Summary
Many bacteria use extracellular signals to coordinate group behaviors, a process referred to as quorum sensing (QS). The bacterium Pseudomonas aeruginosa utilizes a complex QS system to control expression of over 300 genes, including many involved in host colonization and disease. The Pseudomonas Quinolone Signal (PQS) is a component of P. aeruginosa QS, and although it contributes to virulence in some models of infection, the PQS biosynthetic pathway is not fully elucidated. Here, we show that PqsH catalyzes the terminal step in PQS production, synthesizing PQS in vitro using the substrates 2-heptyl-4-quinolone (HHQ), NADH, and oxygen. Structure function studies reveal that the alkyl side chain of HHQ is critical for PqsH activity with the highest activity observed for alkyl chain lengths of 7 and 9 carbons. Due to the PqsH requirement for oxygen, PQS and PQS-controlled virulence factors are not produced by anaerobic P. aeruginosa. Interestingly, anaerobic P. aeruginosa produced PQS in the absence of de novo protein synthesis upon introduction of oxygen, indicating that oxygen is the sole limiting substrate during anaerobic growth. We propose a model in which PqsH poises anaerobic P. aeruginosa to activate PQS-controlled factors immediately upon exposure to molecular oxygen.
Keywords: PqsH, PQS, Pseudomonas, oxygen, quorum sensing
Bacteria are social organisms that display unique behaviors based upon population size. These diverse behaviors include formation of sessile antibiotic-resistant biofilm communities, production of extracellular products involved in disease pathogenesis, biosynthesis and secretion of antimicrobial secondary metabolites, as well as many others (Atkinson & Williams, 2009, Fuqua et al., 2001, Parsek & Greenberg, 2000, Parsek & Greenberg, 2005). These behaviors are often controlled by dedicated signaling pathways that allow bacteria to communicate and coordinate their activities, a process referred to as quorum sensing (QS). A canonical QS system requires the synthesis and secretion of a small signal (< 1 kDa) that increases in concentration as the bacterial population increases. The signal binds to a cognate transcriptional regulator resulting in differential gene expression and consequent behavioral changes (Atkinson & Williams, 2009, Fuqua et al., 2001, Parsek & Greenberg, 2000, Parsek & Greenberg, 2005). While significant advances in understanding the biochemistry of signal production and response have occurred over the last 25 years, recent evidence indicates that environmental cues affect QS behaviors (Farrow & Pesci, 2007, Palmer et al., 2007a, Palmer et al., 2005, Schaefer et al., 2008), suggesting that bacteria do not communicate equivalently in all environments.
The Gram-negative opportunistic pathogen Pseudomonas aeruginosa uses an intricate QS system to control expression of approximately 300 genes (Schuster et al., 2003, Wagner et al., 2003, Whiteley et al., 1999). P. aeruginosa causes a range of infections in immuno-compromised hosts including those with the heritable disease cystic fibrosis (CF). QS is required for P. aeruginosa pathogenesis in many animal and insect models of infection (Pearson et al., 2000, Rumbaugh et al., 1999, Smith et al., 2002, Wu et al., 2001, Jander et al., 2000, Gallagher & Manoil, 2001), and significant effort has been devoted to the discovery of therapeutics that disrupt QS signaling (Njoroge & Sperandio, 2009). P. aeruginosa QS involves at least four signals interwoven into a complex hierarchy (Williams & Camara, 2009). These signals include two classical acyl-homoserine lactone signals as well as the quinolone signaling molecules 2-heptyl-3-hydroxy-4-quinolone (PQS) and 2-heptyl-4-quinolone (HHQ) (Williams, 2007, Williams & Camara, 2009). Each signal interacts with a transcriptional regulator that, when bound to its cognate signal, mediates changes in gene expression. Interestingly, HHQ and PQS both bind the transcriptional regulator MvfR (PqsR), although PQS is approximately 100-fold more potent at stimulating MvfR activity (Xiao et al., 2006).
Aside from their role in cell-cell communication, some QS signals possess additional functions that affect prokaryotic biology (Schertzer et al., 2009). PQS exemplifies this multi-functionality. In addition to serving as a potent signal, PQS chelates iron (Bredenbruch et al., 2006, Diggle et al., 2007) and promotes the formation of membrane vesicles (MVs) that bleb from the P. aeruginosa outer surface (Mashburn & Whiteley, 2005, Mashburn-Warren et al., 2009, Tashiro et al., 2009). Once thought merely to be by-products of cell division, it is now appreciated that MVs traffic molecular cargo between individual bacteria and also from bacteria to eukaryotic cells (Bomberger et al., 2009, Kadurugamuwa & Beveridge, 1999, Kesty et al., 2004). While the cargo transferred has not been fully characterized, MVs are important for P. aeruginosa killing of prokaryotic and eukaryotic cells (Bomberger et al., 2009, Mashburn & Whiteley, 2005).
Despite the central role PQS plays in both expression and trafficking of virulence factors, the details of its biosynthesis have not been fully elucidated. The pathway is predicted to involve condensation of activated anthranilate (anthraniloyl-CoA) with 2-oxo-decanoyl-ACP (from fatty acid biosynthesis) to form HHQ, followed by HHQ hydroxylation to yield PQS (Bredenbruch et al., 2005, Gallagher et al., 2002). Genetic evidence suggests that pqsABCD encodes proteins critical for HHQ formation (Bredenbruch et al., 2005, Calfee et al., 2001, Gallagher et al., 2002), and pqsH encodes the terminal monooxygenase required for HHQ conversion to PQS (Deziel et al., 2004, Gallagher et al., 2002). Of these enzymes, only PqsA has been shown biochemically to be involved in PQS biosynthesis, catalyzing the activation of anthranilate to anthraniloyl-CoA (Coleman et al., 2008). PqsD has been purified and shown to condense anthranilyl-CoA and malonyl-CoA to produce 2,4-dihydroxyquinoline, although this intermediate is not expected to be involved in PQS biosynthesis (Zhang et al., 2008). In this study, we provide biochemical evidence that PqsH utilizes the substrates HHQ, NADH, and molecular oxygen to catalyze the terminal step in PQS biosynthesis, and propose that this enzyme is a biochemical regulator of PQS-controlled social behaviors in P. aeruginosa.
