Abstract
Objective
Warfarin dosing remains challenging because of its narrow therapeutic window and large variability in dose response. We sought to analyze new factors involved in its dosing and to evaluate eight dosing algorithms, including two developed by the International Warfarin Pharmacogenetics Consortium (IWPC).
Methods
we enrolled 108 patients on chronic warfarin therapy and obtained complete clinical and pharmacy records; we genotyped single nucleotide polymorphisms relevant to the VKORC1, CYP2C9, and CYP4F2 genes using integrated fluidic circuits made by Fluidigm.
Results
When applying the IWPC pharmacogenetic algorithm to our cohort of patients, the percentage of patients within 1 mg/d of the therapeutic warfarin dose increases from 54% to 63% using clinical factors only, or from 38% using a fixed-dose approach. CYP4F2 adds 4% to the fraction of the variability in dose (R2) explained by the IWPC pharmacogenetic algorithm (P < 0.05). Importantly, we show that pooling rare variants substantially increases the R2 for CYP2C9 (rare variants: P =0.0065, R2 = 6%; common variants: P= 0.0034, R2 = 7%; rare and common variants: P =0.00018; R2 = 12%), indicating that relatively rare variants not genotyped in genome-wide association studies may be important. In addition, the IWPC pharmacogenetic algorithm and the Gage (2008) algorithm perform best (IWPC: R2 = 50%; Gage: R2 = 49%), and all pharmacogenetic algorithms outperform the IWPC clinical equation (R2 = 22%). VKORC1 and CYP2C9 genotypes did not affect long-term variability in dose. Finally, the Fluidigm platform, a novel warfarin genotyping method, showed 99.65% concordance between different operators and instruments.
Conclusion
CYP4F2 and pooled rare variants of CYP2C9 significantly improve the ability to estimate warfarin dose.
Keywords: algorithms, CYP2C9, CYP4F2, dosing, IWPC, kinetics, pharmacogenetics, rare variants, VKORC1, warfarin
Introduction
Warfarin is a widely used anticoagulant, with over 30 million annual prescriptions [1]. However, it has a narrow therapeutic window and its dose response varies significantly. Warfarin has traditionally been dosed by trial-and-error, as an initial fixed dose is selected, and that dose is adjusted on subsequent visits based upon the international normalized ratio (INR) response until the patient has reached an INR value within the target range. High INRs increase the risk of bleeding, whereas low INRs are associated with recurrent thromboembolism. As a result, warfarin requires close monitoring.
Pharmacogenetics plays a key role in warfarin dosing. Two major genes are associated with the warfarin response, VKORC1 and CYP2C9 [2–5]. Warfarin inhibits vitamin K epoxide reductase (VKOR), an enzyme that converts vitamin K to its biologically active form. There are more than 50 papers in the literature that suggest its association with the warfarin dose [6–9]. The VKORC1 –1639G>A promoter polymorphism is in strong linkage disequilibrium with other VKORC1 polymorphisms [10,11]. One study identified a low dose and a high dose haplotype for VKORC1, which explained 25% of dose variation [12]. Asian–Americans had a greater proportion of the low-dose haplotype, whereas African–Americans had a greater proportion of the high-dose haplotype.
CYP2C9 is a liver enzyme in the cytochrome P 450 family that is responsible for metabolizing warfarin [13]. The two most commonly reported variants, *2 and *3, are less effective than the wild-type *1 variant [14]. Cumulative data on VKORC1 and CYP2C9 compelled the Food and Drug Administration to change the label for warfarin in 2007 to include information from pharmacogenetic tests [15]. Randomized trials in small patient populations have found that pharmacogenetics-guided warfarin therapy decreases the time to stable dose and requires fewer and smaller dosing changes, and multi-center, randomized controlled trials are underway to determine whether pharmacogenetic dosing algorithms improve patient outcomes [16–18]. More recently, the CYP4F2 gene has also been associated with warfarin dosing [19–22].
