direct transport assays, perhaps using a recently established yeast system for expressing ABCD1 (Nyathi et al., 2010), will be necessary to demonstrate which abcd1 phenotypes reflect loss of transport of ABCD1 substrates and which, if any, result from indirect effects. In any case, the subset of abcd1 defects that are rescued by auxin application, such as reduced lateral root formation (Zolman et al., 2001) and delayed stamen filament elongation (Footitt et al., 2007), suggest that the reduced IBA-to-IAA conversion observed when this transporter is impaired (Strader et al., 2010) has developmental consequences for the plant.
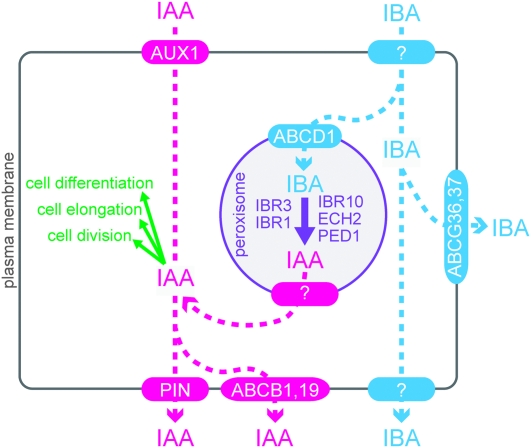
Because peroxisomal enzymes are required for IBA-to-IAA conversion, it follows that an unidentified IAA transporter is necessary to release IBA-derived IAA out of the peroxisome (Figure 4). Intriguingly, several newly characterized members of the PIN family target to internal membranes, including the endoplasmic reticulum (Mravec et al., 2009; Ganguly et al., 2010). Although no PIN proteins have been demonstrated to be peroxisomal, it will be interesting to learn whether the peroxisomal IAA effluxer is a member of this or some other protein family.
Figure 4.
Cellular Model Showing Distinct IAA and IBA Transporters.
The auxin activity of IBA requires its conversion to IAA in peroxisomes, probably by β-oxidation enzymes encoded by IBR1, IBR3, IBR10, ECH2, and PED1.
PERSPECTIVES: MISSING IBA TRANSPORTERS
The available evidence suggests that plants use distinct carriers to move IBA and IAA. AUX1 acts as an influx carrier for IAA, but not IBA. Similarly, PIN2, PIN7, ABCB1, and ABCB19 act as efflux carriers for IAA, but not IBA. Conversely, the PDR family proteins ABCG36 and ABCG37 appear to efflux IBA, but not IAA. These independent transport systems may provide a mechanism to specifically move an inactive precursor, thus avoiding auxin responses during transport (Figure 4). Once in a target cell, IBA β-oxidization to active IAA would allow for auxin responses.
Despite the recent progress, much remains unknown about IBA transport and metabolism. For example, how does IBA enter cells? IBA influx appears to be carrier-assisted. A possible carrier may be the unidentified RIB1, as rib1 mutants are IBA-resistant (Poupart and Waddell, 2000) and display altered IBA transport and normal IAA transport (Poupart et al., 2005). Once in the cell, IBA can be effluxed, conjugated for storage, or β-oxidized to IAA to provide active auxin activity. Peroxisomally localized candidate enzymes have been identified for β-oxidation of IBA–CoA to IAA–CoA (Table 2), but none of these enzymes has been biochemically characterized, and candidates have not emerged for the IBA–CoA ligase or IAA–CoA hydrolysis steps. Also, are there additional IBA efflux carriers? Because ABCG36 and ABCG37 localize to the outer polar domain and appear to function in auxin homeostasis to limit IAA levels, additional IBA efflux carriers may facilitate long-distance IBA transport. It remains to be determined whether these unidentified IBA effluxers will be other members of the ABCG family or members of the PIN or ABCB families that have not yet been tested with IBA. Finally, how is IBA efflux regulated? The intricate regulation of IAA efflux carrier levels and positioning is essential for normal development; it remains to be seen whether disturbing the polarity of IBA efflux carriers, such as the unusual outer polar domain of ABCG36 and ABCG37 in root epidermal cells, has developmental consequences. It seems certain that increasing our understanding of IBA transport and metabolism will continue to yield new insights into the complex mechanisms used by plants to control auxin homeostasis.
FUNDING
The authors’ research is supported by the National Institutes of Health (1K99-GM089987 to L.C.S.; R01GM079177 to B.B.), the National Science Foundation (MCB-0745122 to B.B.), and the Robert A. Welch Foundation (C-1309 to B.B.). Confocal microscopy was performed on equipment obtained through a Shared Instrumentation Grant from the National Institutes of Health (S10RR026399).
