Overview
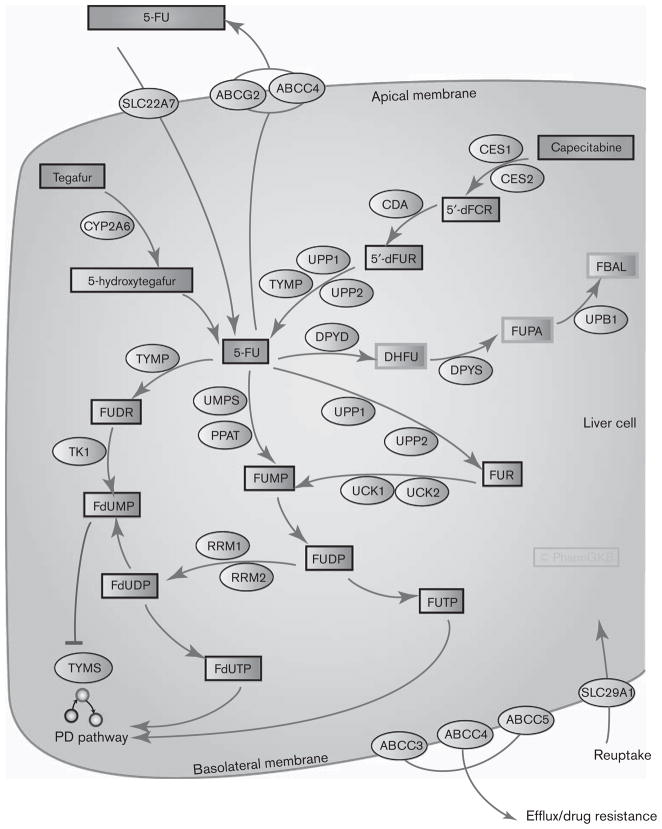
Fluoropyrimidines are antimetabolite drugs widely used in the treatment of solid tumors including colorectal cancer, breast cancer, and cancers of the aerodigestive tract [1–3]. Figure 1 shows candidate genes involved in the pharmacokinetics of three fluoropyrimidine drugs: 5-fluorouracil (5-FU), capecitabine, and tegafur. 5-FU is commonly given intravenously wherein more than 80% of it is metabolized in the liver [1]. Capecitabine is an oral prodrug of 5-FU that passes unaltered through the gut wall and is converted into 5′-deoxy-5-fluorocytidine (5′-dFCR) and then to 5′-deoxy-5-fluorouridine (5′-dFUR) in the liver by carboxylesterase and cytidine deaminase, respectively [4,5]. 5′-dFUR is then converted to 5-FU by thymidine phosphorylase or uridine phosphorylase [4,6]. Tegafur is another prodrug of 5-FU, which is converted by CYP2A6 to an unstable intermediate, 5-hydroxytegafur, which spontaneously breaks down to form 5-FU [5]. Metabolism of 5-FU is discussed further below. Figure 2 shows candidate genes involved in the pharmacodynamics of fluoropyrimidines.
Fig. 1.
Graphic representation of the candidate genes involved in fluoropyrimidine pharmacokinetics. A fully interactive version of this pathway is available online at PharmGKB at http://www.pharmgkb.org/do/serve?objId=PA150653776&objCls=Pathway. 5′-dFCR, 5′-deoxy-5-fluorocytidine; 5′-dFUR, 5′-deoxy-5-fluorouridine; 5-FU, 5-fluorouracil; ABCC, ATP-binding cassette, sub-family C; CDA, cytidine deaminase; CES, carboxylesterase; DHFU, dihydrofluorouracil; DPYD, dihydropyrimidine dehydrogenase’; DPYS, dihydropyrimidinease; FBAL, fluoro-β-alanine; FdUDP, fluorodeoxyuridine diphosphate; FdUMP, fluorodeoxyuridine monophosphate; FdUTP, fluoro-deoxyuridine triphosphate; FUDP, fluorouridine diphosphate; FUDR, fluorodeoxyuridine; FUMP, fluorouridine monophosphate; FUPA, fluoro-β-ureidopropionate; FUR, fluroridine; FUTP, fluorouridine triphosphate; PD, pharmacodynamic; PPAT, phosphoribosyl pyrophosphate amidotransferase; RRM, ribonucleotide reductase M; TK1, thymidine kinase 1; Tp53, tumor protein p53; TYMP, thymidylate phosphorylase; TYMS, thymidylate synthase; UCK, uridine-cytidine kinase; UMPS, uridine monophosphate synthase; UPB1, β-ureidopropionase 1; UPP, uridine phosphorylase.
Fig. 2.