Results
Purification of P. aeruginosa PqsH
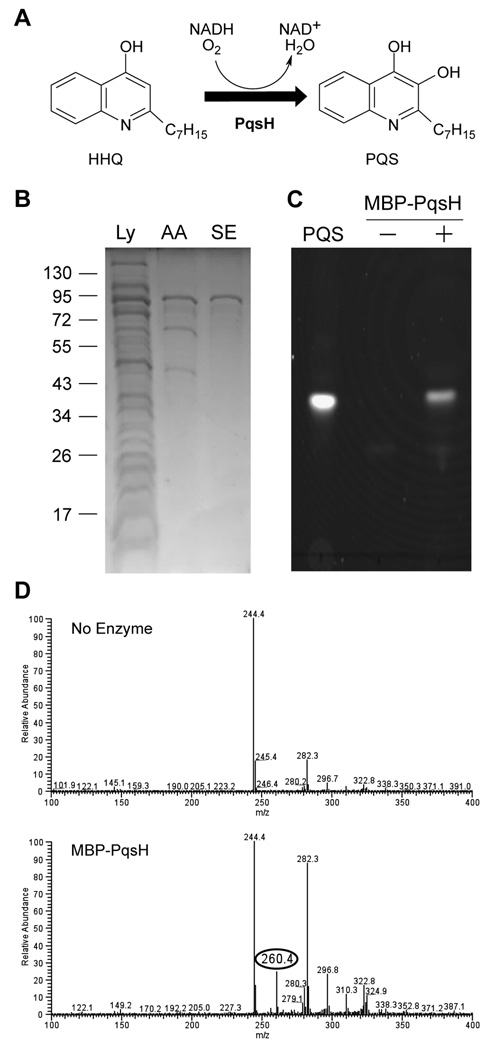
Despite its importance in P. aeruginosa biology, many of the biochemical details of PQS biosynthesis remain unknown. The final step in PQS biosynthesis, hydroxylation of HHQ to PQS (Fig. 1A), is particularly intriguing as this biochemical activity results in a more potent quinolone signal that is thus far unique to P. aeruginosa. Genetic and in silico analyses predict that pqsH (PA2587 on the P. aeruginosa PAO1 chromosome) encodes the monooxygenase responsible for terminal hydroxylation of HHQ (Deziel et al., 2004, Gallagher et al., 2002). To test this hypothesis, we purified and biochemically-characterized PqsH. Initial attempts to purify PqsH produced a poorly soluble enzyme that strongly associated with membrane fractions despite the lack of predicted transmembrane domains. PqsH solubility was significantly improved by fusing the maltose binding protein (MBP) to the N-terminus of PqsH. Soluble lysates from MBP-PqsH overproducing E. coli displayed a prominent band at ~86 kDa on SDS-polyacrylamide gels, the expected size for MBP-PqsH (Fig. 1B). Amylose affinity and size exclusion chromatographies yielded a purified fusion protein (Fig. 1B) that could be stored at −80°C for one week without loss of enzymatic activity. Removal of the MBP-tag from MBP-PqsH did not result in increased enzymatic activity (data not shown); therefore all experiments were performed using MBP-PqsH.
Fig. 1. Purification and activity of MBP-PqsH.
A. The reaction catalyzed by PqsH. B. SDS-polyacrylamide gel showing stages of MBP-PqsH purification. MBP-PqsH was purified through a combination of amylose-affinity and size exclusion chromatography. The lanes contained: Soluble cell lysate (Ly); eluate from amylose affinity column (AA); eluate from size exclusion column (SE). The position of molecular weight standards (kDa) are shown to the left of the gel. C. Straight-phase TLC plate excited with long-wave UV light showing the production of PQS by purified MBP-PqsH. The lanes contain: 250 ng synthetic PQS standard (PQS); products of the reaction mixture lacking purified enzyme (MBP-PqsH −); products of the reaction mixture including purified enzyme (MBP-PqsH +). D. Mass spectra showing the production of PQS by MBP-PqsH. Products of reactions in the absence or presence of enzyme were subjected to positive electrospray ionization mass spectrometry. The highlighted species (circled) corresponds to the [M + H]+ ion of PQS. The peak at 244.4 corresponds to HHQ, and the y-axis designates relative abundance compared to HHQ.
PqsH is an NADH-dependent flavin monooxygenase that oxidizes HHQ to PQS
PqsH is a member of a family of flavin-dependent monooxygenases that utilize NAD(P)H and oxygen to catalyze hydroxylation of aromatic substrates (Massey, 1995). As anticipated, when incubated with NADH and HHQ under aerobic conditions, purified MBP-PqsH consumed HHQ and generated a product that co-migrated with synthetic PQS when analyzed by thin layer chromatography (Figure 1C). Further evidence confirmed this molecule was indeed PQS: the absorbance and fluorescence spectra were identical to that of synthetic PQS (data not shown); and positive electrospray ionization mass spectrometry revealed a reaction product with a mass of 260.4, which corresponds with the [M+H]+ ion of PQS (Fig. 1D).
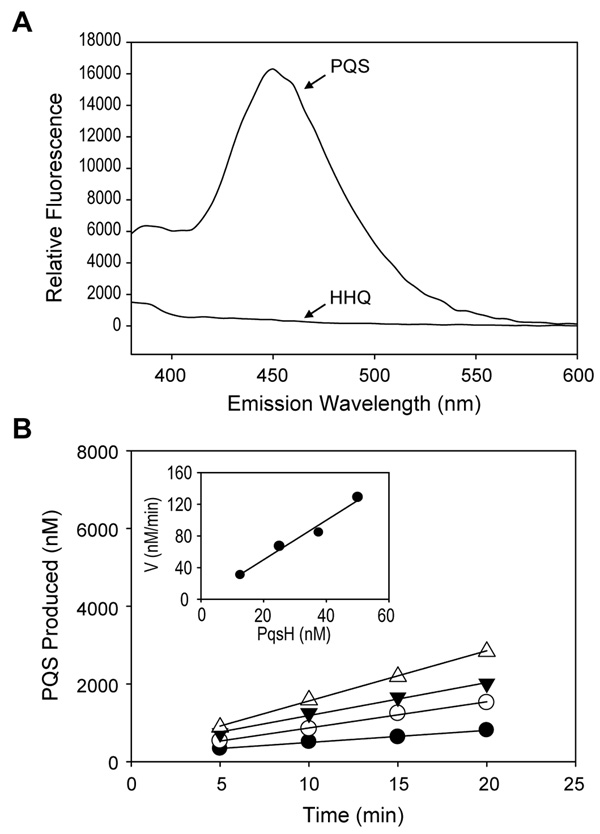
For kinetic characterization of MBP-PqsH, PQS production was monitored using a novel fluorescence assay. In this assay, in vitro reactions were extracted with acidified ethyl acetate, dried down using a continuous stream of N2, resuspended in methanol, and assayed for fluorescence (λexcitation = 342 nm , λemission = 450 nm). While PQS and HHQ are both extracted with ethyl acetate, only PQS displays strong fluorescence under these conditions (Fig. 2A), thus allowing PQS to be quantified without further purification. Kinetic characterization of MBP-PqsH using this assay revealed that formation of PQS was linearly dependent on both time and enzyme concentration (Figure 2B). Estimating enzyme turnover by examining the dependence of the reaction velocity on the enzyme concentration revealed that MBP-PqsH produced PQS with a turnover rate of 2.5 min−1 in vitro (Fig. 2B inset).