Consequently, a number of warfarin dosing algorithms incorporating genetic factors have been proposed in the literature. They include those by Gage et al. [23], Herman et al. [24], Sconce et al. [25], Tham et al. [26], Wu et al. [27], and Zhu et al. [28]. The International Warfarin Pharmacogenetics Consortium (IWPC) recently reported a global warfarin dosing algorithm using genetic and clinical information from 5700 patients over nine countries [29]. The addition of genetic factors was associated with improved performance in dose prediction.
The goals of this study were to validate the IWPC equation on an independent population, determine the local performance of the different dosing algorithms proposed in the literature, assess the additional impact of genetic factors such as the CYP4F2 gene and various CYP2C9 polymorphisms on warfarin dose, and examine whether genes affect the long-term variation in warfarin dosing, using a new method and platform for warfarin clinical genotyping.
Methods
Data collection
The Stanford Oral Anticoagulation Clinic specializes in optimizing the treatment for patients on chronic warfarin therapy. Patients who achieved a stable dose of warfarin for three or more consecutive visits, with a target INR between two and three, were recruited by clinical pharmacists responsible for managing their therapy. Patients provided written informed consent. The clinic actively monitors 400 patients, and 108 of these patients were enrolled in this study. Patients visited the warfarin clinic a median of 40 times (mean=47.6, σ=32.1, min=4, max=135), and they had been on warfarin a median of 174 weeks (mean=190, σ=166, min=6, max=670). DNA was collected through saliva samples, and these samples were genotyped using a novel genotyping method that uses a 48×48 chamber dynamic array that in this configuration, simultaneously genotyped 48 patients at 30 warfarin-related single nucleotide polymorphisms (SNPs). This study was approved by the Institutional Review Board.
Clinical data was obtained using the CoagClinic database (Standing Stone, Westport, Connecticut, USA). This database included a list of medications, the indications for warfarin administration, the INR target range, and the warfarin dose and INR value associated with each patient visit. In addition, a questionnaire was given to each patient (see Fig. S1, Supplemental digital content 1, http://links.lww.com/FPC/A148) to obtain additional clinical information not routinely collected in the medical chart, such as height, weight, supplement use, and vitamin use, and to corroborate data on concomitant medications found in the chart (i.e. amiodarone). The questionnaire was developed in consultation with clinical pharmacists who routinely worked with the patients. Patients provided saliva samples using the Oragene·DNA System (DNA Genotek, Ottawa, Canada). Samples were then genotyped at 30 SNPs relevant to VKORC1, CYP2C9, and CYP4F2 (see Table S1, Supplemental digital content 2, http://links.lww.com/FPC/A149) using ABI TaqMan assays (Life Technologies, Carlsbad, California, USA) on 48.48 Dynamic Arrays (Fluidigm, South San Francisco, California, USA). These SNPs were selected based on suggestions from the FDA for warfarin testing, the availability of an ABI assay for the SNP, and an analysis of common SNPs available on the Drug Metabolizing Enzymes and Transporters Microarray Platform (Affymetrix, Santa Clara, California, USA) [30,31].
Genotyping procedure
DNA was purified as per the Oragene DNA purification protocol (DNA Genotek). DNA concentration was determined using a NanoDrop UV spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The average yield was 241.8±173.0 ng/μl. Samples then underwent preamplification, a multiplexed, locus-specific amplification procedure used to improve signal strength in the final SNP analysis. Forward and reverse primer pairs from all 30 SNPs were pooled together at a final concentration of 90 nmol/l. 1.25 μl of sample, 1.25 μl of the pooled primer mix, and 2.5 μl of Multiplex PCR Master Mix (catalog #206143, Qiagen, Venlo, The Netherlands) were combined and PCR-amplified (95°C for 10 min, followed by 14 cycles of 95°C for 15 s and 60°C for 4min). Amplified samples were diluted five-fold, and a standard SNP analysis was performed on three 48.48 Fluidigm Dynamic Arrays, which were all run on the same instrument on the same day. These arrays use microfluidic channels to simultaneously perform up to 48 assays on 48 individuals with relatively low volumes [32].
