Abstract
Increased complexity of scientific research poses new challenges to scientific data management. Meanwhile, scientific collaboration is becoming increasing important, which relies on integrating and sharing data from distributed institutions. We develop SciPort, a Web-based platform on supporting scientific data management and integration based on a central server based distributed architecture, where researchers can easily collect, publish, and share their complex scientific data across multi-institutions. SciPort provides an XML based general approach to model complex scientific data by representing them as XML documents. The documents capture not only hierarchical structured data, but also images and raw data through references. In addition, SciPort provides an XML based hierarchical organization of the overall data space to make it convenient for quick browsing. To provide generalization, schemas and hierarchies are customizable with XML-based definitions, thus it is possible to quickly adapt the system to different applications. While each institution can manage documents on a Local SciPort Server independently, selected documents can be published to a Central Server to form a global view of shared data across all sites. By storing documents in a native XML database, SciPort provides high schema extensibility and supports comprehensive queries through XQuery. By providing a unified and effective means for data modeling, data access and customization with XML, SciPort provides a flexible and powerful platform for sharing scientific data for scientific research communities, and has been successfully used in both biomedical research and clinical trials.
Keywords: Scientific Database, Data Integration, XML Database, Scientific Data Modeling, Data Sharing
1. INTRODUCTION
With increased complexity of medical problems, medical research is increasingly a collaborative effort across multiple institutions and disciplines. Scientific researchers need first an effective system to manage their complex data, results, and the experiments that generate the results, and then a platform to integrate, share and search these across multiple institutions. Therefore, researchers are able to reuse experiments, pool expertise and validate approaches. As stated in the NIH roadmap and blueprint initiatives,1 to achieve the need to develop new partnerships of research, we need to represent and record clinical research information, exchange and share such information through standard information protocol, provide a modern information technology platform.
While there is a strong demand of managing and sharing scientific data, many challenges exist. Scientific data have high complexity and diversity, and data are often in large scales. New technology advances bring diverse instruments and dynamic computation tools, leading to heterogenous data formats such as medical images, spreadsheets, PDF files, XML documents, and many others. Especially, for biomedical research, the majority of the data is stored as images and files. Thus, the data are a mix of (often hierarchically) structural data and files (images, raw data, etc). The mixture of all types of scientific data demands an adaptable platform that can provide general data modeling and management of scientific data.
While the need for collaboration and integration keeps on increasing, scientific data tend to be isolated, especially when the size of data is in large scale. Meanwhile, researchers would rather have the best control of their data on a server located on their own labs, instead of “outsourcing” them somewhere else. Each researcher and its collaborators will naturally form a unit of data source of themselves. Thus, a distributed scientific data integration platform is essential for such scenarios.
To meet these demands, we develop SciPort, a general scientific data management and integration platform, which provides: i) a general and comprehensive scientific data model to represent any scientific data, so researchers can quickly organize their data and experiments; ii) properly organization of data, thus data can be presented and browsed in a hierarchical way; iii) convenient tools to help data providers to author such data; iv) a system architecture that provides transparent integration of data across all institutions for sharing, and v) high adaptability of the system thus it can be quickly customized for different applications and domains.