Fig. 2. Development of the PqsH biochemical assay.
A. Fluorescence emission spectra of PQS and HHQ after excitation at 340 nm. Emission at 450 nm was used to quantify PQS after ethyl acetate extraction of enzyme reactions. B. PQS production is linearly dependent to both time and enzyme concentration. MBP-PqsH (● 12.5 nM, ○ 25 nM, ▼ 37.5 nM, △ 50 nM) was mixed with all substrates and production of PQS was monitored over time. The inset shows reaction velocity vs. enzyme concentration, and the slope of this line reveals an enzyme turnover of 2.5 min−1.
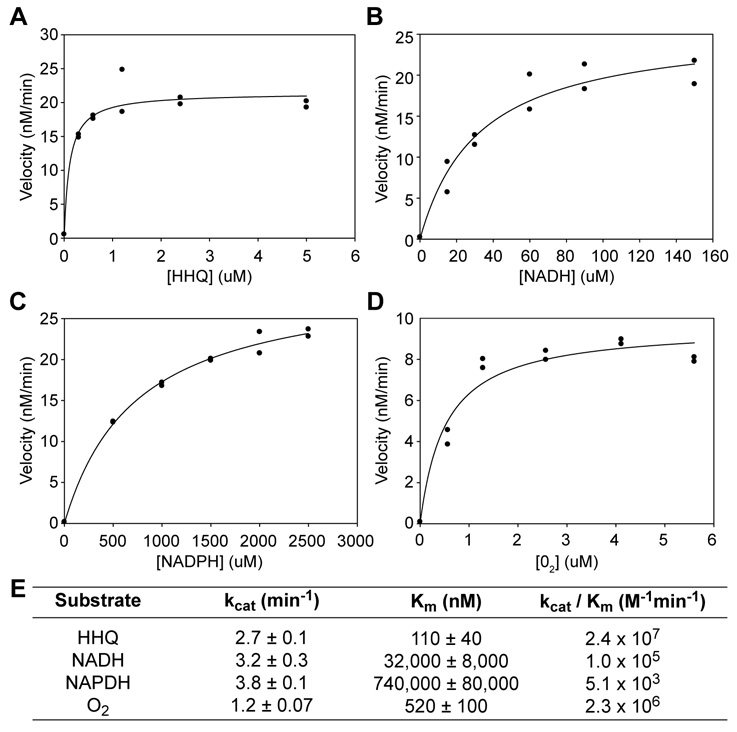
Steady state kinetic analysis confirmed HHQ, NADH, and oxygen as substrates of MBP-PqsH, consistent with its predicted role as an HHQ hydroxylating monooxygenase (Figure 3A–D). Kinetic constants were derived by varying the concentration of each substrate in the presence of constant concentrations of the other two substrates and fitting the data to the Michaelis-Menten equation (Fig. 3A–D). Apparent Kms for HHQ and oxygen were high nanomolar while the apparent Km for NADH was low micromolar (Fig. 3E). The apparent kcat in each case was in good agreement with the turnover rate (Fig 2B inset). These results demonstrate that PqsH is indeed the final enzyme in PQS biosynthesis and uses HHQ, NADH and oxygen as substrates. MBP-PqsH was also capable of HHQ oxidation using NADPH as the electron donor (Fig. 3C), though the catalytic efficiency (kcat/Km) was approximately 20-fold lower than for NADH (Fig. 3C & E).
Fig. 3. Steady-state kinetic analysis of MBP-PqsH activity.
Dependence of MBP-PqsH reaction velocity on the concentration of A. HHQ, B. NADH, C. NADPH and D. oxygen. The concentration of each substrate was varied in the presence of > 5× Km concentrations of the other two substrates. Each reaction was carried out in duplicate and all data points are plotted. E. Apparent steady-state kinetic parameters derived from A–D. Errors are standard error of the estimate from the fit to the Michaelis-Menten equation.
Alkyl side-chain length contributes to substrate specificity
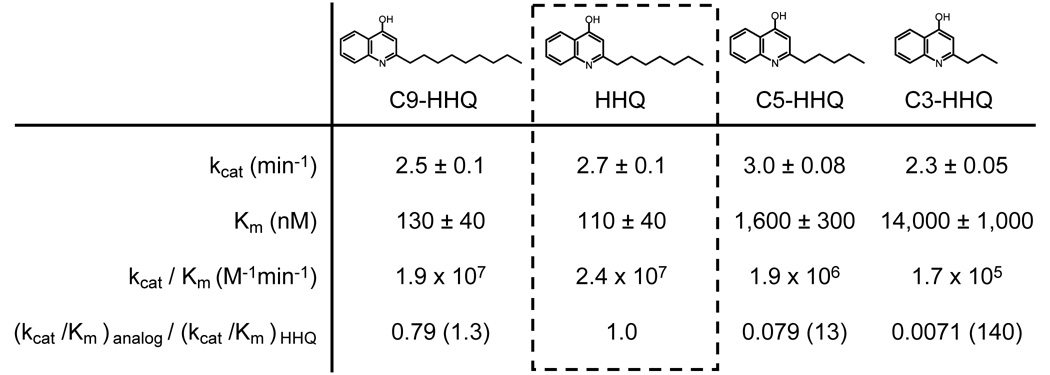
P. aeruginosa naturally produces PQS analogs with alkyl chain lengths of 5, 7 (authentic PQS), and 9 carbons, although molecules with the 7- and 9-carbon alkyl chains predominate (Deziel et al., 2004). To examine if PqsH was responsible for synthesis of these PQS analogs, we tested the ability of MBP-PqsH to hydroxylate HHQ analogs having alkyl side-chain lengths of 3 (C3-HHQ), 5 (C5-HHQ), and 9 (C9-HHQ) carbons. Interestingly, MBP-PqsH does not discriminate between authentic HHQ and C9-HHQ as the catalytic efficiencies (kcat/Km) were nearly identical (Fig. 4); however catalytic efficiencies were significantly reduced for substrates with shorter alkyl chains (Fig. 4). This reduction in catalytic efficiency for C5-HHQ (~13 fold) and C3-HHQ (~140 fold) was almost entirely due to elevated Km values, reflecting reduced affinity of MBP-PqsH for these substrates. In all cases, the identity of the reaction products were confirmed by positive electrospray ionization mass spectrometry which yielded [M + H]+ ions of mass 288.4, 232.3 and 204.3, for C9-PQS, C5-PQS, and C3-PQS respectively. These data indicate that PqsH is capable of synthesizing multiple PQS analogs in vitro, with the highest catalytic efficiency corresponding to the most abundant products produced by P. aeruginosa during in vitro growth.