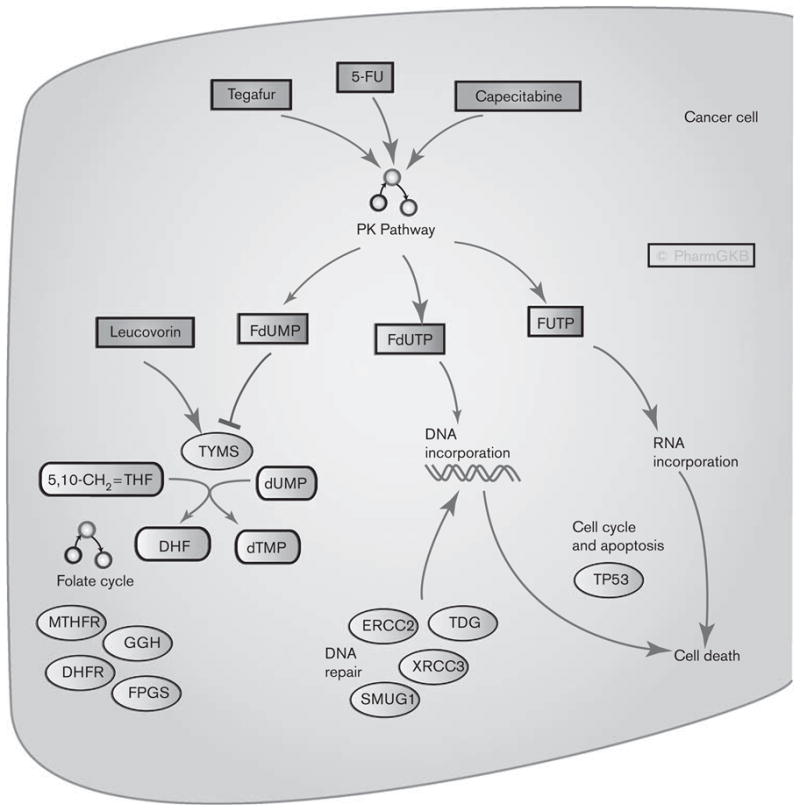
Graphic representation of the candidate genes involved in fluoropyrimidine pharmacodynamics. A fully interactive version of this pathway is available online at PharmGKB at http://www.pharmgkb.org/search/pathway/5fu/5fu-pd.jsp. DHF, dihydrofolate; DHFR, dihydrofolate reductase; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; ERCC, excision repair cross complementing; FdUMP, fluorodeoxyuridine monophosphate; FdUTP, fluoro-deoxyuridine triphosphate; FPGS, folylpolyglutamate synthase; FUTP, fluorouridine triphosphate; GGH, Gammaglutamyl hydrolase; MTHFR, methylenetetrahydrofolate reductase; PK, pharmacokinetics; SMUG1, Single-strand selective monofunctional uracil DNA glycosylase 1; TDG, thymidine DNA glycosylase; THF, 5,10-methylene tetrahydrofolate; TYMS, thymidylate synthase.
Pharmacokinetics and transport
There are several routes for the metabolism of 5-FU, some of which lead to activation and pharmacodynamic actions of the drug. The rate-limiting step of 5-FU catabolism is dihydropyrimidine dehydrogenase (DPYD) conversion of 5-FU to dihydrofluorouracil (DHFU) [7,8]. DHFU is then converted to fluoro-β-ureidopropionate (FUPA) and subsequently to fluoro-β-alanine (FBAL) by dihydropyrimidinease (DPYS) and β-ureidopropionase (UPB1), respectively [7]. Deficiency in enzymes in this pathway can result in severe and even fatal 5-FU toxicity. Several variants in DPYD have been associated with toxicity (Table 1) To modulate the activity of fluoropyrimidines, inhibitors of DPYD, such as uracil and eniluracil can be coadministered. This slows the degradation of 5-FU and improves the response rate [2].
Table 1.