2. XML-BASED DATA MODELING OF SCIENTIFIC DATA
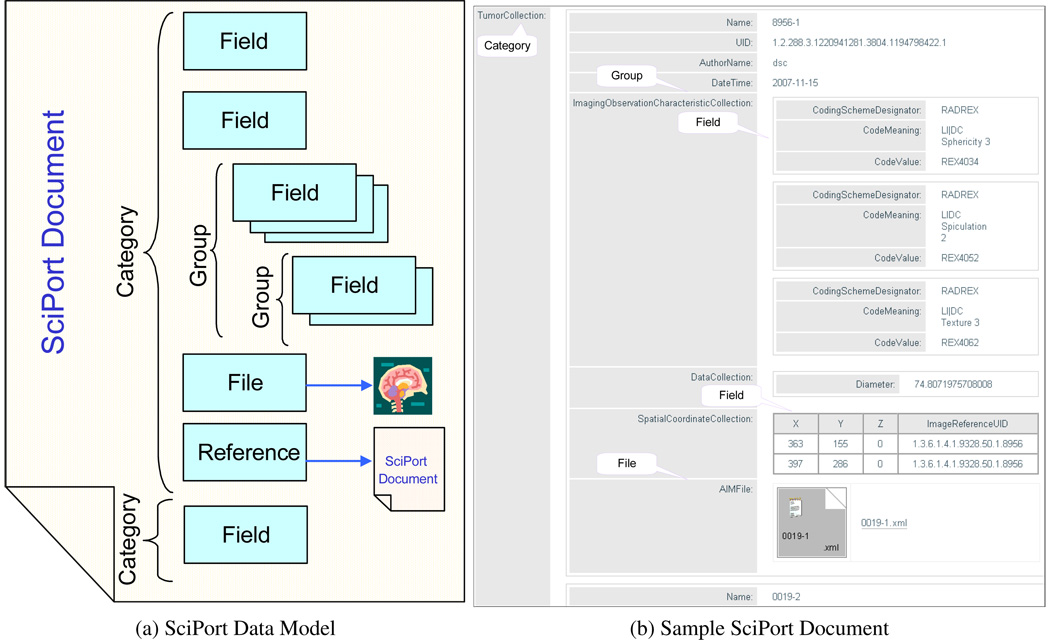
There are two main characteristics of scientific data: i) complex data structures, often highly nested; ii) heterogenous data formats, including both structural data and files. We propose a general scientific data model, as shown in Figure 1.
Figure 1.
SciPort Data Modeling
2.1 The Data Model
The central fundamental entity of the data model is SciPort Document, which captures both metadata and content. Metadata include title, description, author information, creation date, modification date, publication date, access permission information, etc. The content is the body of the document that captures both structural data and files/images. The latter are referenced through file links. There are several primitive objects in the content of the document:
Category. A category relates a list of fields.
Field. Fields are used to represent structural data, including integer, float, date, datetime, text, textarea, and radiobox, etc.
File. Files are used to link files together into the document. For common image files and DICOM files, the system will also automatically generate previews of the images.
Reference. A reference type links to another SciPort document.
Group. A group is similar to a table, which groups a collection of fields, and there can be multiple instances for a group, like rows of a table.
Figure 1b shows a sample SciPort Document (the content part) of annotations on lung images.
Implementation with XML
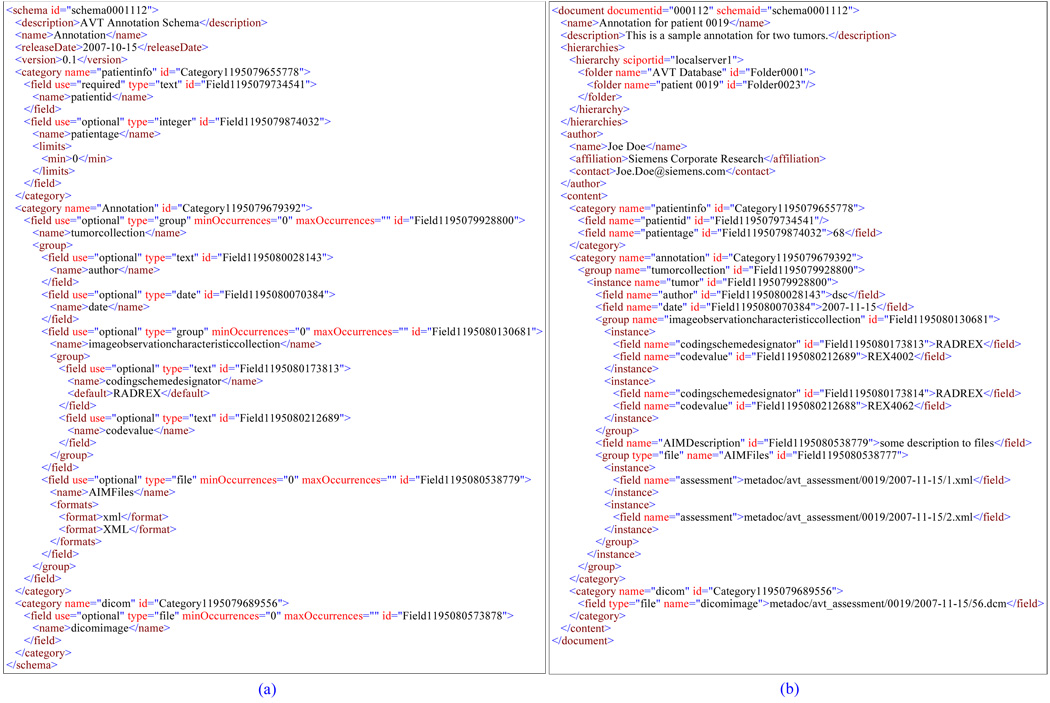
The model can be best implemented as XML – we call it SciPort Exchange Document. The hierarchy nature of the data model fits very well with the tree based XML data model. Figure 2 (b) shows the sample XML document. The XML based implementation provides many advantages: i) the self-describing and rich structure of XML makes it possible to represent arbitrary complex scientific data; ii) Powerful queries can be supported directly on top of XML documents with standard XML query language XQuery;2 iii) XML documents can be easily exchanged and shared on the Web; and iv) The XML represented scientific data can be easily converted into different formats through XSLT or XQuery to support users’ different needs.