Fig. 4. The HHQ alkyl chain length is critical for MBP-PqsH binding.
MBP-PqsH activity was analyzed using synthetic HHQ analogs as substrates. The concentration of each substrate analog was varied in the presence of > 55× Km concentrations of NADH and O2. Kinetic parameters were derived by fitting the data to the Michaelis-Menten equation, and errors represent the standard error of the estimate from this fit. The catalytic efficiency (kcat/Km) is shown for each substrate. (kcat/Km)analog/(kcat/Km)HHQ represents the ratio of the catalytic efficiency of each analog to the catalytic efficiency of HHQ. Numbers in parentheses represent fold reduction in catalytic efficiency for each analog.
P. aeruginosa does not produce detectable PQS anaerobically
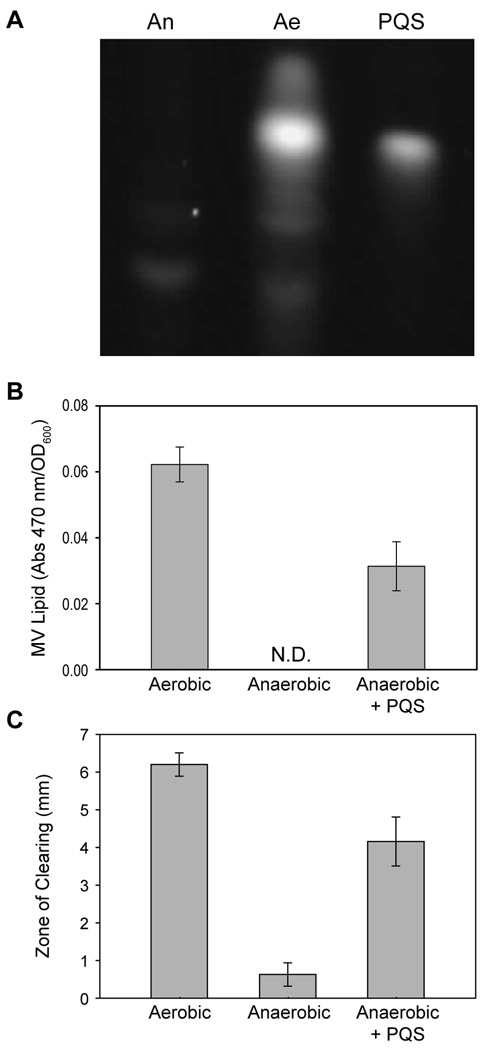
P. aeruginosa is a facultative anaerobe capable of fermentative and respiratory growth in the absence of oxygen. While P. aeruginosa pathogenesis studies have historically focused on bacteria grown aerobically, recent studies have proposed that P. aeruginosa may encounter regions of very low oxygen during in vivo growth (Beckmann et al., 2005, Jewes & Spencer, 1990, Mashburn et al., 2005, Rogers et al., 2004). Based on our steady-state kinetic analysis demonstrating that PQS production was dependent on oxygen levels (Fig. 3C), we reasoned that P. aeruginosa would produce significantly less PQS under low oxygen conditions. Examination of PQS production revealed that anaerobic P. aeruginosa produces undetectable levels of PQS (Fig. 5A), confirming recent results by Toyufuku et al. (Toyofuku et al., 2008). Consequently, anaerobic P. aeruginosa exhibited deficiencies in PQS-controlled behaviors, including production of outer membrane vesicles (MVs) (Fig. 5B). Addition of exogenous PQS to anaerobic cultures stimulated MV production indicating that the lack of MVs was attributable to the absence of PQS in anaerobic conditions and not the lack of oxygen per se (Fig. 5B). It is critical to point out that these results are not due to differential growth yields, as bacteria were grown to similar population densities and MV levels were normalized to culture optical density.
Fig. 5. P. aeruginosa requires oxygen for PQS production.
A. Aerobic and anaerobic P. aeruginosa cultures were extracted and analyzed for PQS production. Shown is a straight-phase TLC plate excited under long-wave UV light. The lanes contain: anaerobic culture extract (An); aerobic culture extract (Ae): 150 ng synthetic PQS standard (PQS). Although present in these extracts, HHQ is not visualized on this TLC due to its extremely low fluorescence emission. B. P. aeruginosa was grown aerobically, anaerobically, and anaerobically in the presence of 25 uM exogenous synthetic PQS. Outer membrane vesicles were collected by ultracentrifugation and analyzed for lipid content as described in Methods. Values are standardized to an OD600 of 1.0, and error bars represent standard error of the mean, n ≥ 3. N.D. = Not Detectable. C. P. aeruginosa was spotted onto a plate of S. epidermidis, and the radius of the zones of clearing was measured after overnight growth aerobically, anaerobically, and anaerobically with exogenous addition of PQS. Although clearly noted, zones of clearing for anaerobic P. aeruginosa grown in the presence of exogenous PQS were hazy and not completely devoid of S. epidermidis. Error bars represent standard error of the mean, n ≥ 11.
Oxygen depletion reduces P. aeruginosa virulence and antimicrobial activity
MVs are a key component involved in P. aeruginosa killing of prokaryotic and eukaryotic cells (Bomberger et al., 2009, Mashburn & Whiteley, 2005). As anaerobic P. aeruginosa do not produce detectable PQS or MVs, we hypothesized that the killing activity of P. aeruginosa supernatants would be reduced in anaerobically-grown bacteria. To test this hypothesis, aerobic and anaerobic P. aeruginosa were examined for the ability to kill the prokaryote Staphylococcus epidermidis. For these experiments, S. epidermidis was spread on the surface of agar Petri dishes, and P. aeruginosa was added to paper discs on the agar surface. Petri dishes were incubated under aerobic and anaerobic conditions, and P. aeruginosa antimicrobial activity was assessed by measuring the zone of clearing around the discs. Aerobic P. aeruginosa exhibited significant antimicrobial activity; however anaerobic cultures displayed markedly reduced activity (Fig. 5C). Addition of PQS to the agar medium restored P. aeruginosa antimicrobial activity under anaerobic conditions demonstrating that the reduction in this activity anaerobically was primarily due to the lack of PQS. This decreased killing activity was not restricted to prokaryotic cells as anaerobic supernatants also demonstrated reduced toxicity to human lung epithelial cells (Fig. S1).