Variants that have been associated with fluoropyrimidine effects
| Gene | Rs# | Common names | Variant | Phenotype | PMIDs |
|---|---|---|---|---|---|
| DPYD | rs3918290 | DPYD*2A, DPYD:67887533G > A, DPYD:IVS14 +1G>A | A | Toxicity: neurotoxicity, mucositis, leukopenia | 10071185 [25]; 18299612 [29]; 17165084 [26]; 15858133 [45] |
| DPYD | rs1801159 | DPYD*5, DPYD:I543V, DPYD:A1627G | G | Toxicity: nausea, vomiting, leukopenia | 17848752 [46] |
| DPYD | rs1801265 | DPYD*9A, DPYD:C29R, DPYD:T85C | C | Toxicity: nausea, vomiting | 17848752 [46] |
| DPYD | rs2297595 | DPYD:496A >G, DPYD:Met166Val | G | Toxicity | 19104657 [47] |
| DPYS | NA | DPYS:833G > A, DPYS:Gly278Asp | Toxicity | 14555507 [7] | |
| ERCC2 | rs13181 | ERCC2:2251A >C, ERCC2:Lys751Gln | C | Efficacy: early relapse | 18267032 [48] |
| GSTP1 | rs1695 | GSTP1:Ile105Val | G | Toxicity (chemoradiotherapy) | 18540691 [49] |
| MTHFR | rs1801133 | MTHFR:677C>T | T/C | Efficacy: response; survival Toxicity: diarrhea | 19465420 [34]; 12738713 [36] |
| MTHFR | rs1801131 | MTHFR:1298A >C | A/C | Efficacy: response; survival | 15608557 [50]; 17704422 [51] |
| TP53 | rs1042522 | P53:Arg72Pro, TP53Arg72Pro, p53 codon 72 | Arg | Efficacy: survival | 18357466 [52] |
| TYMS | rs34743033 | 28bp tandem repeat, TSER, TYMS:(CCGCGCCACTTGGC CTGCCTCCGTCCCG)2/3/4/7/8/9 | TSER*3 | Toxicity: neutropenia, diarrhea Efficacy: response; survival |
16818689 [53]; 11913730 [54]; 11556832 [55]; 14522928 [56] |
| TYMS | rs34489327 | TYMS:1494del TTAAAG, TYMS:6bp- | Deletion | Efficacy: survival | 16575011 [57]; 16141798 [58] |
| TYMS | NA | TYMS:TSER*3G>C | *3C | Efficacy: survival | 15386371 [59]; 19082493 [60] |
| UMPS | rs1801019 | OPRT: Gly213Ala | C | Toxicity: neutropenia, diarrhea | 16818689 [53] |
DPYD, dihydropyrimidine dehydrogenase; DPYS, dihydropyrimidinease; ERCC2, excision repair cross-complementing group 2; GSTP1, glutathione S-transferase pi 1; MTHFR, methylenetetrahydrofolate reductase; NA, not available; OPRT, orotate phosphoribosyltransferase; PMID, pubMed identifier; TP53, tumor protein p53; TSER, TYMS enhancer region; TYMS, thymidylate synthase; UMPS, uridine monophosphate synthase.
The main mechanism of 5-FU activation is the conversion to fluorodeoxyuridine monophosphate (FdUMP), which inhibits the enzyme thymidylate synthase (TYMS), an important part of the folate–homocysteine cycle and purine and pyrimidine synthesis (see Pharmacodynamics) [2]. The conversion of 5-FU to FdUMP can occur via thymidylate phosphorylase (TYMP) to fluorodeoxyuridine and then by the action of thymidine kinase to FdUMP or indirectly by fluorouridine monophosphate (FUMP), or fluroridine (FUR) to fluorouridine diphosphate (FUDP) and then ribonucleotide reductase action to fluorodeoxyuridine diphosphate and FdUMP [2].
An important consideration in the use of 5-FU and related drugs is the development of drug resistance by the tumor. Some resistance mechanisms involve expression changes in pharmacodynamic gene candidates [TYMS and tumor protein p53 (TP53)]. Drug resistance can also involve changes in drug transport. There is conflicting data about the transporters involved in the pharmacokinetics of 5-FU. SLC29A1 expression was not associated with survival in one study of pancreatic tumors [9], but resistance/sensitivity was associated with its expression in another study of pancreatic tumor cell lines [10]. Transport of 5-FU has been reported in an in-vitro expression system of SLC22A7 [11]. Several transporters have been implicated in 5-FU resistance including ABCG2 [12,13], ATP-binding cassette, sub-family C 3 (ABCC3), ABCC4, and ABCC5 [14].
Pharmacodynamics
The principal mechanism of action of fluoropyrimidines has been considered to be the inhibition of TYMS, but recent evidence has also shown alternative pharmacodynamic pathways acting through the incorporation of drug metabolites into the DNA and RNA [2,15,16]. The fluoropyrimidines are broken down into three metabolites that have pharmacodynamic effects, FdUMP, fluoro-deoxyuridine triphosphate (FdUTP), and fluorouridine triphosphate (FUTP) (see pharmacokinetics pathway for more details) that act through these different mechanisms. In the clinic, 5-FU is commonly given either as bolus injection with leucovorin [LV; 5-formyl tetrahydrofolate (THF)] or as a continuous infusion. The mechanism of action of 5-FU may differ with different modes of administration, with bolus treatment favoring RNA damage and continuous treatment favoring DNA damage [16,17].
