Abstract
The NIH initiated the PharmGKB in April 2000. The primary mission was to create a repository of primary data, tools to track associations between genes and drugs, and to catalog the location and frequency of genetic variations known to impact drug response. Over the past 10 years, new technologies have shifted research from candidate gene pharmacogenetics to phenotype-based pharmacogenomics with a consequent explosion of data. PharmGKB has refocused on curating knowledge rather than housing primary genotype and phenotype data, and now, captures more complex relationships between genes, variants, drugs, diseases and pathways. Going forward, the challenges are to provide the tools and knowledge to plan and interpret genome-wide pharmacogenomics studies, predict gene–drug relationships based on shared mechanisms and support data-sharing consortia investigating clinical applications of pharmacogenomics.
Keywords: biocuration, bioinformatics, consortia, gene variant, pathway, pharmacogenomics, PharmGKB
Past
The Pharmacogenomics Knowledge Base (PharmGKB) began in 2000 as one of the first ‘post-genomic’ databases [1]. At that time, there was no standard format for the description and storage of genotype and phenotype data from pharmacogenetic studies. An important challenge was to maintain the quality of data without compromising the privacy of subjects [2]. As data collection methods advanced and pharmacogenomics outpaced pharmacogenetics, the PharmGKB adapted to increasing volumes of data and new ways to present this. Relationships were built with other resources, such as the University of California Santa Cruz (CA, USA) Genome Browser [3], Drugbank [4] and Biopax [5], to enhance the knowledge by selecting, aggregating and annotating data relevant to pharmacogenomics (Table 1).
Table 1.
Relationships and exchange of information with other resources allows PharmGKB to focus on the subset of data relevant for pharmacogenomics.
| Data type | Resources |
|---|---|
| Genes | HUGO Gene Nomenclature Committee, University of California Santa Cruz, Genome Browser, NCBI, Comparative Toxicogenomics Database, GeneCards, Source, MutDB, Gen Atlas, Ensembl, UniProt |
| Drugs | Drugbank, WHO, OpenEye, Comparative Toxicogenomics Database, Kyoto Encyclopedia of Genes and Genomes, Chemical Entities of Biological Interest |
| Diseases | Medical Subject Headings, Systematized Nomenclature of Medicine |
| Literature | NCBI |
| Pathways | BioPax, Pathways Interaction Database, Reactome, Biocarta |
| Variants | dbSNP, Alfred, HuGENet, JSnp, HapMap, Seattle SNPs, Affymetrix |
| Structures | Protein Data Bank, NCBI |
| Other | Pharmacogenetics Research Network, US FDA, Centers for Disease Control and Prevention, GAPPNet |
Initially, the main aim was gathering highly detailed primary data from the community at large and specifically, the Pharmacogenetics Research Network [6]; more than ten groups across the USA spanning a variety of different gene, drug and disease interests from asthma to thiopurine S-methyltransferase. The PharmGKB team worked with these groups to define gene variant data in detail, and how it was obtained, which formed the PharmGKB XML schema [7]. These schema allowed many useful comparisons, such as sequencing data from one researcher with RFLPs to another group working on the same gene and to see frequencies of variants in and across sample sets.
Defining phenotypes in a computationally robust manner was another challenge. Several vocabularies and ontologies were tested for describing clinical entities for the electronic medical record, but these lacked the kind of molecular detail needed for many of the Pharmacogenetics Research Network studies. The idea was to be able to integrate data across several studies, such as combining irinotecan area under the curve data from patients in a study at the University of Chicago (IL, USA) with those from Washington University (MO, USA). The computational challenges in combining such data are substantial; data may be provided in different units and collected under different conditions. The challenge of standardization requires trained curators who understand the relevant phenotypes. Data standardization can be different for every dataset, and is expensive and not possible for all phenotype data. Therefore, the PharmGKB adopted a two-level procedure. Curators capture and tag metadata for all submitted studies without a default effort in standardization. If there is a dataset of particular importance and impact, it is curated to enable comparisons across studies, for example, international normalized ratios and genotypes across the datasets from the International Warfarin Pharmacogenetics Consortium [8].