Figure 2.
XML Based Representations: (a) Schema defined in XML; (b) Document defined in XML
XML-based Definition of Scientific Document Schemas
To make it possible for users to customize their own schemas, schemas of scientific XML documents are defined in another XML format – SciPort Schema Document, as shown in an example (Figure 2a).
2.2 Authoring
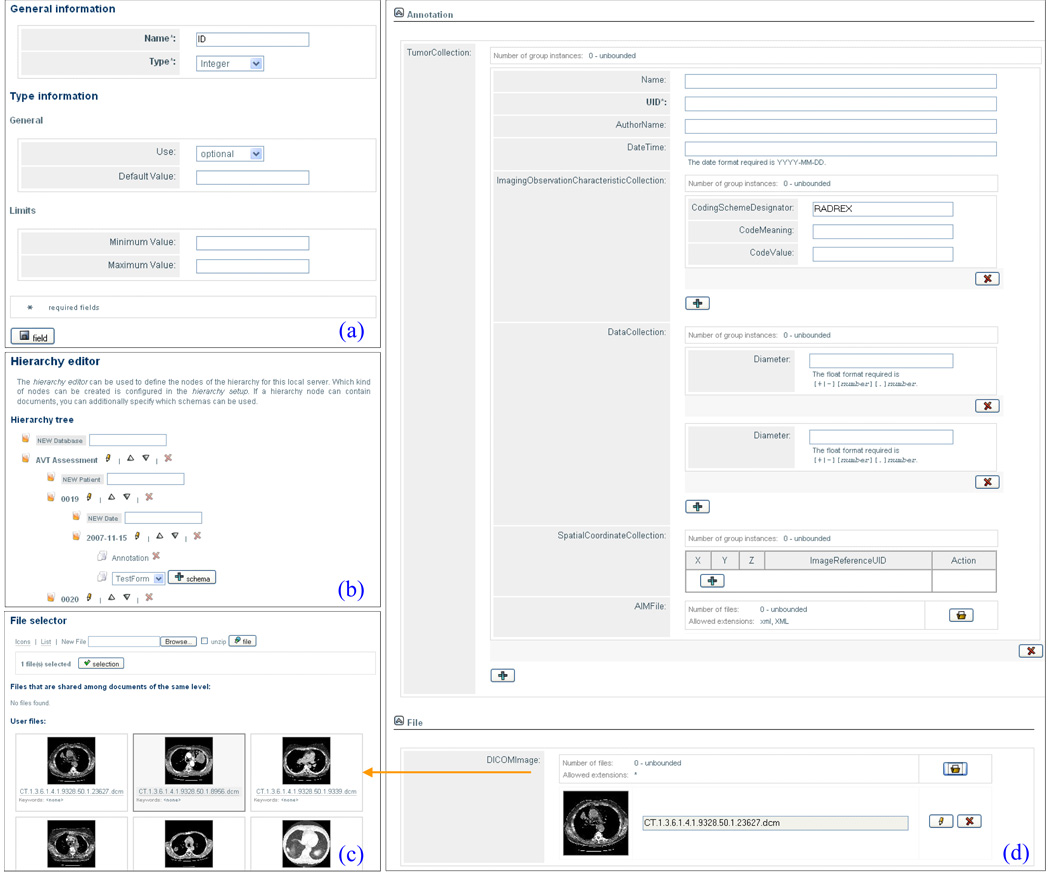
SciPort is a Web-based platform and provides Web-based authoring tools. With XML-defined schema, Web forms can be automatically generated. As shown in Figure 3, users can customize a schema (Figure 3a), define and edit a hierarchy (Figure 3b, discussed in Section 2.3), author a document (Figure 3d), and link images to a document through file cabinet (Figure 3c).