FdUMP forms a covalent complex with TYMS [18] and prevents the binding and conversion of dUMP to dTMP, necessary for pyrimidine and DNA synthesis, and blocks the simultaneous conversion of 5,10-methylene tetrahydrofolate to dihydrofolate, a key component of the folate pathway that recycles methyl groups and synthesizes methionine. The inhibition of TYMS leads to an imbalance of deoxyuridine triphosphate (dUTP) and deoxythymidine triphosphate (dTTP) and a rise in the misincorporation of dUTP into DNA [19]. The complex of FdUMP and TYMS is stabilized by the coadministration of folate analogs that can bind in place of 5,10-methylene tetrahydrofolate, such as LV [18].
Owing to its involvement in the metabolism of endogenous folates, the administration of LV, folates, and the activity of other folate cycle enzymes can impact the activity of TYMS. Gamma-glutamyl hydrolase (GGH) and folylpolyglutamate (FPGS) synthase expression affect the levels of reduced folate in human colon cancer cells in vitro and thus determines the LV enhancement of 5-FU cytotoxicity [20]. The expression of dihydrofolate reductase (DHFR) has been shown to be altered in tumor cells compared with normal cells [21], and although not a direct target for fluoropyrimidines as it is for methotrexate, it may affect fluoropyrimidine pharmacodynamics through changes in folate availability. The phenotype of the tumor may also be important with respect to the folate and methylation side of this pathway, for example, the CpG island methylator phenotype of colorectal cancer, in which gene promoters are hypermethylated, has been associated with positive outcomes for 5-FU-based treatments [22].
There is some debate as to whether DNA damage caused by the incorporation of dUTP or FdUTP into DNA is the cause of cytotoxicity of fluoropyrimidines [19]. Whether it is uracil or 5-FU incorporation into DNA, the resultant damage occurs due to the increased base-excision repair causing DNA fragmentation and ultimately cell death. Single-strand selective monofunctional uracil DNA glycosylase 1 (SMUG1), a uracil-DNA glycosylase excises 5-FU from DNA and protects against the cell death in vitro [23]. A recent study gives in-vitro evidence that thymidine DNA glycosylase is the main base-excision enzyme responsible for 5-FU excision related to DNA strand breaks [19].
Pharmacogenomics
The most commonly studied genes in the pharmacogenomics (PGx) of fluoropyrimidines are DPYD, TYMS, and methylenetetrahydrofolate reductase (MTHFR) (discussed in detail below). In general, those studies that have examined the toxicity have focused on DPYD and those studies looking at efficacy consider TYMS and MTHFR. The additional variants associated with fluoropyrimidine PGx are summarized in Table 1.
Some of the most severe and fatal fluoropyrimidine toxicities observed were due to DPYD deficiencies [24]. The most common DPYD variant associated with fluoropyrimidine toxicity is DPYD*2A, a G>A single nucleotide polymorphism in the splice site of intron 14 [25] (for more details and mapping information see http://www.pharmgkb.org/search/annotatedGene/dpyd/index.jsp). Although observed at a frequency of less than 1% in the Caucasian population DPYD*2A is the most common variant associated with fluoropyrimidine toxicity [26,27]. However, this variant does not always correlate with reduced DPYD activity in vivo [28]. A recent prospective study showed that DPYD*2A had a small predictive capability for severe FU toxicity and this was more pronounced in male patients and in particular treatment regimens [29]. Other variants in DPYD have also been associated with fluoropyrimidine toxicity (see Table 1) and these are found at higher frequencies than DPYD*2A [27].
TYMS variants have been associated with TYMS expression and response to FU chemotherapy [30,31]. The most commonly studied variants are a 28 bp repeat in the 5′ untranslated region (5′-UTR) also known as TYMS enhancer region (TSER), a 6 bp deletion in the 3′-UTR and a G>C single nucleotide polymorphism within the third repeat of TSER (for more details and mapping information on these variants see http://www.pharmgkb.org/search/annotatedGene/tyms/index.jsp). Despite many studies examining the effects of these variants, contradictory findings from heterogeneous studies have shown that a clear predictive strategy has not been developed for clinical use [29,32]. A recent study of copy number variation in TYMS in colorectal tumor samples showed that high copy numbers were associated with disease relapse and death [33] indicating that simple genotyping may not provide the whole picture.
There have been several small studies of MTHFR and fluoropyrimidine PGx, mostly in colorectal cancer patients, with study sizes ranging from 43 to 331 patients and highly conflicting results [34]. The most commonly studied variants are MTHFR:677C>T and MTHFR: 1298C>A (for more details and mapping information on these variants see http://www.pharmgkb.org/search/annotatedGene/mthfr/index.jsp). For MTHFR:677C>T the T allele has been associated with worse response or shorter survival [35], better response [36,37] and had no effect on response or survival in other studies [38,39]. Similarly, the MTHFR:1298C>A A allele has been associated with shorter survival [40] and had no effect in other studies [37,39], and the TA haplotype of both the variants was associated with worse response [35].