As a central knowledge-sharing site, PharmGKB noticed an opportunity to facilitate data-sharing consortia, in which investigators with complementary data create a collaboration based on a common scientific interest and the ability to combine datasets. PharmGKB then uses its curatorial staff to integrate, aggregate and annotate the contributing datasets. We have facilitated the formation of several consortia (for the pharmacogenomics of warfarin, tamoxifen and irinotecan), bringing together groups to create diverse sample sets that provide greater statistical power to detect complex associations [9]. The success of these consortia relies on a trustworthy framework for collaboration, as participants are often scientific competitors in other venues. PharmGKB involvement ensures high-quality curation, including the development of a standard template to capture the data integrating and recoding, formatting to allow comparison across the many groups and annotating with metadata to allow for computational searching. Most importantly, PharmGKB acts as an independent party and has developed a reputation as a dependable and scientifically neutral collaborator.
Present
Over the last decade, PharmGKB has collected and annotated pharmacogenomic data from a variety of sources. The published literature is a major source of knowledge, but the volume of papers is so vast that finding the information is cumbersome. We have developed structures to tag and describe relationships in the literature such that they can be found by search mechanisms but also still understood by readers. Gene, drug, disease and variant relationships have been identified and labeled with categories of interest (clinical outcomes; pharmacodynamics [PD]; pharmacokinetics [PK]; cellular and molecular functional assays and genotype data) [10]. The data are accessed from the related gene, drug and disease tabs on individual gene, drug and disease pages. Top gene pages include CYP2D6, ABCB1 and CYP2C9; the top drug pages visited are warfarin, amiodarone and clopidogrel and top diseases include Torsades de pointes, breast neoplasms and epilepsy (Table 2). We have over 4000 literature annotations (as of 17th November 2009) that link gene, drug and disease relationships. Natural language processing is used to streamline the identification of articles of interest to annotate and we are developing tools to speed up the annotation process [11].
Table 2.
Top ten PharmGKB gene, drug, disease and pathway pages for 2009.
| Rank | Genes | Drugs | Diseases | Pathways |
|---|---|---|---|---|
| 1 | CYP2D6 | Warfarin | Torsades de pointes | Platelet aggregation pathway (PD) |
| 2 | ABCB1 | Amiodarone | Breast neoplasms | Codeine and morphine pathway (PK) |
| 3 | CYP2C9 | Clopidogrel | Epilepsy | Nicotine pathway (PK) |
| 4 | CYP2C19 | Oxaliplatin | Precursor cell lymphoblastic, leukemia lymphoma | EGFR inhibitors pathway (PD) |
| 5 | CYP3A4 | Fluoxetine | Cardiovascular diseases | Antiplatelet drug clopidogrel pathway (PK) |
| 6 | VKORC1 | Capecitabine | Asthma | Bisphosphonate pathway |
| 7 | TPMT | Metoprolol | Stroke | ACE-inhibitor pathway (PD) |
| 8 | UGT1A1 | Imatinib | Hypertension | Warfarin pathway (PD) |
| 9 | CYP1A2 | Salbutamol | Type 2 diabetes mellitus | Warfarin pathway (PK) |
| 10 | CYP2B6 | Phenylephrine | Depression | Selective serotonin reuptake inhibitors pathway |
ACE: Angiotensin-converting enzyme; EGFR: EGR receptor; PD: Pharmacodynamics; PK: Pharmacokinetics.
From knowledge of the literature, PharmGKB scientists develop and maintain drug pathways with production-quality graphics and supporting scientific evidence. The PharmGKB currently has 60 curated pathways (as of 17th November 2009) illustrating PD and/or PK aspects for over 180 drugs. The top pathways viewed include platelet aggregation PD pathway, codeine and morphine PK pathway and nicotine PK pathway (Table 2). The repository of relationships built from the literature annotations now allows us to generate automated networks that can be used to start new PharmGKB pathways or downloaded for users to explore with their own methods.