Figure 3.
Sample Screenshots of Authoring: (a) Editing a Field of a Schema; (b) Editing a Hierarchy; (c) File Selection for a Document; (d) Document Authoring
2.3 Hierarchical Organization of Scientific Data
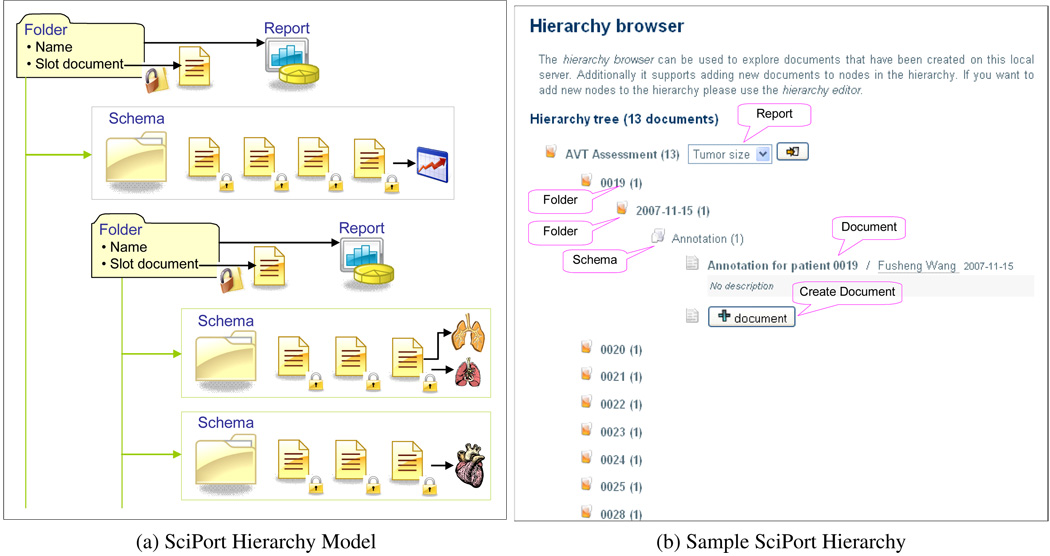
To provide an intuitive way to present data besides searching, we develop a hierarchical model to organize scientific data. This provides a vertical organization of scientific data across multiple institutions or groups. Indeed, a research collaboration can be naturally modeled as a tree structured hierarchy, thus documents can be quickly browsed and identified through this hierarchy.
As shown in Figure 4a, a hierarchy provides a folder-like organization of data. There can be multiple levels defined, and each folder can also be associated with a slot document to describe the folder. Each folder can have subfolders or schemas with which documents are created. Thus under a schema, a list of documents from this schema is grouped together. At each folder, a report can be defined that provides data analysis of all data under the folder. Such hierarchy organization provides a quick browsing of all documents in the system (Figure 4b).
Figure 4.
SciPort Hierarchy Modeling
2.4 Managing Scientific Documents
By modeling scientific data as XML documents, we can take advantage of existing XML database technologies. A native XML database provides native interfaces to store and query XML documents, and there is no transformation of data formats and queries between a client and the database. Here we take native XML database approach to manage scientific documents, thus we can avoid complex data model and query translation between XML and RDBMS, this is especially beneficial when users can define arbitrary structured and nested schemas for their scientific data. We provide two options: Oracle Berkeley DB XML,3 and DB2 Express-C4 with Pure XML support. The former is an open source embedded XML database, and the latter is a free version of DB2. Both are in very low cost (free if no support needed).