Thirty SNPs relevant to warfarin dosing were analyzed, and each SNP was run once per chip. The data turn-around time for the entire procedure was 30 h, with 9 h of hands-on time. Samples 50–66 were repeated on the second and third chip, and they showed 100% concordance. A fourth chip retested 48 samples from the first chip. However, it was operated by a different person on a different day, using a different BioMark instrument (Fluidigm), and it displayed 99.65% concordance. The average call rate for all samples was 98.1% (see Table S2, Supplemental digital content 3, http://links.lww.com/FPC/A150). The no-call rates are primarily because of a few samples with a lower amount of starting DNA and are highly correlated between VKORC1, CYP2C9, and CYP4F2 SNPs. Two assays accounted for 30% of the no-calls, yet even these assays had a call rate greater than 90%.
Statistical analysis
A patient was determined to have a stable warfarin dose if he or she remained on the same dose for three or more consecutive visits and had a target INR between two to three. A computer algorithm was written to determine the most recent occurrence of three or more consecutive warfarin doses of the same value. No additional INR criteria were necessary since if the INR was out-of-range, then it would have naturally warranted a change in dose. Moreover, this definition of stable dose was used by one site in the IWPC study [29].
Next, the IWPC pharmacogenetic equation was used to predict the therapeutic dose for each patient using both clinical and pharmacogenetic data, the IWPC clinical equation was used to predict the therapeutic dose using clinical data alone, and the fixed-dose model assumed that every patient took a dose of 35 mg/week, a standard starting dose. Dosing algorithms proposed in the literature were also used to predict the therapeutic dose. Mean absolute error (MAE), which is the average of the absolute values of the differences between the actual doses and predicted doses, was used to compare model performance. The coefficient of determination (R2) for each model was calculated using R (version 2.6.2). Using the IWPC dosing models, three values were calculated: the percent of patients within 1 mg/day of the actual dose, the percent more than 1 mg/day over the actual dose, and the percent more than 1 mg/day under the actual dose. The IWPC had earlier determined that the 1mg/day cut-off was clinically significant.
Furthermore, the additional role of the CYP4F2 gene was assessed. Linear regression was performed with the actual warfarin dose as a function of the dose predicted by the IWPC pharmacogenetic equation. Next, linear regression was performed with the actual warfarin dose as a function of both CYP4F2 and the IWPC pharmacogenetic predicted dose. Finally, a likelihood ratio test was used to compare the pharmacogenetic model both with and without CYP4F2.
Moreover, the effects of the various genetic variants were assessed by comparing differences in the mean dose between different groups (i.e. CYP2C9 *18 A/A vs. T/A) using two-tailed t-tests. For two-tailed t-tests, a study with 108 patients had 80% power to detect an effect size of 0.55 (Cohen’s d) or above (P<0.05), which corresponds to a medium effect size. When there were three or more groups, analysis of variance (ANOVA) was performed using the R statistical program. This study analyzed 14 SNPs that showed variation in the population and 19 clinical factors, leading to a Bonferroni-adjusted P value of 0.05/33=0.0015 because of multiple comparisons. All SNPs in this study were in Hardy–Weinberg equilibrium using Pearson’s χ2 test at the 5% significance level.
Results
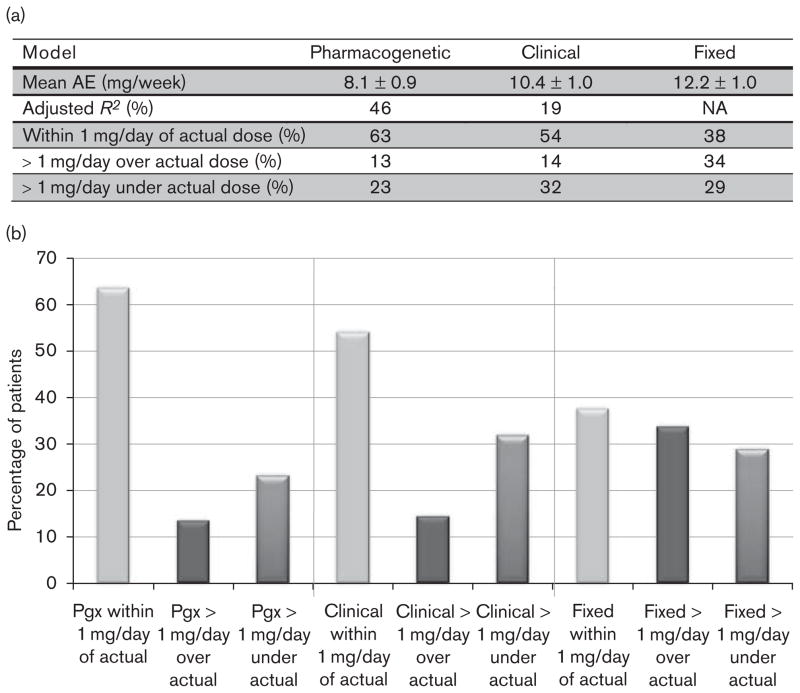
One hundred and six samples were of sufficient volume to undergo genomic DNA isolation, while 104 patients achieved a stable warfarin dose. The genetic and the clinical characteristics of these 104 patients are shown in Table 1. The MAE for the three IWPC dosing models was 8.1 mg/week for the pharmacogenetic equation, 10.4 mg/week for the clinical equation, and 12.2 mg/week for the fixed-dose model. The percentage of patients within 1 mg/day of the actual dose or deviating greater than 1 mg/day is shown in Fig. 1.