Concluding remarks
Although there are good PGx candidates for predicting fluoropyrimidine toxicity and efficacy they have yet to be developed for routine clinical application. The impact of DPYD on toxicity is clear, however, because of the low frequencies of these variants, the unclear relationship of genotype to phenotype, and the lack of the diagnostic tools for prospective testing, it is not currently clinically relevant [41,42]. More prospective studies that address the roles of sex, treatment regimen on additional unidentified variants on DPYD genotype related toxicity [29] are needed.
Many studies of the impact of variants on tumor response, disease progression, have been confounded by cotreatment with additional antineoplastic drugs, different treatment regimens, and small study sizes. A recent review of MTHFR variants noted that those that showed no effect were often fluoropyrimidines in combination with other drugs, whereas those that showed correlations involved treatment with fluoropyrimidines alone or with LV. This suggests a pathway-based multivariant approach that may prove most effective for predicting fluoropyrimidine drug response [21,43,44].
Acknowledgments
The authors thank Fen Liu for assistance with the graphics and to Connie Oshiro for critical reading of the manuscript. This work is supported by the NIH/NIGMS (U01GM61374).
References
- 1.Diasio RB, Harris BE. Clinical pharmacology of 5-fluorouracil. Clin Pharmacokinet. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 2.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 3.Hwang JJ, Marshall JL. Capecitabine: fulfilling the promise of oral chemotherapy. Expert Opin Pharmacother. 2002;3:733–743. doi: 10.1517/14656566.3.6.733. [DOI] [PubMed] [Google Scholar]
- 4.Miwa M, Ura M, Nishida M, Sawada N, Ishikawa T, Mori K, et al. Design of a novel oral fluoropyrimidine carbamate, capecitabine, which generates 5-fluorouracil selectively in tumours by enzymes concentrated in human liver and cancer tissue. Eur J Cancer. 1998;34:1274–1281. doi: 10.1016/s0959-8049(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 5.Yen-Revollo JL, Goldberg RM, McLeod HL. Can inhibiting dihydropyrimidine dehydrogenase limit hand-foot syndrome caused by fluoropyrimidines? Clin Cancer Res. 2008;14:8–13. doi: 10.1158/1078-0432.CCR-07-1225. [DOI] [PubMed] [Google Scholar]
- 6.Cao D, Russell RL, Zhang D, Leffert JJ, Pizzorno G. Uridine phosphorylase (–/–) murine embryonic stem cells clarify the key role of this enzyme in the regulation of the pyrimidine salvage pathway and in the activation of fluoropyrimidines. Cancer Res. 2002;62:2313–2317. [PubMed] [Google Scholar]
- 7.Van Kuilenburg AB, Meinsma R, Zonnenberg BA, Zoetekouw L, Baas F, Matsuda K, et al. Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin Cancer Res. 2003;9:4363–4367. [PubMed] [Google Scholar]
- 8.Kopper L, Lapis K, Institoris L. Incorporation of 3H-dibromodulcitol and 3H-dianhydrodulcitol into ascites tumor cells. Autoradiographic study. Neoplasma. 1976;23:47–52. [PubMed] [Google Scholar]
- 9.Farrell JJ, Elsaleh H, Garcia M, Lai R, Ammar A, Regine WF, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–195. doi: 10.1053/j.gastro.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 10.Tsujie M, Nakamori S, Nakahira S, Takahashi Y, Hayashi N, Okami J, et al. Human equilibrative nucleoside transporter 1, as a predictor of 5-fluorouracil resistance in human pancreatic cancer. Anticancer Res. 2007;27:2241–2249. [PubMed] [Google Scholar]
- 11.Kobayashi Y, Ohshiro N, Sakai R, Ohbayashi M, Kohyama N, Yamamoto T. Transport mechanism and substrate specificity of human organic anion transporter 2 (hOat2 [SLC22A7]) J Pharm Pharmacol. 2005;57:573–578. doi: 10.1211/0022357055966. [DOI] [PubMed] [Google Scholar]
- 12.Yuan J, Lv H, Peng B, Wang C, Yu Y, He Z. Role of BCRP as a biomarker for predicting resistance to 5-fluorouracil in breast cancer. Cancer Chemother Pharmacol. 2009;63:1103–1110. doi: 10.1007/s00280-008-0838-z. [DOI] [PubMed] [Google Scholar]