Acknowledgments
We are grateful to Jiří Friml for pis1-1 carrying 35S:GFP-ABCG37, Shauna Somerville for pen3-1 carrying PEN3:PEN3-GFP (ABCG36-GFP), and to Sarah Christensen, Lisa Farmer, Wendell Fleming, and Jerrad Stoddard for critical comments on the manuscript. No conflict of interest declared.
References
- Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M. Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 2008;283:21817–21826. doi: 10.1074/jbc.M709655200. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, et al. Interactions of PIN and PGP auxin transport mechanisms. Biochem. Soc. Trans. 2007;35:137–141. doi: 10.1042/BST0350137. [DOI] [PubMed] [Google Scholar]
- Bednarek P, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19:131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaert K. Growth- and inhibiting-substances in relation to the rest period of the potato tuber. Nature. 1954;174:970–972. [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx IA, Anderson JP, Maclean DJ, Cammue BP, Ebert PR, Manners JM. Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 2003;133:1272–1284. doi: 10.1104/pp.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande WZ, Ray PM. Nature of cell-to-cell transfer of auxin in polar transport. Planta. 1976;129:43–52. doi: 10.1007/BF00390912. [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J, Trombik T, Fraysse AS, Boutry M. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 2006;580:1123–1130. doi: 10.1016/j.febslet.2005.12.043. [DOI] [PubMed] [Google Scholar]
- Epstein E, Ludwig-Müller J. Indole-3-butyric acid in plants: occurrence, synthesis, metabolism, and transport. Physiol. Plant. 1993;88:382–389. [Google Scholar]
- Fawcett CH, Wain RL, Wightman F. The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proc. Royal Soc. London, Series B. 1960;152:231–254. doi: 10.1098/rspb.1960.0035. [DOI] [PubMed] [Google Scholar]
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL. The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 2007;144:1467–1480. doi: 10.1104/pp.107.099903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Syōno K. PIS1, a negative regulator of the action of auxin transport inhibitors in Arabidopsis thaliana. Plant J. 1997;12:583–595. doi: 10.1046/j.1365-313x.1997.00583.x. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho HT. Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 2010;153:1046–1061. doi: 10.1104/pp.110.156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Schiefelbein J. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. Root Hairs: April 4, 2002. doi: 10.1199/tab.0060:), www.aspb.org/publications/Arabidopsis/ [Google Scholar]
- Grunewald W, Friml J. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 2010;29:2700–2714. doi: 10.1038/emboj.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiol. 2002;43:1–11. doi: 10.1093/pcp/pcf023. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks MA, Turner JE, Murphy EC, Graham IA. Acetate non-utilizing mutants of Arabidopsis: evidence that organic acids influence carbohydrate perception in germinating seedlings. Mol. Genet. Genomics. 2004;271:249–256. doi: 10.1007/s00438-004-0985-9. [DOI] [PubMed] [Google Scholar]
- Ito H, Gray WM. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 2006;142:63–74. doi: 10.1104/pp.106.084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell. 2001;13:1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl Acad. Sci. U S A. 2010;107:2355–2360. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50:207–218. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- Kobae Y, Sekino T, Yoshioka H, Nakagawa T, Martinoia E, Maeshima M. Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol. 2006;47:309–318. doi: 10.1093/pcp/pcj001. [DOI] [PubMed] [Google Scholar]
- Łangowski L, Růžička K, Naramoto S, Kleine-Vehn J, Friml J. Trafficking to the outer polar domain defines the root–soil interface. Curr. Biol. 2010;20:904–908. doi: 10.1016/j.cub.2010.03.059. [DOI] [PubMed] [Google Scholar]
- Lee M, Lee K, Lee J, Noh EW, Lee Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 2005;138:827–836. doi: 10.1104/pp.104.058107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold AC, Lam SL. Polar transport of three auxins. In: Klein RM, editor. 4th International Conference on Plant Growth Regulators. 1961. pp. 411–418. Iowa. [Google Scholar]
- Linka N, Weber AP. Intracellular metabolite transporters in plants. Mol Plant. 2010;3:21–53. doi: 10.1093/mp/ssp108. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 2000;32:219–230. [Google Scholar]
- Ludwig-Müller J, Raisig A, Hilgenberg W. Uptake and transport of indole-3-butyric acid in Arabidopsis thaliana: comparison with other natural and synthetic auxins. J. Plant Physiol. 1995;147:351–354. [Google Scholar]