Table 1.
Characteristics of the patient population (N=104)
| Variable | Value (%) |
|---|---|
| Warfarin dose | |
| Median | 32.5 mg/week |
| Interquartile range | 24.0–42.5 mg/week |
| VKORC1 genotype | |
| G/G | 38 (37) |
| A/G | 46 (44) |
| A/A | 16 (15) |
| NA | 4 (4) |
| CYP2C9 genotype | |
| *1/*1 | 61 (59) |
| *1/*2 | 17 (16) |
| *3/*18 | 10 (10) |
| *2/*2 | 4 (4) |
| *1/*6 | 2 (2) |
| *1/*11 | 2 (2) |
| *1/*12 | 2 (2) |
| *6/*7 | 1 (1) |
| *1/*9 | 1 (1) |
| *2/*12 | 1 (1) |
| *3/*12/*18 | 1 (1) |
| NA | 2 (2) |
| CYP4F2 genotype | |
| C/C | 55 (53) |
| T/C | 40 (38) |
| T/T | 6 (6) |
| NA | 3 (3) |
| Age | |
| Mean, median | 64 years, 67 years |
| Interquartile range | 53–75.3 years |
| Height | |
| Mean, median | 172 cm, 173 cm |
| Interquartile range | 163–180 cm |
| Weight | |
| Mean, median | 81 kg, 78.8 kg |
| Interquartile range | 64.9–90.7 kg |
| Race | |
| White | 78 (75) |
| Asian | 18 (17) |
| Black | 8 (8) |
| Sex | |
| Male | 60 (58) |
| Female | 44 (42) |
| Inducers use | |
| Number (percent) | 1 (1) |
| Amiodarone use | |
| Number (percent) | 5 (5) |
Inducers included carbamazepine, rifampin, and phenytoin.
Fig. 1.
(a) Performance of the three different dosing models: pharmacogenetic, clinical, and fixed. Includes all patients who reached stable dose (N=104). (b) Percentage of patients who were either within 1 mg/day of the actual dose, more than 1 mg/day over the actual dose, or more than 1 mg/day under the actual dose for the three different dosing models: pharmacogenetic, clinical, and fixed.
The potential role of CYP4F2 in providing additional predictive information was evaluated. The coefficient of determination (R2) between the stable warfarin dose and the prediction of the pharmacogenetic equation was 46%, and the R2 value between the stable warfarin dose and the dose predicted using the pharmacogenetic equation combined with CYP4F2 was 51%. T-tests revealed that the mean dose for individuals with the CYP4F2 ‘C/C’ genotype (32.2±1.5mg/week) was significantly lower than the mean dose for individuals with the ‘T/C’ genotype (40.5±3.0 mg/week) (P<0.01). CYP4F2 ‘C/C’ was not significantly different from CYP4F2 ‘T/T’ (P=0.61), as only six patients had the ‘T/T’ genotype. The overall test for trend, assuming an additive model, was at the cusp of significance (P=0.055), whereas ANOVA for the CYP4F2 SNP was significant (P=0.042). Finally, a likelihood ratio test confirmed that CYP4F2 provides additional predictive information (P=2.4×10 −9).