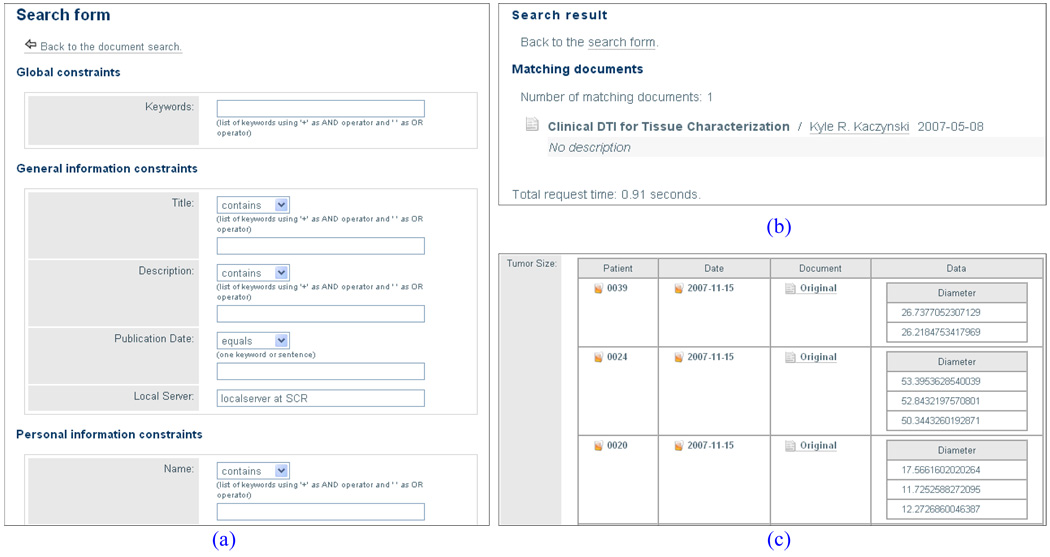
Figure 5a, 5b and 5c show sample screenshots of search form, searching result, and report result respectively.
Figure 5.
Sample Search Screenshots: (a)Search Form; (b) Search Result; (c) Report Result
3. METADATA BASED INTEGRATION
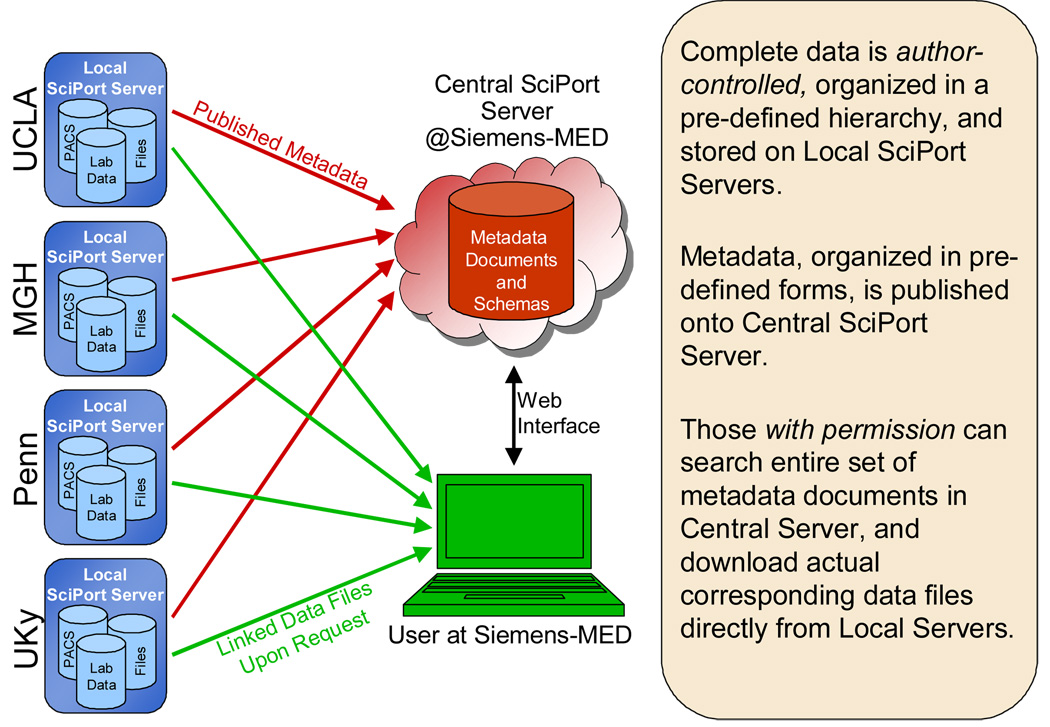
SciPort uses a Central Server based Peer-to-Peer (“peer” as database) architecture to manage and integrate scientific data: each Local Server itself is a symmetric independent scientific database server, and multiple Local Servers at distributed locations integrate and share data with the Central Server through publishing metadata to the Central Server while linking files to the Local Servers (Figure 6).