- Maher EP, Martindale SJB. Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem. Genet. 1980;18:1041–1053. doi: 10.1007/BF00484337. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int. J. Plant Sci. 1996;157:280–295. [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development. 2008;135:3345–3354. doi: 10.1242/dev.021071. [DOI] [PubMed] [Google Scholar]
- Normanly J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. 2010 doi: 10.1101/cshperspect.a001594. Perspect Biol. 2, a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y, De Marcos Lousa C, van Roermund CW, Wanders RJ, Johnson B, Baldwin SA, Theodoulou FL, Baker A. The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2Δ mutant for metabolism of long-chain fatty acids and exhibits fatty acyl–CoA-stimulated ATPase activity. J. Biol. Chem. 2010;285:29892–29902. doi: 10.1074/jbc.M110.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a001446. a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupart J, Waddell CS. The rib1 mutant is resistant to indole-3-butyric acid, an endogenous auxin in Arabidopsis. Plant Physiol. 2000;124:1739–1751. doi: 10.1104/pp.124.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupart J, Rashotte AM, Muday GK, Waddell CS. The rib1 mutant of Arabidopsis has alterations in indole-3-butyric acid transport, hypocotyl elongation, and root architecture. Plant Physiol. 2005;139:1460–1471. doi: 10.1104/pp.105.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Poupart J, Waddell CS, Muday GK. Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 2003;133:761–772. doi: 10.1104/pp.103.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, et al. Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc. Natl Acad. Sci. U S A. 2010;107:10749–10753. doi: 10.1073/pnas.1005878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CC, Fleming AJ. Hormonal and environmental regulation of a plant PDR5-like ABC transporter. J. Biol. Chem. 1996;271:19351–19357. doi: 10.1074/jbc.271.32.19351. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sanchez-Rodriguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell. 2009;21:1992–2007. doi: 10.1105/tpc.109.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Hendrickson Culler A, Cohen JD, Bartel B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010;153:1577–1586. doi: 10.1104/pp.110.157461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Monroe-Augustus M, Rogers KC, Lin GL, Bartel B. Arabidopsis iba response5 (ibr5) suppressors separate responses to various hormones. Genetics. 2008;180:2019–2031. doi: 10.1534/genetics.108.091512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. 2011 doi: 10.1105/tpc.111.083071. Plant Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Swarup R, et al. Structure–function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell. 2004;16:3069–3083. doi: 10.1105/tpc.104.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants: implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005;137:835–840. doi: 10.1104/pp.105.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson K-S, Hertel R, Müller S. 1-N-naphthylphthalamic acid and 2,3,4-triiodobenzoic acid. Planta. 1973;109:337–352. doi: 10.1007/BF00387102. [DOI] [PubMed] [Google Scholar]
- Tognetti VB, et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22:2660–2679. doi: 10.1105/tpc.109.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuno K, Shikanai T, Yamada Y, Hashimoto T. AGR, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 1998;39:1111–1118. doi: 10.1093/oxfordjournals.pcp.a029310. [DOI] [PubMed] [Google Scholar]
- van den Brule S, Smart CC. The plant PDR family of ABC transporters. Planta. 2002;216:95–106. doi: 10.1007/s00425-002-0889-z. [DOI] [PubMed] [Google Scholar]
- van den Brule S, Muller A, Fleming AJ, Smart CC. The ABC transporter SpTUR2 confers resistance to the antifungal diterpene sclareol. Plant J. 2002;30:649–662. doi: 10.1046/j.1365-313x.2002.01321.x. [DOI] [PubMed] [Google Scholar]
- Verrier PJ, et al. Plant ABC proteins: a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Went F, White R. Experiments of the transport of auxin. Bot. Gaz. 1938;100:465–484. [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yamamoto KT. Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol. 1998;39:660–664. doi: 10.1093/oxfordjournals.pcp.a029419. [DOI] [PubMed] [Google Scholar]
- Yang T, Davies PJ. Promotion of stem elongation by indole-3-butyric acid in intact plants of Pisum sativum L. Plant Growth Regul. 1999;27:157–160. [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Zimmerman PW, Wilcoxon F. Several chemical growth substances which cause initiation of roots and other responses in plants. Contrib. Boyce Thompson Inst. 1935;7:209–229. [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics. 2008;180:237–251. doi: 10.1534/genetics.108.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B. IBR3, a novel peroxisomal acyl–CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 2007;64:59–72. doi: 10.1007/s11103-007-9134-2. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]