The potential contribution of rare variants of CYP2C9 polymorphisms in predicting warfarin dosing was studied by looking at their respective genotypes in relationship to average warfarin dose. For CYP2C9 *12, individuals with the ‘T/C’ genotype had a lower average dose than individuals with the ‘C/C’ genotype (P=0.034, R2=4%) (Table 2). For CYP2C9 *18, individuals with the ‘T/A’ genotype had a lower average dose than people with the ‘A/A’ genotype, a difference that was not quite significant (P=0.064, R2=3%) (Table 2). The *18 allele was the only one that showed significant linkage disequilibrium with another SNP (*3) (Table 1). For all the rare CYP2C9 polymorphisms that exhibited variation in our sample (*6, *7, *9, *11, *12, and *18), individuals who were heterozygous at that CYP2C9 polymorphism had a lower average dose than individuals who were homozygous for the wild-type genotype (Table 2, Table S3, Supplemental digital content 4, http://links.lww.com/FPC/A151). Moreover, individuals heterozygous at any of these six alleles had a lower average dose than individuals who were homozygous wild-type for all six alleles (27.0 mg/week vs. 37.6 mg/week, P=0.0065, R2=6%) (Table 3). For comparison, individuals with a variant allele at either *2 or *3, more common alleles, also had a lower average dose than individuals who were homozygous wild-type for both alleles (28.9 mg/week vs. 38.6 mg/week, P=0.0034, R2=7%) (Table 4). The aforementioned calculations for CYP2C9, including those for CPY2C9 *2 and *3, were significant at the P value of less than 0.05 level, but not at the more stringent P value of less than 0.0015 cut-off. However, individuals who varied at either *2, *3, *6, *7, *9, *11, *12, or *18 had a significantly lower average dose than individuals who were homozygous wild-type for all eight alleles (28.5 mg/week vs. 40.2 mg/week, P=0.00018, R2=12%) (Table 5). CYP2C9 was a significant factor in this study at the more stringent P value of less than 0.0015 level only after pooling rare and common variants, which decreased the P value and increased R2. Moreover, a likelihood ratio test comparing using either common CYP2C9 alleles alone or both common and rare alleles showed that rare alleles offer additional predictive information (P=0.01).
Table 2.
Analysis of rare CYP2C9 variants. Analysis of the *12 and *18 alleles alone
| CYP2C9 SNP | Number | Average dose ± SE |
|---|---|---|
| *12 C/C | 87 | 36.3 ± 1.7 |
| *12 T/C | 4 | 19.6 ± 2.6 |
| *12 NA | 13 | 34.1 ± 5.1 |
| *18 A/A | 87 | 36.5 ± 1.7 |
| *18 T/A | 11 | 27.2 ± 3.3 |
| *18 NA | 6 | 35.3 ± 8.1 |
| CYP2C9 regression | Adjusted R2 | P value |
| *12 | 3.9% | 0.034 |
| *18 | 2.5% | 0.064 |
SNP, single nucleotide polymorphism.
Table 3.
Comparison of individuals who were either wild-type for *6, *7, *9, *11, *12, and *18, or had a variant genotype at any of these alleles
| CYP2C9 (*6–*18) | Number | Average dose ± SE | Comparison | Value |
|---|---|---|---|---|
| Homozygous WT at all SNPs | 82 | 37.6 ± 1.8 | P value | 0.0065 |
| Heterozygous at any | 20 | 27.0 ± 2.1 | Adjusted R2 | 6% |
SNP, single nucleotide polymorphism; WT, wild-type.
Table 4.
Comparison of individuals who were either wild-type for *2 and *3, or had a variant genotype at either *2 or *3
| CYP2C9 (*2–*3) | Number | Average dose ± SE | Comparison | Value |
|---|---|---|---|---|
| Homozygous WT at all SNPs | 69 | 38.6 ± 2.1 | P value | 0.0034 |
| Heterozygous at any | 33 | 28.9 ± 1.7 | Adjusted R2 | 7% |
SNP, single nucleotide polymorphism; WT, wild-type.