Figure 6.
Central Server Based Data Integration
Each Local Server is a peer node installed with exactly the same software, which itself can work independently as a server for managing scientific data for a local institution or project. The Central Server is first a directory server and keeps a directory of all scientific data by indexing all XML documents, and also keeps a single hierarchical organization of all documents from multiple Local Servers, with links to their storage locations (URL) at Local Servers. This central directory architecture provides an integrated view of scientific data across the entire collaborative institutions.
4. SYSTEM ARCHITECTURE
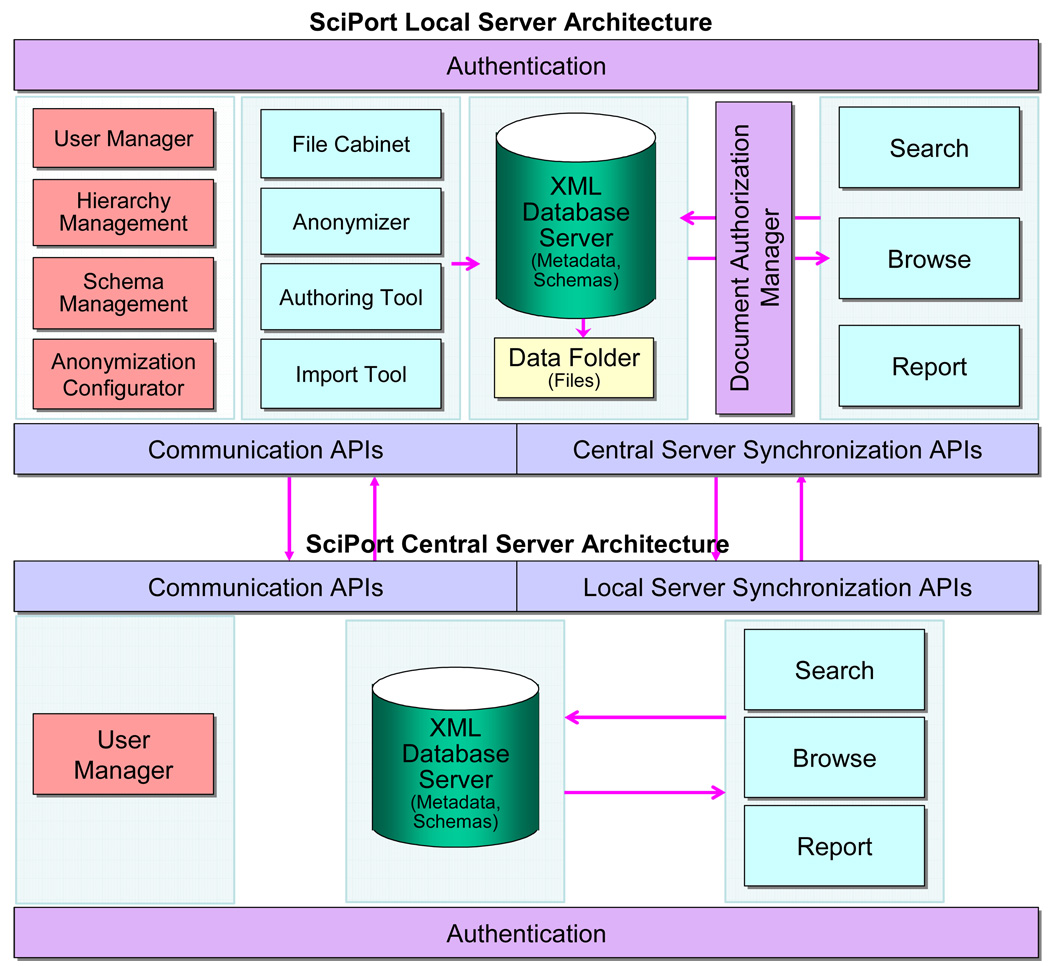
In the following, we briefly explain each component in the architecture of SciPort (Figure 7).