Oxygen is the only PqsH substrate absent in anaerobic P. aeruginosa
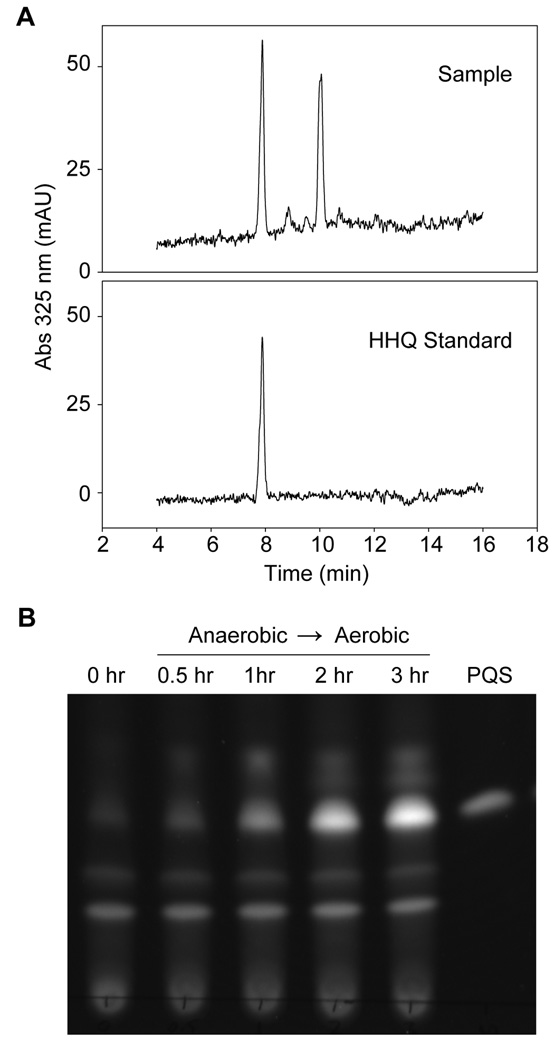
Although PQS is not produced anaerobically due to the lack of the substrate oxygen, it was not clear if the other PqsH substrates were also absent in anaerobic P. aeruginosa populations. While P. aeruginosa clearly produces NADH via central metabolism anaerobically, HHQ production in the absence of oxygen had not been examined. To test this, HHQ was quantified from anaerobic P. aeruginosa cultures by HPLC using synthetic HHQ as a standard. The presence of an absorbance peak at 325 nm that eluted with the same retention time as the standard confirmed the presence of HHQ in anaerobic cultures (Fig. 6A). By integrating the sample peak and comparing the data to HHQ standards, we calculated the concentration of HHQ present in the anaerobic culture to be 1.5 ± 0.5 µM.
Fig. 6. Oxygen is the sole limiting PQS substrate in anaerobic P. aeruginosa.
A. P. aeruginosa was grown anaerobically and quinolones were extracted from cultures with acidified ethyl acetate. HHQ levels within culture extracts were determined by reverse-phase HPLC over a Varian C-8 column using a discontinuous water:methanol gradient. Sample and HHQ (synthetic standard) chromatograms are shown. Both traces show a characteristic peak at ~ 8 min corresponding to HHQ. By comparison to known concentrations of standard, the concentration of HHQ in anaerobic culture was calculated to be 1.5 ± 0.5 µM. B. TLC analysis showing that anaerobic cells produce PQS upon a shift to aerobic conditions despite addition of the protein synthesis inhibitor tetracycline. P. aeruginosa was grown anaerobically and analyzed for PQS production (t = 0 hr). Tetracycline was added, and the culture was moved to aerobic conditions. Samples were analyzed for PQS production at 0.5, 1, 2 and 3 hours. Extracts were analyzed by straight-phase TLC excited with long-wave UV light.
Transcriptome studies (Alvarez-Ortega & Harwood, 2007, Filiatrault et al., 2005, Palmer et al., 2007b, Platt et al., 2008) suggest that PqsH is produced in the presence and absence of oxygen. Based on these data, along with our observations that all substrates other than oxygen are present in anaerobic cultures, we hypothesized that anaerobic P. aeruginosa was poised to immediately begin synthesis of PQS upon introduction of oxygen. To test this hypothesis, tetracycline was added to anaerobic P. aeruginosa to inhibit protein synthesis. Bacteria were then shifted to an aerobic atmosphere, and samples were removed at defined intervals to assess PQS production. Within 30 minutes, cells produced functional levels of PQS (~1 µM, (Xiao et al., 2006) implicating oxygen as the sole limiting substrate for PQS biosynthesis in anaerobic P. aeruginosa (Fig. 6B). These results reveal that anaerobic P. aeruginosa produces active PqsH along with the substrates HHQ and NADH, despite the fact that a critical substrate, oxygen, is absent.
Discussion
Many aspects of P. aeruginosa biology are coupled to the ability of this bacterium to communicate and coordinate its activities via QS. As our understanding of QS has evolved, it has become clear that bacteria do not communicate equivalently in all environments. Indeed, recent evidence from our laboratory and others reveals that P. aeruginosa quinolone communication is impacted by the nutritional environment, allowing this bacterium to initiate PQS-controlled group activities at lower cell densities in the presence of specific nutrients (Farrow & Pesci, 2007, Palmer et al., 2007a). Results from the current study demonstrate that PQS signaling also requires oxygen; however, in contrast to previous studies in which specific nutritional cues were shown to modulate PQS levels, oxygen is essential for PQS production. Based on these data, we propose that oxygen is an environmental trigger that controls whether P. aeruginosa will engage in PQS-controlled behaviors.
Although P. aeruginosa is a facultative anaerobe, it displays a strong preference for aerobic environments, possessing the ability to constantly sense and move toward higher oxygen concentrations (Hong et al., 2004, Nichols & Harwood, 2000). Within aerobic environments, P. aeruginosa displays several beneficial characteristics, including enhanced growth rates. Our results reveal that PQS-mediated social behaviors are also restricted to aerobic environments. This has important implications, as PQS is an inducer of numerous traits important for P. aeruginosa virulence and fitness in polymicrobial environments (Bomberger et al., 2009, Calfee et al., 2001, Cugini et al., 2007, Mashburn et al., 2005, Mashburn & Whiteley, 2005). As expected, anaerobic P. aeruginosa produce undetectable levels of MVs (Fig. 5B) and display reduced killing of prokaryotic and eukaryotic cells (Fig. 5C, Fig. S1).