Table 5.
Comparison of individuals who were either wild-type for *2, *3, *6, *7, *9, *11, *12, and *18, or had a variant genotype at any of these alleles
| CYP2C9 (*2–*18) | Number | Average dose ± SE | Comparison | Value |
|---|---|---|---|---|
| Homozygous WT at all SNPs | 61 | 40.2 ± 2.2 | P value | 0.00018 |
| Heterozygous at any | 41 | 28.5 ± 1.5 | Adjusted R2 | 12% |
SNP, single nucleotide polymorphism; WT, wild-type.
Other than VKORC1 – 1639, three other VKORC1 SNPs were associated with therapeutic dose – VKORC1 1173 (ANOVA, P<10−3), VKORC1 2255 (ANOVA, P<10−4), and VKORC1 3730 (ANOVA, P=0.003) – but pair-wise combinations of VKORC1 SNPs did not have a greater predictive ability than VKORC1 – 1639 alone (Table S1, Supplemental digital content 2, http://links.lww.com/FPC/A149). VKORC1 497 was not significant (R2<1%). The other 16 SNPs collected by this study showed no variation in the patient population (Table S1, Supplemental digital content 2, http://links.lww.com/FPC/A149). Overall, examination of a discrete, targeted set of SNPs for VKORC1, CYP2C9, and CYP4F2 sufficiently explained 33% of the dose variance, which improved to 50% when clinical factors were added.
We compared the performance of the IWPC equations relative to six models in the literature (Table 6). The IWPC pharmacogenetic equation had a performance that was very similar to the Gage (2008) equation, both of which outperformed all other models. Although the other pharmacogenetic dosing algorithms had less predictive ability, they all outperformed the IWPC clinical equation in terms of MAE and R2.
Table 6.
Comparison of model performance
| Model | MAE ± SE (mg/week) | Adjusted R2 (%) |
|---|---|---|
| IWPC Pgx. (2009) [29] | 7.80 ± 0.84 | 50 |
| IWPC clinical (2009) [29] | 10.14 ± 1.04 | 22 |
| Gage et al. [23] | 7.79 ± 0.83 | 49 |
| Sconce et al. [25] | 8.98 ± 0.94 | 47 |
| Zhu et al. [28] | 8.98 ± 0.95 | 39 |
| Tham et al. [26] | 9.72 ± 0.95 | 33 |
| Herman et al. [24] | 9.73 ± 0.89 | 47 |
| Wu et al. [27] | 9.98 ± 0.95 | 35 |
This analysis was restricted to the 95 patients who had all the variables necessary for the eight different dosing equations.
IWPC, International Warfarin Pharmacogenetics Consortium; MAE, mean absolute error.
Although recent research has studied the kinetics of warfarin dosing [33–37], we examined how genetics could affect long-term variability in dose response. Neither VKORC1 nor CYP2C9 genotypes were a determinant of long-term dose stability.
Discussion
Overall, this study validates the performance of the IWPC pharmacogenetic algorithm in predicting warfarin dosing, also confirming that it compares very favorably with other algorithms published in the literature. Moreover, genetic factors, such as CYP4F2 and rare CYP2C9 polymorphisms, provide additional information in the prediction of warfarin dose, beyond the current variables in the model.
We were able to validate the performance of the IWPC pharmacogenetic equation in our independent patient population. The MAE values in this population for the pharmacogenetic, clinical, and fixed-dose algorithms (Fig. 1) compare favorably with the values of 8.3, 10.0, and 13.3 mg/week respectively from the derivation cohort of the IWPC paper. This observation holds despite the different ethnic makeup of the two populations, as whites and Asians comprised 75 and 17% of the patient population respectively in this study (Table 1), as opposed to 55 and 30%, respectively in the IWPC study. Thus, the variability of warfarin dosing among individuals of different ethnic backgrounds can largely be explained by the genotypic variants of VKORC1 and CYP2C9.