PharmGKB curators have not only annotated gene–drug relationships, but have annotated specific human variations of importance to pharmacogenomics in the ‘variant annotation project’ [12]. Curators summarize the findings of pharmacogenomic relevance regarding a genomic variant and associate these with the appropriate genes, drugs and diseases. Mapping the genomic variants is not as trivial a task; many papers either do not include dbSNP identifiers or have them hidden within the methods sections, or, when they are used, the authors often neglect to specify which base is associated with the phenotype [13]. We have built a dictionary to attempt to cross-reference the various names for variants used in the literature and databases (3000 variant annotations as of 17th November, 2009). We are participating in efforts being made by the biocuration community to require inclusion of standard identifiers for variants, such as dbSNP rs number, in publications. We also write detailed online summaries of very important pharmaco genes (41 to date) and their variants, many of which have also been published [14–23].
Our user interface (Figure 1) now makes searching easier for people to get directly to genes, variants, drugs and pathways of interest. For example, users can easily find genes related to their drug of interest by entering the drug name in the gene search box or view annotations and frequency data for variants related to their drug of interest by searching with the variant box.
Figure 1. The PharmGKB homepage has targeted searching.
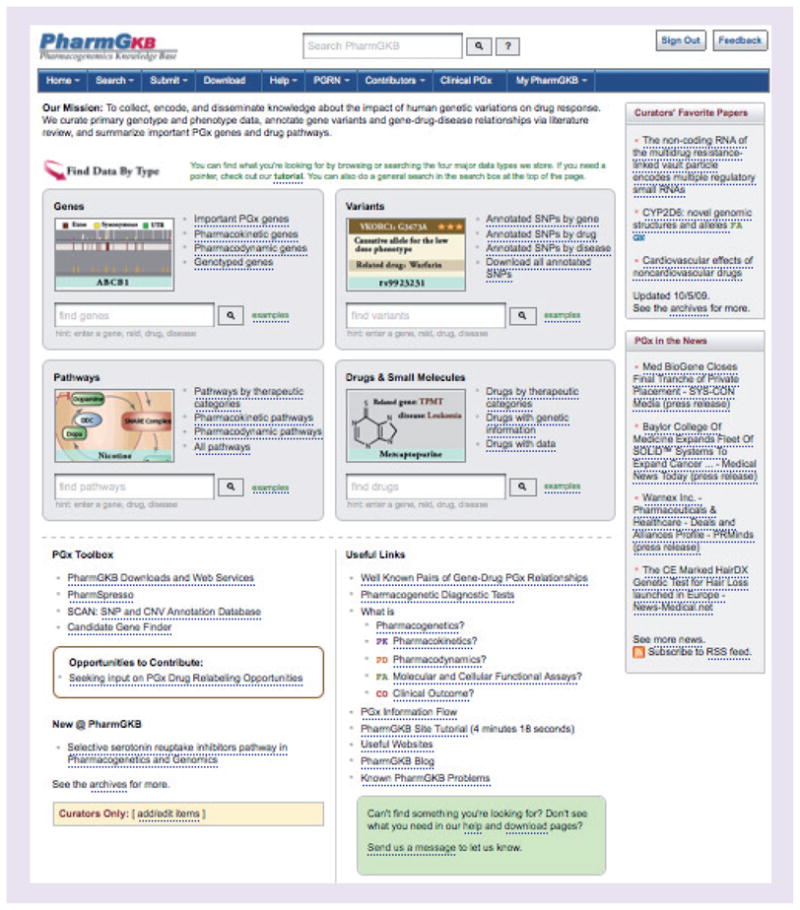
The gene search box returns links to genes associated with the search term. Example queries can be viewed and include: search by gene symbol, drug, disease, drug/disease combination, Entrez Gene ID, PubMed ID and protein name [102].