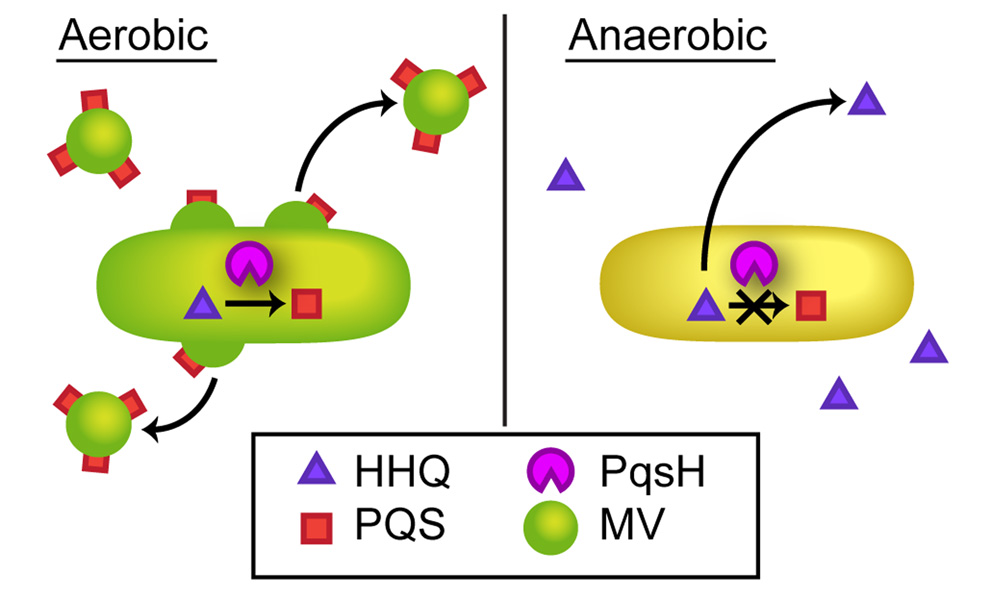
The most intriguing observation from this work is that anaerobic P. aeruginosa produce PQS in the absence of de novo protein synthesis upon shifting to an aerobic environment (Fig. 6B). This leads to a model in which the lack of PQS-mediated behaviors displayed by anaerobic P. aeruginosa are due to substrate limitation of existing PqsH enzyme, and that the limiting substrate is oxygen. Thus it appears that P. aeruginosa is poised to immediately produce PQS upon exposure to aerobic conditions (Fig. 7). This has implications for P. aeruginosa residing in environments where oxygen levels may fluctuate, but also for bacteria that are actively moving from anaerobic to aerobic environments. The model predicts that as P. aeruginosa transits from anaerobic to aerobic environments, it will not only carry active PqsH, but also the PqsH substrates HHQ and NADH. It is likely that both substrates would be available as cells move to aerobic environments since NADH is produced intracellularly via central metabolism and a portion of HHQ produced remains associated with cells, likely through interaction with membrane lipids (Mashburn & Whiteley, 2005). This model may also provide new insight into the importance of HHQ as a signaling molecule. Although HHQ is 100-fold less potent at stimulating transcription of pqsABCDE than PQS, its presence in anaerobic environments may ensure basal transcription of this HHQ-generating operon. In support of this model, the levels of HHQ produced anaerobically (~1.5 µM) are sufficient to induce pqsABCDE transcription (Xiao et al., 2006), although they are ~10-fold less than those observed in aerobic stationary phase cultures (Lepine et al., 2004).
Fig. 7. Substrate limitation of PqsH controls PQS-mediated behaviors.
Oxygen is required for the activity of PqsH. When grown aerobically, P. aeruginosa synthesizes PQS, which induces several phenotypes including generation of membrane vesicles (MVs). Under anaerobic conditions, PQS is not produced due to the absence of oxygen; however PqsH, as well as the substrates HHQ and NADH, are present. Anaerobic P. aeruginosa is thus poised to aggressively produce PQS upon re-introduction of oxygen even in the absence of de novo protein synthesis. We propose that PqsH acts as a biochemical regulator of PQS-controlled phenotypes during the transition between anaerobic and aerobic environments.
Aside from its requirement for PQS biosynthesis, oxygen is the preferred terminal electron acceptor for P. aeruginosa respiration. As such, P. aeruginosa is extremely adept at localizing to aerobic environments and utilizing oxygen as a terminal electron acceptor, even when present at extremely low levels. Of the collection of terminal oxygenases available to P. aeruginosa, two are believed to be most important for respiration in low oxygen environments. Cbb3-2 and the cyanide insensitive oxygenase CIO belong to enzyme families with extraordinarily high affinity for oxygen (Km = 3–8 nM) (Cunningham et al., 1997, D'Mello et al., 1996, Preisig et al., 1996). Although PqsH also has high affinity for oxygen (apparent Km = 520 nM), it is approximately 100-fold lower than high affinity respiratory oxygenases. These data suggest that, despite the importance of PQS, P. aeruginosa would preferentially utilize oxygen for respiration, in lieu of PQS production, in very low oxygen environments. Thus PqsH is a member of a hierarchy of high-affinity enzymes allowing P. aeruginosa to prioritize management of environmental oxygen.
Results from this study also have implications for P. aeruginosa disease. Biofilms are surface-attached assemblages of microorganisms that display a variety of unique phenotypes, including enhanced resistance to antibiotics. This enhanced resistance has broad medical consequences, as estimates of the frequency of infections caused by biofilms are as high as 65 – 80% (Centers for Disease Control and Prevention / NIH). Interestingly, dissolved oxygen concentrations within in vitro-grown biofilms have been observed to drop below 300 nM (Xu et al., 1998). Steep oxygen gradients have also been observed within the viscous mucous layer of the cystic fibrosis lung, an environment P. aeruginosa is known to colonize and often numerically dominate (Worlitzsch et al., 2002). In fact, several groups have argued that P. aeruginosa grows within anaerobic niches in the CF lung (Beckmann et al., 2005, Jewes & Spencer, 1990, Rogers et al., 2004), though this view has recently been challenged (Alvarez-Ortega & Harwood, 2007). Regardless, it can be surmised from our studies that P. aeruginosa growing in biofilms or the CF lung is transiently exposed to oxygen levels below the apparent PqsH Km, suggesting that PqsH could serve to biochemically regulate P. aeruginosa social behaviors by modulating the rate of PQS production. It is important to point out that PQS has been shown to be dispensable for virulence in a mouse model of acute infection (Xiao et al., 2006); thus the need for PQS, and therefore PqsH, may depend upon the pathogenic context including the site of infection, the number of infecting bacteria, and the P. aeruginosa strain studied (Lee et al., 2006).
Here, we demonstrate that PqsH is indeed responsible for the final step in PQS biosynthesis. PqsH appears positioned at the intersection of ecology, metabolism and quorum sensing to allow rapid activation/quenching of PQS-controlled group activities without the need for additional oxygen sensors. It is also notable that PqsH is produced under conditions in which its substrates are unavailable, suggesting that P. aeruginosa prepares signal generating machinery in anticipation of an environmental cue.