Compared with other earlier published algorithms, the IWPC pharmacogenetic equation compares favorably with the Gage (2008) equation. More importantly, they outperform all other pharmacogenetic equations (Table 6). The Gage algorithm was developed in a cohort of 1015 individuals, 83% of whom were white, in comparison with a cohort of 5700 individuals, 55% of whom were white in the IWPC study. Despite these differences in ethnic backgrounds, both equations were developed using large cohorts of patients, which may explain their superior performance.
Using the IWPC pharmacogenetic equation markedly increases the percent of patients with an ideal dose (within 1 mg/day of the actual dose) and leads to the smallest number of dose overestimates and underestimates (Fig. 1). Although the clinical equation leads to substantially fewer overestimates of warfarin dose than the fixed-dose model, it actually leads to slightly more underestimates of dose. This, too, is consistent with the findings of the IWPC study and likely occurred because the clinical equation is more conservative.
The CYP4F2 gene variant showed additional predictive ability beyond that offered by the IWPC pharmacogenetic algorithm. In our cohort, 4% of the variation in warfarin dose could be attributed to CYP4F2, improving the accuracy of the estimate of warfarin dose. Earlier estimates of the effect of CYP4F2 range from 1 to 2% to as high as 7% [19,20,22]. The relatively high percentage of patients of European descent may account for the utility of CYP4F2 in this population. VKORC1 alleles other than the – 1639 polymorphism were also significant, yet they did not offer any additional predictive ability beyond VKORC1 – 1639, likely as they are in high-linkage disequilibrium with that SNP.
Finally, by pooling rare CYP2C9 variants, we showed that individuals who varied at any of the rare CYP2C9 SNPs had a lower therapeutic warfarin dose than individuals who were wild-type at all six (Table 2, Table S3, Supplemental digital content 4, http://links.lww.com/FPC/A151). By pooling all eight SNPs, both common and rare, into a composite variable that evaluated whether patients were heterozygous at any SNP or homozygous wild-type at all, the combined R2 value was additive and the resultant P value was highly significant at the P value of less than 0.0015 level for multiple comparisons. The results of a likelihood ratio test were also consistent with this data. Hence, the effect of the rare CYP2C9 alleles is not merely because of the linkage disequilibrium with *2 and *3, indicating that ‘nontraditional’ CYP2C9 polymorphisms may have an important role to play in warfarin dosing. Our ability to pool these variants is based on the observation that they all tend to reduce the activity of the enzyme.
In conclusion, this study validates the favorable performance of the IWPC pharmacogenetic equation, while confirming that CYP4F2 increases model performance. Our results also indicate that additional candidate variants, such as rare CYP2C9 variants, add to the predictive ability of warfarin dosing models. We can expect to see a growing number of additional candidate SNPs that could be associated with warfarin dose in the near future.
Supplementary Material
Acknowledgments
This study was supported by NIH grant U01GM61374 (NIH/NIGMS Pharmacogenetics Research Network and Database) (Russ B. Altman), Microsoft Corporation (Russ B. Altman), the Howard Hughes Medical Institute Research Training Fellowship (Hersh Sagreiya), grants from the Stanford Medical Scholars Research Program (Hersh Sagreiya), and genotyping support was provided by Fluidigm Corporation. The authors thank Dr Teri Klein and members of the IWPC (http://www.pharmgkb.org/views/project.jsp?pId=56) for useful discussions about the pharmacogenomics of warfarin. Dr Jun Wang provided expert advice on genotyping analysis and acted as an independent reviewer of the data at Fluidigm.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Website (www.pharmacogeneticsandgenomics.com).