- 13.Yuan JH, Cheng JQ, Jiang LY, Ji WD, Guo LF, Liu JJ, et al. Breast cancer resistance protein expression and 5-fluorouracil resistance. Biomed Environ Sci. 2008;21:290–295. doi: 10.1016/S0895-3988(08)60044-6. [DOI] [PubMed] [Google Scholar]
- 14.Hagmann W, Jesnowski R, Faissner R, Guo C, Lohr JM. ATP-binding cassette C transporters in human pancreatic carcinoma cell lines. Upregulation in 5-fluorouracil-resistant cells. Pancreatology. 2009;9:136–144. doi: 10.1159/000178884. [DOI] [PubMed] [Google Scholar]
- 15.Brody JR, Hucl T, Costantino CL, Eshleman JR, Gallmeier E, Zhu H, et al. Limits to thymidylate synthase and TP53 genes as predictive determinants for fluoropyrimidine sensitivity and further evidence for RNA-based toxicity as a major influence. Cancer Res. 2009;69:984–991. doi: 10.1158/0008-5472.CAN-08-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobrero AF, Aschele C, Bertino JR. Fluorouracil in colorectal cancer–a tale of two drugs: implications for biochemical modulation. J Clin Oncol. 1997;15:368–381. doi: 10.1200/JCO.1997.15.1.368. [DOI] [PubMed] [Google Scholar]
- 17.Humeniuk R, Menon LG, Mishra PJ, Gorlick R, Sowers R, Rode W, et al. Decreased levels of UMP kinase as a mechanism of fluoropyrimidine resistance. Mol Cancer Ther. 2009;8:OF1–OF8. doi: 10.1158/1535-7163.MCT-08-0716. [DOI] [PubMed] [Google Scholar]
- 18.Gmeiner WH. Novel chemical strategies for thymidylate synthase inhibition. Curr Med Chem. 2005;12:191–202. doi: 10.2174/0929867053363432. [DOI] [PubMed] [Google Scholar]
- 19.Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J, et al. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakamoto E, Tsukioka S, Oie S, Kobunai T, Tsujimoto H, Sakamoto K, et al. Folylpolyglutamate synthase and γ-glutamyl hydrolase regulate leucovorin-enhanced 5-fluorouracil anticancer activity. Biochem Biophys Res Commun. 2008;365:801–807. doi: 10.1016/j.bbrc.2007.11.043. [DOI] [PubMed] [Google Scholar]
- 21.Kidd EA, Yu J, Li X, Shannon WD, Watson MA, McLeod HL. Variance in the expression of 5-fluorouracil pathway genes in colorectal cancer. Clin Cancer Res. 2005;11:2612–2619. doi: 10.1158/1078-0432.CCR-04-1258. [DOI] [PubMed] [Google Scholar]
- 22.Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup? Int J Clin Oncol. 2008;13:498–503. doi: 10.1007/s10147-008-0854-3. [DOI] [PubMed] [Google Scholar]
- 23.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the SMUG1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–945. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 24.Milano G, Etienne MC, Pierrefite V, Barberi-Heyob M, Deporte-Fety R, Renee N. Dihydropyrimidine dehydrogenase deficiency and fluorouracil-related toxicity. Br J Cancer. 1999;79:627–630. doi: 10.1038/sj.bjc.6690098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Kuilenburg AB, Vreken P, Abeling NG, Bakker HD, Meinsma R, Van Lenthe H, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum Genet. 1999;104:1–9. doi: 10.1007/pl00008711. [DOI] [PubMed] [Google Scholar]
- 26.Saif MW, Ezzeldin H, Vance K, Sellers S, Diasio RB. DPYD*2A mutation: the most common mutation associated with DPD deficiency. Cancer Chemother Pharmacol. 2007;60:503–507. doi: 10.1007/s00280-006-0392-5. [DOI] [PubMed] [Google Scholar]
- 27.McLeod HL, Collie-Duguid ES, Vreken P, Johnson MR, Wei X, Sapone A, et al. Nomenclature for human DPYD alleles. Pharmacogenetics. 1998;8:455–459. doi: 10.1097/00008571-199812000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Collie-Duguid ES, Etienne MC, Milano G, McLeod HL. Known variant DPYD alleles do not explain DPD deficiency in cancer patients. Pharmacogenetics. 2000;10:217–223. doi: 10.1097/00008571-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, Dippon J, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 30.Marsh S. Thymidylate synthase pharmacogenetics. Invest New Drugs. 2005;23:533–537. doi: 10.1007/s10637-005-4021-7. [DOI] [PubMed] [Google Scholar]
- 31.Gusella M, Padrini R. G>C SNP of thymidylate synthase with respect to colorectal cancer. Pharmacogenomics. 2007;8:985–996. doi: 10.2217/14622416.8.8.985. [DOI] [PubMed] [Google Scholar]