PharmGKB has received overwhelmingly positive feedback from users regarding the usefulness of PharmGKB in research, as well as educational programs and presentations. PharmGKB is used to introduce the concept of pharmacogenomics to students in medicine, pharmacy, genetics, toxicology and public health, as well as for the continuing education of medical professionals, including physicians, pharmacists and nurses. We have also used expertise from PharmGKB to pilot a pharmaco genomics project for high school students and teachers. DNATwist [101] is an interactive website that introduces basic concepts of pharmacogenomics that is also being adapted for use at the Tech Museum of Innovation in California (CA, USA) [24].
Future
The field of pharmacogenomics is at a critical juncture. Some have criticized the slow pace at which pharmacogenomic interventions are entering routine clinical practice. These critics fail to appreciate the important advances made in understanding the genetic basis of drug response, an important prerequisite for using genetics to intervene. For example, genome-wide association studies focused on drug response are just now emerging, with only a handful published to date [25]. The criticisms associated with genome-wide association studies for complex disease [26] may be less relevant for many drug responses, where common variants may have more explanatory power [27]. At the same time, our ability to sequence entire genes, exomes and even human genomes is giving us unprecedented access to rare variations, whose interpretation will be critical [28]. The key underlying need is to move from the observation of an association to an understanding of the mechanism. Only with a mechanistic understanding can we find the causative common variants in genome-wide association studies, and only with mechanistic models can we determine which rare variants (and in which combinations) explain drug-response phenotypes. PharmGKB will continue to provide the platform to examine the relationships between variants and drug response, adding new tools as new data is gathered and disseminating it to researchers and educators.
Acknowledgments
The authors would like to thank PharmGKB team members past and present without whom none of this would be possible: Dorit Berlin, John Conroy, Katrina Easton, Ray Fergerson, Li Gong, Mei Gong, Winston Gor, Joan Hebert, Tina Hernandez- Boussard, Micheal Hewett, Amy Hodge, Laura Hodges, Daniel Holbert, Mark Kiuchi, Steve Lin, Feng Liu, Xing Jian Lou, Charity Lu, Andrew MacBride, Diane Oliver, Connie Oshiro, Ryan Owen, Daniel Rubin, Katrin Sangkuhl, Farhad Shafa, Ravi Shankar, Rebecca Tang, TC Truong, Ryan Whaley, Michelle Whirl Carrillo, Mark Woon and Tina Zhou.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work is supported by the NIH/NIGMS (U01GM61374). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Caroline F Thorn, Department of Genetics, Stanford University Medical Center, Stanford, CA, USA.
Teri E Klein, Department of Genetics, Stanford University Medical Center, Stanford, CA, USA.
Russ B Altman, Email: feedback@pharmgkb.org, Department of Genetics, Stanford university Medical Center, 300 Pasteur Drive, Lane L301, Mail Code 5120, Stanford, CA 94305-5120, USA, Tel.: +1 650 725 0659, Fax: +1 650 725 3863 and Department of Bioengineering, Stanford University Medical Center, Stanford, CA, USA.
Bibliography
- 1.Klein TE, Chang JT, Cho MK, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1(3):167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 2.Lin Z, Owen AB, Altman RB. Genetics. Genomic research and human subject privacy. Science. 2004;305(5681):183. doi: 10.1126/science.1095019. [DOI] [PubMed] [Google Scholar]
- 3.Rhead B, Karolchik D, Kuhn RM, et al. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2009;38(Database issue):D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luciano JS. PAX of mind for pathway researchers. Drug Discov Today. 2005;10(13):937–942. doi: 10.1016/S1359-6446(05)03501-4. [DOI] [PubMed] [Google Scholar]
- 6.Giacomini KM, Brett CM, Altman RB, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81(3):328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whirl-Carrillo M, Woon M, Thorn CF, Klein TE, Altman RB. An XML-based interchange format for genotype–phenotype data. Hum Mutat. 2008;29(2):212–219. doi: 10.1002/humu.20662. [DOI] [PubMed] [Google Scholar]