Experimental Procedures
Bacterial strains and media
P. aeruginosa strain PA14 was obtained from the PA14 Non-Redundant Transposon Mutant Library (http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi; (Liberati et al., 2006)). P. aeruginosa was routinely cultured on Brain Heart Infusion broth/agar (BHI). Anaerobic cultures were grown overnight in BHI broth supplemented with KNO3 (100 mM) and cysteine (2 mM), while aerobic cultures were grown overnight in quarter-strength BHI supplemented with KNO3 (100 mM) and cysteine (2 mM) to achieve comparable cell yields. Escherichia coli DH5α was used as the recipient for transformation and cultured on LB Miller broth/agar (Fisher Scientific). Cultures were grown at 37°C with shaking at 250 rpm. Staphylococcus epidermidis strain ATCC 14990 was used for antimicrobial studies. Antibiotics were used at the following concentrations unless otherwise noted: ampicillin, 75 µg/ml for E. coli; chloramphenicol, 20 µg/ml for E. coli.
DNA manipulations
Standard molecular biology methods were used (Ausubel et al., 1997). Restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs. Chromosomal DNA from P. aeruginosa was isolated using DNeasy Tissue kits (Qiagen), and plasmid isolations were performed using QIAprep spin miniprep kits (Qiagen). DNA fragments were purified using QIAquick mini-elute PCR purification kits (Qiagen), and PCR was performed using the Expand Long Template PCR System (Roche).
Plasmid Construction
The pqsH gene was amplified from P. aeruginosa PA14 chromosomal DNA using primers PET-21a–For (5’-GTAGTACATATGACCGTTCTTATCCAGGGGGCC-3’) and PET-21a–Rev2 (5’-CCTACTACGCGGCCGCCTGTGCGGCCATCTCACCG-3’). The resulting amplicon was digested with NdeI and NotI and ligated into pET21a to create pET21a–pqsH. The pqsH gene was then amplified from pET21a–pqsH with primers pMAL-For2 (5’-GTAGTATCTAGAACCGTTCTTATCCAGGGGGCCGGG-3’) and pMAL-Rev2 (5’-GTAGTAAAGCTT TCAGTGGTGGTGGTGGTGGTGC-3’). The resulting amplicon was digested with XbaI and HindIII and ligated into pMAL-C4x to create pMAL-C4x–pqsH. The pqsH sequence in each plasmid was confirmed by DNA sequencing.
Overexpression and Purification of PqsH
E. coli BL21(DE3) Codon Plus RP cells were freshly transformed with pMAL-C4x–pqsH for each purification. For large-scale purification, one colony was picked into 3 L Terrific Broth containing 0.25% glucose (w/v), ampicillin, and chloramphenicol. Cultures were grown shaking at 37°C to OD600 ~ 0.7 before addition of IPTG (20 µM) and riboflavin (100 µM). The culture was moved to 16°C (250 rpm) for 15 hr. Cells were harvested by centrifugation at 7000 × g for 10 min and lysed by French press (20,000 psi) in Lysis Buffer (20 mM Tris, 1 mM DTT, 0.5 mM EDTA, 10 µM FAD, 100 µM NADH, 200 mM NaCl, 10% glycerol, pH 7.5) plus Benzonase™ nuclease (Novagen) and Complete™ (−EDTA) protease inhibitor cocktail (Roche) as per manufacturer’s instructions. The lysate was clarified by centrifugation at 110,000 × g for 40 min. The supernatant fraction was loaded onto a 5 mL MBP-trap column (GE biosciences) equilibrated with Lysis buffer (minus Benzonase, protease inhibitors cocktail, and glycerol) and eluted over a discontinuous maltose gradient from 0 to 2 mM. Fractions containing the expressed 86,000 Da protein band when analyzed by SDS-PAGE were pooled and concentrated before being loaded onto a HiLoad 16/60 Superdex 200 size exclusion column (GE biosciences). Elution was isocratic in GF buffer (20 mM HEPES, 1 mM DTT, 0.5 mM EDTA, 10 µM FAD, 100 µM NADH, 200 mM NaCl, pH 8.0). Fractions containing only the expressed 86,000 Da band when analyzed by SDS-PAGE were pooled, concentrated, diluted and concentrated again to ~ 1 mL in storage buffer (20 mM HEPES, 1 mM DTT, 0.5 mM EDTA, 10 µM FAD, 100 µM NADH, 20% glycerol, pH 8.0) before being stored in appropriate aliquots at −80°C. Identity of the fusion protein was confirmed by Western blot using anti-MBP antibody (NEB).
Steady State Kinetics
750 µL reactions containing HHQ (0.3 – 5 µM), NADH (15 – 150 µM) and oxygen (0.56 µM – 5.6 µM) in reaction buffer (40 mM HEPES, 10 µM FAD, 8% methanol (v/v), pH 8.0) were initiated upon addition of purified MBP-PqsH to 8 nM. Reactions were allowed to progress for various times before being quenched with 3 volumes of acidified ethyl acetate (0.1 mL acetic acid/L ethyl acetate). The organic phase was removed and dried under a stream of nitrogen gas. Dried reaction products were resuspended in 250 µL methanol (optima grade, Fisher) and assayed for PQS fluorescence (λexcitation = 340 nm, λemission = 450 nm). Results were compared to a standard curve generated using synthetic PQS dissolved in methanol. Experiments varying the concentration of oxygen were performed in an anaerobic hood (atmosphere: 10% CO2, 5% H2, balance N2). Desired oxygen concentrations were achieved by mixing deoxygenated components with oxygen-saturated water (273 µM at 25°C) immediately before initiation. Steady state kinetic parameters were determined by varying the concentration of each substrate while the concentrations of the other two substrates were kept constant (at > 5 times their respective Km) and fitting the data to the Michaelis-Menten equation (SimaPlot 10.0.1, Systat Software Inc., San Jose, CA).
Extraction and Visualization of PQS
PQS was extracted from cultures grown under aerobic and anaerobic conditions. 5 mL or 9 mL of stationary phase culture was extracted 1:1 with acidified ethyl acetate (0.1 mL / L acetic acid). The organic phase was removed and dried under nitrogen gas. Dried samples were resuspended in 250 µL or 100 µL methanol (Optima grade, Fisher), and 5 µL were spotted onto a straight-phase phosphate-impregnated TLC plate which had been activated for 1 hr at 100°C. The sample was resolved using a 95:5 dichloromethane:methanol mobile phase. PQS was visualized on the plate by photography after excitation by long-wave UV light.
Analysis of Reaction Products by Mass Spectrometry
Large scale production of PQS and PQS derivatives (using HHQ analogs as substrates) was carried out to confirm the mass of the reaction products. 1.5 – 3.0 mL reactions containing 25 nM – 80 nM MBP-PqsH and > 5× Km concentrations of all substrates were incubated at room temperature for 1 – 2 hours. Reactions were quenched 1:1 with acidified ethyl acetate, dried under nitrogen gas and resuspended in methanol. Analysis was performed by the University of Texas Analytical Instrumentation Facility Core using an LCQ mass analyzer with a positive ESI ion source.