- 32.Marsh S. Impact of pharmacogenomics on clinical practice in oncology. Mol Diagn Ther. 2007;11:79–82. doi: 10.1007/BF03256226. [DOI] [PubMed] [Google Scholar]
- 33.Jensen SA, Vainer B, Witton CJ, Jorgensen JT, Sorensen JB. Prognostic significance of numeric aberrations of genes for thymidylate synthase, thymidine phosphorylase and dihydrofolate reductase in colorectal cancer. Acta Oncol. 2008;47:1054–1061. doi: 10.1080/02841860801942158. [DOI] [PubMed] [Google Scholar]
- 34.Afzal S, Jensen SA, Vainer B, Vogel U, Matsen JP, Sorensen JB, et al. MTHFR polymorphisms and 5-FU-based adjuvant chemotherapy in colorectal cancer. Ann Oncol. 2009;20:1660–1666. doi: 10.1093/annonc/mdp046. [DOI] [PubMed] [Google Scholar]
- 35.Terrazzino S, Agostini M, Pucciarelli S, Pasetto LM, Friso ML, Ambrosi A, et al. A haplotype of the methylenetetrahydrofolate reductase gene predicts poor tumor response in rectal cancer patients receiving preoperative chemoradiation. Pharmacogenet Genomics. 2006;16:817–824. doi: 10.1097/01.fpc.0000230412.89973.c0. [DOI] [PubMed] [Google Scholar]
- 36.Cohen V, Panet-Raymond V, Sabbaghian N, Morin I, Batist G, Rozen R. Methylenetetrahydrofolate reductase polymorphism in advanced colorectal cancer: a novel genomic predictor of clinical response to fluoropyrimidine-based chemotherapy. Clin Cancer Res. 2003;9:1611–1615. [PubMed] [Google Scholar]
- 37.Jakobsen A, Nielsen JN, Gyldenkerne N, Lindeberg J. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphism in normal tissue as predictors of fluorouracil sensitivity. J Clin Oncol. 2005;23:1365–1369. doi: 10.1200/JCO.2005.06.219. [DOI] [PubMed] [Google Scholar]
- 38.Marcuello E, Altes A, Menoyo A, Rio ED, Baiget M. Methylenetetrahydrofolate reductase gene polymorphisms: genomic predictors of clinical response to fluoropyrimidine-based chemotherapy? Cancer Chemother Pharmacol. 2006;57:835–840. doi: 10.1007/s00280-005-0089-1. [DOI] [PubMed] [Google Scholar]
- 39.Ruzzo A, Graziano F, Loupakis F, Santini D, Catalano V, Bisonni R, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFIRI chemotherapy. Pharmacogenomics J. 2008;8:278–288. doi: 10.1038/sj.tpj.6500463. [DOI] [PubMed] [Google Scholar]
- 40.Capitain O, Boisdron-Celle M, Poirier AL, Abadie-Lacourtoisie S, Morel A, Gamelin E. The influence of fluorouracil outcome parameters on tolerance and efficacy in patients with advanced colorectal cancer. Pharmacogenomics J. 2008;8:256–267. doi: 10.1038/sj.tpj.6500476. [DOI] [PubMed] [Google Scholar]
- 41.Yen JL, McLeod HL. Should DPD analysis be required prior to prescribing fluoropyrimidines? Eur J Cancer. 2007;43:1011–1016. doi: 10.1016/j.ejca.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 42.Maitland ML, Vasisht K, Ratain MJ. TPMT, UGT1A1 and DPYD: genotyping to ensure safer cancer therapy? Trends Pharmacol Sci. 2006;27:432–437. doi: 10.1016/j.tips.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 43.De Mattia E, Toffoli G. C677T and A1298C MTHFR polymorphisms, a challenge for antifolate and fluoropyrimidine-based therapy personalisation. Eur J Cancer. 2009;45:1333–1351. doi: 10.1016/j.ejca.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Gu J, Wu TT, Swisher SG, Liao Z, Correa AM, et al. Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol. 2006;24:3789–3798. doi: 10.1200/JCO.2005.03.6640. [DOI] [PubMed] [Google Scholar]
- 45.Steiner M, Seule M, Steiner B, Bauer I, Freund M, Köhne CH, et al. 5-Fluorouracil/irinotecan induced lethal toxicity as a result of a combined pharmacogenetic syndrome: report of a case. J Clin Pathol. 2005;58:553–555. doi: 10.1136/jcp.2004.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Li YM, Jin X. DPYD*5 gene mutation contributes to the reduced DPYD enzyme activity and chemotherapeutic toxicity of 5-FU: results from genotyping study on 75 gastric carcinoma and colon carcinoma patients. Med Oncol. 2007;24:251–258. doi: 10.1007/BF02698048. [DOI] [PubMed] [Google Scholar]