- 8.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen RP, Altman RB, Klein TE. PharmGKB and the International Warfarin Pharmacogenetics Consortium: the changing role for pharmacogenomic databases and single-drug pharmacogenetics. Hum Mutat. 2008;29(4):456–460. doi: 10.1002/humu.20731. [DOI] [PubMed] [Google Scholar]
- 10.Altman RB, Flockhart DA, Sherry ST, Oliver DE, Rubin DL, Klein TE. Indexing pharmacogenetic knowledge on the World Wide Web. Pharmacogenetics. 2003;13(1):3–5. doi: 10.1097/00008571-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sangkuhl K, Berlin DS, Altman RB, Klein TE. PharmGKB: understanding the effects of individual genetic variants. Drug Metab Rev. 2008;40(4):539–551. doi: 10.1080/03602530802413338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu W, Ned R, Wulf A, Liu T, Khoury MJ, Gwinn M. The need for genetic variant naming standards in published abstracts of human genetic association studies. BMC Res Notes. 2009;2:56. doi: 10.1186/1756-0500-2-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owen RP, Sangkuhl K, Klein TE, Altman RB. Cytochrome P450 2D6. Pharmacogenet Genomics. 2009;19(7):559–562. doi: 10.1097/FPC.0b013e32832e0e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt M, Adjei A, Weinshilboum R, et al. Very important pharmacogene summary: sulfotransferase 1A1. Pharmacogenet Genomics. 2009;19(6):404–406. doi: 10.1097/FPC.0b013e32832e042e. [DOI] [PubMed] [Google Scholar]
- 16.Thorn CF, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for angiotensin-converting enzyme. Pharmacogenet Genomics. 2009;20(2):143–146. doi: 10.1097/FPC.0b013e3283339bf3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodges LM, Markova SM, Chinn LW, et al. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein) Pharmacogenet Genomics. 2010 doi: 10.1097/FPC.0b013e3283385a1c. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Pelleymounter L, Weinshilboum R, et al. Very important pharmacogene summary: thiopurine S-methyltransferase. Pharmacogenet Genomics. 2010 doi: 10.1097/FPC.0b013e3283352860. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Booven D, Marsh S, McLeod H, et al. Cytochrome P450 2C9–CYP2C9. Pharmacogenet Genomics. 2010;20(4):277–281. doi: 10.1097/FPC.0b013e3283349e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshiro C, Thorn CF, Roden DM, Klein TE, Altman RB. KCNH2 pharmacogenomics summary. Pharmacogenet Genomics. 2010 doi: 10.1097/FPC.0b013e3283349e9c. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina MW, Sangkuhl K, Klein TE, Altman RB. PharmGKB: very important pharmacogene –. HMGCR Pharmacogenet Genomics. 2010 doi: 10.1097/FPC.0b013e328336c81b. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen RP, Gong L, Sagrieya H, Klein TE, Altman RB. VKORC1 pharmacogenomics summary. Pharmacogenet Genomics. 2009 doi: 10.1097/FPC.0b013e32833433b6. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litonjua AA, Gong L, Duan QL, et al. Very important pharmacogene summary ADRB2. Pharmacogenet Genomics. 2010;20(1):64–69. doi: 10.1097/FPC.0b013e328333dae6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berlin DS, Person MG, Mittal A, et al. DNATwist: a web-based tool for teaching middle and high school students about pharmacogenomics. Clin Pharmacol Ther. 2010 doi: 10.1038/clpt.2009.303. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crowley JJ, Sullivan PF, McLeod HL. Pharmacogenomic genome-wide association studies: lessons learned thus far. Pharmacogenomics. 2009;10(2):161–163. doi: 10.2217/14622416.10.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 27.Nelson MR, Bacanu SA, Mosteller M, et al. Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J. 2009;9(1):23–33. doi: 10.1038/tpj.2008.4. [DOI] [PubMed] [Google Scholar]
- 28.Tabone T. Mutations, structural variations, and genome-wide resequencing: where to from here in our understanding of disease and evolution? Hum Mutat. 2008;29(6):886–890. doi: 10.1002/humu.20781. [DOI] [PubMed] [Google Scholar]
Websites
- 101.DNA Twist. http://DNATwist.org.
- 102.PharmGKB. www.pharmgkb.org.