Quantification of MVs
MV production was assessed for stationary phase aerobic and anaerobic P. aeruginosa. MV production was quantified by phospholipid analysis of purified vesicles (Stewart, 1980). Briefly, cells were removed from the culture by centrifugation (5100 × g for 15 min) followed by filtration through a 0.45 µm membrane. Cell-free supernatants were then centrifuged at 265,000 × g for 1 hr to pellet vesicles. Pellets were washed in MV buffer (50 mM Tris, 5 mM NaCl, 1 mM MgSO4, pH 7.4), re-pelleted and resuspended once more in MV buffer. MVs were extracted 1:1 with chloroform. The organic layer was removed, combined with an equal volume of ammonium ferrothiocyanate solution (27.03 g/L FeCl3•6H2O, 30.4 g/L NH4SCN), and vortexed. The organic layer was then removed, dried down under N2 gas, resuspended in chloroform, and analyzed for absorbance at 470 nm. The resulting absorbance value was then normalized to the OD600 of the extracted culture.
S. epidermidis antimicrobial assay
Lysis of Staphylococcus epidermidis on Petri plates was performed by thoroughly swabbing a BHI + 100 mM KNO3 agar plate with an overnight culture of S. epidermidis. After drying, sterile test discs (7 mm diameter, Schleicher and Schuell) were placed onto the surface of the agar. 5 µl of an overnight culture of P. aeruginosa was added to the disc. Plates were incubated aerobically or anaerobically overnight, and zones of clearing were measured. When necessary, PQS was added at 25 µM to BHI agar before pouring the plates.
Tetracycline inhibition of protein synthesis
A 100 mL culture of P. aeruginosa was grown anaerobically overnight and 9 mL was removed and analyzed for PQS production as described above (t = 0 hr). 9 mL anaerobic BHI broth was then replaced and tetracycline (25 µg/ml) was added. After 5 min, the culture was moved to aerobic conditions and 9 mL samples were extracted at 0.5, 1, 2 and 3 hours. Extracts were analyzed by TLC with a 95:5 dichloromethane:methanol mobile phase and excitation was under long-wave UV light.
Acknowledgements
This work was funded by a grant from the NIH (5R01AI075068 to MW). MW is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease. JWS is supported by a Canadian Cystic Fibrosis Foundation postdoctoral fellowship.
References
- Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface. 2009;6:959–978. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. New York, N.Y.: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- Beckmann C, Brittnacher M, Ernst R, Mayer-Hamblett N, Miller SI, Burns JL. Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infect Immun. 2005;73:444–452. doi: 10.1128/IAI.73.1.444-452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS pathogens. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol. 2006;8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- Bredenbruch F, Nimtz M, Wray V, Morr M, Muller R, Haussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol. 2005;187:3630–3635. doi: 10.1128/JB.187.11.3630-3635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee MW, Coleman JP, Pesci EC. Interference with Pseudomonas Quinolone Signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2001;98:11633–11637. doi: 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JP, Hudson LL, McKnight SL, Farrow JM, 3rd, Calfee MW, Lindsey CA, Pesci EC. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol. 2008;190:1247–1255. doi: 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugini C, Calfee MW, Farrow JM, 3rd, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- Cunningham L, Pitt M, Williams HD. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol Microbiol. 1997;24:579–591. doi: 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- D'Mello R, Hill S, Poole RK. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 1996;142(Pt 4):755–763. doi: 10.1099/00221287-142-4-755. [DOI] [PubMed] [Google Scholar]
- Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci of the USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Farrow JM, 3rd, Pesci EC. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J Bacteriol. 2007;189:3425–3433. doi: 10.1128/JB.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault MJ, Wagner VE, Bushnell D, Haidaris CG, Iglewski BH, Passador L. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect Immun. 2005;73:3764–3772. doi: 10.1128/IAI.73.6.3764-3772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Gallagher LA, Manoil C. Pseudomonas aeruginosa PA01 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2004;231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewes LA, Spencer RC. The incidence of anaerobes in the sputum of patients with cystic fibrosis. J Med Microbiol. 1990;31:271–274. doi: 10.1099/00222615-31-4-271. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology. 1999;145(Pt 8):2051–2060. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepine F, Milot S, Deziel E, He J, Rahme LG. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Amer Soc Mass Spec. 2004;15:862–869. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Howe J, Brandenburg K, Whiteley M. Structural requirements of the Pseudomonas quinolone signal for membrane vesicle stimulation. J Bacteriol. 2009;191:3411–3414. doi: 10.1128/JB.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- Massey V. Introduction: flavoprotein structure and mechanism. FASEB J. 1995;9:473–475. doi: 10.1096/fasebj.9.7.7737454. [DOI] [PubMed] [Google Scholar]
- Nichols NN, Harwood CS. An aerotaxis transducer gene from Pseudomonas putida. FEMS Microbiol Lett. 2000;182:177–183. doi: 10.1111/j.1574-6968.2000.tb08893.x. [DOI] [PubMed] [Google Scholar]
- Njoroge J, Sperandio V. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007a;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Brown SA, Whiteley M. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J Bacteriol. 2007b;189:4449–4455. doi: 10.1128/JB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci USA. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68:4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica-Ibrulj I, Potvin E, Levesque RC, Fedynak A, Brinkman FS, Schurr J, Hwang SH, Lau GW, Limbach PA, Rowe JJ, Lieberman MA, Barraud N, Webb J, Kjelleberg S, Hunt DF, Hassett DJ. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J Bacteriol. 2008;190:2739–2758. doi: 10.1128/JB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, Thony-Meyer L, Appleby CA, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16s ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- Schertzer JW, Boulette ML, Whiteley M. More than a signal: non-signaling properties of quorum sensing molecules. Trends Microbiol. 2009;17:189–195. doi: 10.1016/j.tim.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Harris SG, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002;184:1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JC. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem. 1980;104:10–14. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Sakai R, Toyofuku M, Sawada I, Nakajima-Kambe T, Uchiyama H, Nomura N. Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa. J Bacteriol. 2009;191:7509–7519. doi: 10.1128/JB.00722-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol. 2008;190:7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- Williams P, Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Song Z, Givskov M, Doring G, Worlitzsch D, Mathee K, Rygaard J, Hoiby N. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology. 2001;147:1105–1113. doi: 10.1099/00221287-147-5-1105. [DOI] [PubMed] [Google Scholar]
- Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Frank MW, Zhu K, Mayasundari A, Rock CO. PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by Pseudomonas aeruginosa. J Biol Chem. 2008;283:28788–28794. doi: 10.1074/jbc.M804555200. [DOI] [PMC free article] [PubMed] [Google Scholar]