References
- 1.Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. Arch Intern Med. 2007;167:1414–1419. doi: 10.1001/archinte.167.13.1414. [DOI] [PubMed] [Google Scholar]
- 2.D’Andrea G, D’Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 3.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 4.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 5.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 6.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 7.Lal S, Jada SR, Xiang X, Lim WT, Lee EJ, Chowbay B. Pharmacogenetics of target genes across the warfarin pharmacological pathway. Clin Pharmacokinet. 2006;45:1189–1200. doi: 10.2165/00003088-200645120-00004. [DOI] [PubMed] [Google Scholar]
- 8.Jonas DE, McLeod HL. Genetic and clinical factors relating to warfarin dosing. Trends Pharmacol Sci. 2009;30:375–386. doi: 10.1016/j.tips.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Wallin R, Wajih N, Hutson SM. VKORC1: a warfarin-sensitive enzyme in vitamin K metabolism and biosynthesis of vitamin K-dependent blood coagulation factors. Vitam Horm. 2008;78:227–246. doi: 10.1016/S0083-6729(07)00011-8. [DOI] [PubMed] [Google Scholar]
- 10.Yuan HY, Chen JJ, Lee MT, Wung JC, Chen YF, Charng MJ, et al. A novel functional VKORC1 promoter polymorphism is associated with interindividual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadee W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112:1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi H, Echizen H. Pharmacogenetics of warfarin elimination and its clinical implications. Clin Pharmacokinet. 2001;40:587–603. doi: 10.2165/00003088-200140080-00003. [DOI] [PubMed] [Google Scholar]
- 14.Oldenburg J, Bevans CG, Fregin A, Geisen C, Muller-Reible C, Watzka M. Current pharmacogenetic developments in oral anticoagulation therapy: the influence of variant VKORC1 and CYP2C9 alleles. Thromb Haemost. 2007;98:570–578. [PubMed] [Google Scholar]
- 15.Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis. 2008;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 16.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 17.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28:1084–1097. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 19.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borgiani P, Ciccacci C, Forte V, Sirianni E, Novelli L, Bramanti P, et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10:261–266. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- 21.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman D, Peternel P, Stegnar M, Breskvar K, Dolzan V. The influence of sequence variations in factor VII, gamma-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost. 2006;95:782–787. [PubMed] [Google Scholar]
- 25.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 26.Tham LS, Goh BC, Nafziger A, Guo JY, Wang LZ, Soong R, et al. A warfarin-dosing model in Asians that uses single-nucleotide polymorphisms in vitamin K epoxide reductase complex and cytochrome P450 2C9. Clin Pharmacol Ther. 2006;80:346–355. doi: 10.1016/j.clpt.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multiethnic population: comparison with other equations. Pharmacogenomics. 2008;9:169–178. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Shennan M, Reynolds KK, Johnson NA, Herrnberger MR, Valdes R, Jr, et al. Estimation of warfarin maintenance dose based on VKORC1 ( – 1639 G> A) and CYP2C9 genotypes. Clin Chem. 2007;53:1199–1205. doi: 10.1373/clinchem.2006.078139. [DOI] [PubMed] [Google Scholar]
- 29.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumaual C, Miao X, Daly TM, Bruckner C, Njau R, Fu DJ, et al. Comprehensive assessment of metabolic enzyme and transporter genes using the Affymetrix Targeted Genotyping System. Pharmacogenomics. 2007;8:293–305. doi: 10.2217/14622416.8.3.293. [DOI] [PubMed] [Google Scholar]
- 31.Daly TM, Dumaual CM, Miao X, Farmen MW, Njau RK, Fu DJ, et al. Multiplex assay for comprehensive genotyping of genes involved in drug metabolism, excretion, and transport. Clin Chem. 2007;53:1222–1230. doi: 10.1373/clinchem.2007.086348. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Lin M, Crenshaw A, Hutchinson A, Hicks B, Yeager M, et al. High-throughput single nucleotide polymorphism genotyping using nanofluidic dynamic arrays. BMC Genomics. 2009;10:561. doi: 10.1186/1471-2164-10-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millican EA, Lenzini PA, Milligan PE, Grosso L, Eby C, Deych E, et al. Genetic-based dosing in orthopedic patients beginning warfarin therapy. Blood. 2007;110:1511–1515. doi: 10.1182/blood-2007-01-069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Schwarz UI, Ritchie MD, Roden DM, Stein CM, Kurnik D. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2008;113:3925–3930. doi: 10.1182/blood-2008-09-176859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spreafico M, Lodigiani C, van Leeuwen Y, Pizzotti D, Rota LL, Rosendaal F, et al. Effects of CYP2C9 and VKORC1 on INR variations and dose requirements during initial phase of anticoagulant therapy. Pharmacogenomics. 2008;9:1237–1250. doi: 10.2217/14622416.9.9.1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.