Figure 7.
The Architecture of SciPort
4.1 Local Server Architecture
Authentication. Each Local Server has its own authentication management, thus a login is required before accessing information on the Server.
Administrative and customization. These include User Manager to manage users, roles and groups, Hierarchy Management to define and edit the hierarchy, and Schema Management to create and edit schemas. There is also an Anoymization Configurator to configure what information to be anonymized and in which way.
Authoring Tools. These include File Cabinet, a temporary user space on the Local Server to store uploaded files that can later be used for authoring documents; Anonymizer to anonymize private information; Authoring Tool to create, update, delete a document; and Import Tool to import a document.
XML Database. This is the repository of scientific documents and schemas, and provides storage, indexing and searching capabilities.
File Folder. Files linked to documents are stored in the file folders and linked to the database.
Document Authorization Manager. When a document is authored, permission to access the document is assigned, and this manager will control the access for each document access. Documents which are published to the Central Server will automatically be accessible from the Central Server where authentication is already checked.
Search. The search tool provides keywords based search on documents, and returns a list of qualified documents. Each document includes an Export function to download the document together with its linked files as a single zip file.
Browse. This tool browses documents through their hierarchy.
Report. A report will extract information and aggregate them into a table format, and can be exported into different formats such as Excel, PDF, etc.
Communication APIs. The Local Server provides Web Services based APIs that can be used to access the server remotely.
Central Server Synchronization APIs. The Local Server provides Web Services based APIs to synchronize metadata documents (automatically), schemas and reports to the Central Server.
4.2 Central Server Architecture
The Central Server has its own user management, authentication, database server and query tools. The major components are discussed as follows:
Authentication. The Central Server has its own authentication management, thus a login is required before accessing information on the Server. The user accounts are managed with the User Manager tool.
XML Database to effectively manage XML-based documents.
Search tool to provide comprehensive search (both keyword based and structure based).
Browse tool to browse published data and their hierarchy on the Central Server.
Report tool to view and evaluate reports.
Communication APIs to provide Web Services based APIs that can be used to access the server remotely.
Local Server Synchronization APIs. The Central Server provides Web Services based APIs to synchronize documents, schemas and reports to Local Servers.
4.3 SciPort Workflow
Before SciPort is first used, administrative users on each Local Server will perform the following initial setup:
setup user accounts and assign privileges for each user.
Setup hierarchy levels.
Define schemas with the Schema Management Tool.
Configure anonymization rules with the Anonymization Configurator.
Configure Central Server address and security key information to setup Central Server to be used for data sharing.
Once the setup is ready, a Local Server is ready to use by normal users.
A user can upload files of interest into a personal file cabinet on the Local SciPort Server through File Cabinet. Uploaded DICOM files will be automatically anonymized with the Anonymizer.
The user can edit the hierarchy by creating hierarchy nodes through the Hierarchy Edit Tool. Through the Authoring Tool the user can create a new document by selecting a schema in the hierarchy. Once a document is created, if anonymization is configured, the data will be automatically be anonymized. After that, the document is automatically sent to the XML database and the files are stored in the file folder and linked to the database.
After submitting the authored document, the user has the option to publish documents to a Central SciPort Server.
New documents can then be searched or browsed at the Local SciPort Server; reports ( if already created) can be evaluated on the documents.
The Central Server needs to be setup first by setup user accounts and their privileges. After that, the Central Server is ready to use for the Central Server users to search, browse documents, and evaluate reports.
4.4 Implementation
Open system architecture
SciPort is built with J2EE and XML, running on Apache Tomcat servers and Berkeley DB XML database server (or DB2 Express-C). The system is OS neutral and can run on any machines. It uses standard protocols, including XML, XSLT, XPath and XQuery, and Web Services, and uses Web programming languages Java Servlets, Java Server Pages, JavaScripts and Ajax.