- 47.Gross E, Busse B, Riemenschneider M, Neubauer S, Seck K, Klein HG, et al. Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients. PLoS One. 2008;3:e4003. doi: 10.1371/journal.pone.0004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang MY, Fang WY, Lee SC, Cheng TL, Wang JY, Lin SR. ERCC2 2251A >C genetic polymorphism was highly correlated with early relapse in high-risk stage II and stage III colorectal cancer patients: a preliminary study. BMC Cancer. 2008;8:50. doi: 10.1186/1471-2407-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agostini M, Pasetto LM, Pucciarelli S, Terrazzino S, Ambrosi A, Bedin C, et al. Glutathione S-transferase P1 Ile105Val polymorphism is associated with haematological toxicity in elderly rectal cancer patients receiving preoperative chemoradiotherapy. Drugs Aging. 2008;25:531–539. doi: 10.2165/00002512-200825060-00006. [DOI] [PubMed] [Google Scholar]
- 50.Etienne MC, Formento JL, Chazal M, Francoual M, Magné N, Formento P, et al. Methylenetetrahydrofolate reductase gene polymorphisms and response to fluorouracil-based treatment in advanced colorectal cancer patients. Pharmacogenetics. 2004;14:785–792. doi: 10.1097/00008571-200412000-00001. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W, Press OA, Haiman CA, Yang DY, Gordon MA, Fazzone W, et al. Association of methylenetetrahydrofolate reductase gene polymorphisms and sex-specific survival in patients with metastatic colon cancer. J Clin Oncol. 2007;25:3726–3731. doi: 10.1200/JCO.2007.11.4710. [DOI] [PubMed] [Google Scholar]
- 52.Huang ZH, Hua D, Li LH, Zhu JD. Prognostic role of p53 codon 72 polymorphism in gastric cancer patients treated with fluorouracil-based adjuvant chemotherapy. J Cancer Res Clin Oncol. 2008;134:1129–1134. doi: 10.1007/s00432-008-0380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ichikawa W, Takahashi T, Suto K, Sasaki Y, Hirayama R. Orotate phosphoribosyltransferase gene polymorphism predicts toxicity in patients treated with bolus 5-fluorouracil regimen. Clin Cancer Res. 2006;12:3928–3934. doi: 10.1158/1078-0432.CCR-05-2665. [DOI] [PubMed] [Google Scholar]
- 54.Pullarkat ST, Stoehlmacher J, Ghaderi V, Xiong YP, Ingles SA, Sherrod A, et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 55.Iacopetta B, Grieu F, Joseph D, Elsaleh H. A polymorphism in the enhancer region of the thymidylate synthase promoter influences the survival of colorectal cancer patients treated with 5-fluorouracil. Br J Cancer. 2001;85:827–830. doi: 10.1054/bjoc.2001.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawakami K, Watanabe G. Identification and functional analysis of single nucleotide polymorphism in the tandem repeat sequence of thymidylate synthase gene. Cancer Res. 2003;63:6004–6007. [PubMed] [Google Scholar]
- 57.Dotor E, Cuatrecases M, Martínez-Iniesta M, Navarro M, Vilardell F, Guinó E, et al. Tumor thymidylate synthase 1494del6 genotype as a prognostic factor in colorectal cancer patients receiving fluorouracil-based adjuvant treatment. J Clin Oncol. 2006;24:1603–1611. doi: 10.1200/JCO.2005.03.5253. [DOI] [PubMed] [Google Scholar]
- 58.Hitre E, Budai B, Adleff V, Czeglédi F, Horváth Z, Gyergyay F, et al. Influence of thymidylate synthase gene polymorphisms on the survival of colorectal cancer patients receiving adjuvant 5-fluorouracil. Pharmacogenet Genomics. 2005;15:723–730. doi: 10.1097/01.fpc.0000175598.42141.59. [DOI] [PubMed] [Google Scholar]
- 59.Marcuello E, Altés A, del Rio E, César A, Menoyo A, Baiget M. Single nucleotide polymorphism in the 5′ tandem repeat sequences of thymidylate synthase gene predicts for response to fluorouracil-based chemotherapy in advanced colorectal cancer patients. Int J Cancer. 2004;112:733–737. doi: 10.1002/ijc.20487. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez-Contreras ME, Sánchez-Hernández JJ, González E, Herráez B, Domínguez I, Lozano M, et al. Combination of polymorphisms within 5′ and 3′ untranslated regions of thymidylate synthase gene modulates survival in 5 fluorouracil-treated colorectal cancer patients. Int J Oncol. 2009;34:219–229. [PubMed] [Google Scholar]