Rich Web Based Application
SciPort is a Web-based application, thus it is possible for users to use it at any place and at any time. Taking advantage of Web 2.0 technologies such as Ajax, SciPort provides rich capabilities as desktop applications.
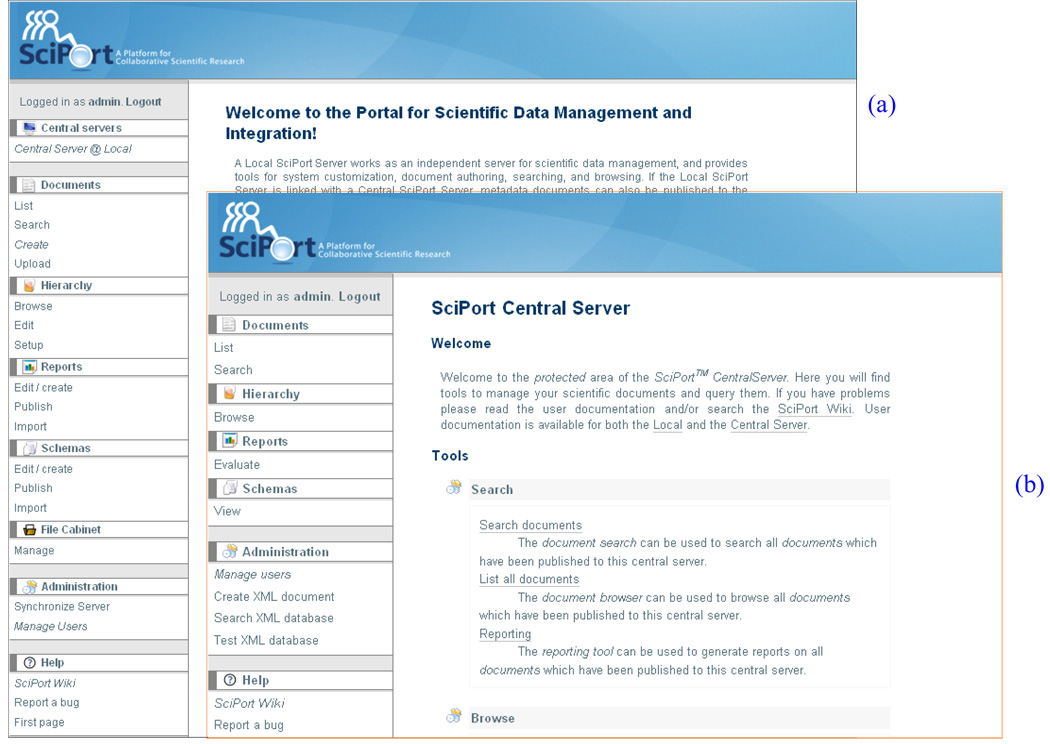
Figure 8a and Figure 8b show the sceenshots of Local Server and Central Server Interfaces respectively.
Figure 8.
Screenshots of Servers: (a) Local Server; (b) Central Server
5. APPLICATIONS
SciPort provides a highly adaptable and flexible scientific data management and integration platform without requiring the expensive and time-consuming services of database administrators or programmers. SciPort has been successfully used for scientific data management and sharing in NIH research consortia, clinical trials, and large scale research collaboration inside Siemens.
6. CONCLUSION
By providing an XML based unified and effective means for data modeling and data management, customization, and integration, SciPort provides a flexible and powerful platform for managing and sharing scientific data for scientific research communities, and has been successfully used in biomedical research and clinical trials.
Acknowledgement
The project is funded in part by the National Institutes of Health, under Grant No. 1U54CA105480-01.
REFERENCES
- 1.NIH Roadmap Initiatives. http://nihroadmap.nih.gov/initiatives.asp.
- 2.XQuery 1.0: An XML Query Language. http://www.w3.org/TR/xquery/.
- 3.Oracle Berkeley DB XML. http://www.oracle.com/database/berkeley-db/xml/.
- 4.DB2 Express-C Overview. http://www-306.ibm.com/software/data/db2/express/