Abstract
Vibrio vulnificus is an opportunistic human pathogen commonly found in estuarine environments. Infections are associated with raw oyster consumption and can produce rapidly fatal septicemia in susceptible individuals. Standard enumeration of this organism in shellfish or seawater is laborious and inaccurate; therefore, more efficient assays are needed. An oligonucleotide probe derived from the cytolysin gene, vvhA, was previously used for colony hybridizations to enumerate V. vulnificus. However, this method requires overnight growth, and vibrios may lack culturability under certain conditions. In the present study, we targeted the same locus for development of a TaqMan real-time PCR assay. Probe specificity was confirmed by amplification of 28 V. vulnificus templates and by the lack of a PCR product with 22 non-V. vulnificus strains. Detection of V. vulnificus in pure cultures was observed over a 6-log-unit linear range of concentration (102 to 108 CFU ml−1), with a lower limit of 72 fg of genomic DNA μl of PCR mixture−1 or the equivalent of six cells. Similar sensitivity was observed in DNA extracted from mixtures of V. vulnificus and V. parahaemolyticus cells. Real-time PCR enumeration of artificially inoculated oyster homogenates correlated well with colony hybridization counts (r2 = 0.97). Numbers of indigenous V. vulnificus cells in oysters by real-time PCR showed no significant differences from numbers from plate counts with probe (t test; P = 0.43). Viable but nonculturable cells were also enumerated by real-time PCR and confirmed by the BacLight viability assay. These data indicate that real-time PCR can provide sensitive species-specific detection and enumeration of V. vulnificus in seafood.
Vibrio vulnificus produces a rapidly fatal septicemia, which is primarily associated with the ingestion of raw oysters(3, 16). Contact of wounds with seawater or shellfish can also lead to serious infections that can progress to septicemia or require limb amputation. Methods for examination of seafood safety currently rely on fecal coliform analysis; however, V. vulnificus is not associated with fecal contamination (30). Although current standards do not regulate the number of V. vulnificus organisms in shellfish, the Food and Drug Administration supports postharvest treatments that will greatly reduce or eliminate the numbers of this organism. Therefore, quantitative methods are needed to accurately and efficiently validate application of these treatments to the seafood industry.
V. vulnificus is indigenous to estuaries worldwide and can be readily isolated from the environment (24, 30, 37); however, standard detection methods require enrichment and selective plating media to reduce the growth of background organisms (10). Species identification requires additional time-consuming assays that are frequently inaccurate and laborious. Standard plate count based on DNA probe hybridization for colony identification can also be used for enumeration in oyster tissues (19, 36, 37), but this assay still requires overnight growth of bacteria and usually involves several days of processing time for large numbers of samples. Numerous studies have indicated that, under conditions of reduced temperatures (4 to 5°C) and nutrient availability, vibrios become nonculturable on standard media while retaining viability (8, 13, 26, 32, 33). Although the contribution of a viable but nonculturable (VBNC) population to V. vulnificus disease is unknown, human infections have been reported with VBNC V. cholerae (8). Therefore, enumeration assays that do not require cultivation may be useful for risk assessment. Several PCR assays (5, 6, 23), as well as reverse transcription-PCR (RT-PCR) (9) for detection of vibrios without culture have been described; unfortunately, these methods are not inherently quantitative and require post-PCR analysis for enumeration of V. vulnificus.
Real-time PCR offers rapid, quantitative analysis for detection of food-borne pathogens (1, 2, 7, 18, 21, 22). The TaqMan system (PE Applied Biosystems Inc., Foster City, Calif.) uses fluorogenic probes to detect PCR products as they form; the exonuclease activity of Taq polymerase releases a labeled reporter dye at the 5′ end of the probe from the quencher dye at the 3′ end with each cycle of amplification. Thus, increased fluorescence is directly proportional to the formation of PCR products. Plotting the increase in fluorescence versus cycle number gives a comprehensive picture of the PCR process, and quantification of initial template concentration can be calculated from data on the exponential phase of amplification.
The most frequent target of species-specific V. vulnificus DNA probes and PCR assays is the hemolysin/cytolysin gene, vvhA (39). The present study also employed this sequence to develop a quantitative real-time PCR assay with TaqMan technology. Assay sensitivity and specificity were examined in pure or mixed cultures, and enumeration of V. vulnificus by real-time PCR, in either artificially inoculated or naturally contaminated oysters, was compared to plate count determinations with a gene probe. VBNC cells were also examined by real-time PCR analysis.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
A total of 50 bacterial strains, including V. vulnificus (n = 28) and non-V. vulnificus strains (n = 22), were used to evaluate the specificity of the TaqMan probe and primers (Table 1). V. vulnificus and V. cholerae strains were obtained from Glenn Morris, Jr., University of Maryland School of Medicine, Baltimore, Md. V. parahaemolyticus strains were obtained from Angelo Depaola at Gulf Coast Seafood Laboratory, Food and Drug Administration, Dauphin Island, Ala. The strains of Salmonella enterica, Listeria monocytogenes, Escherichia coli, Pseudomonas aeruginosa, and Shigella flexneri were provided by ABC Research Corporation, Gainesville, Fla. Bacterial strains were grown in an agitating incubator (New Brunswick Scientific, Edison, N.J.) overnight at appropriate growth temperatures in Luria broth (LB) prepared with 1.0% tryptone, 0.5% yeast extract, and 1.0% NaCl. Unless stated otherwise, all media were purchased from Difco (Sparks, Md.) and reagents were from Sigma Chemicals (St. Louis, Mo.). Strains were stored in LB with 50% glycerol at −70°C. Serial dilutions of cultures in artificial seawater (ASW), prepared with 18-ppt synthetic sea salt (Instant Ocean; Aquarium System; Mentor), were used to enumerate cells by spread plating on LB agar (LA) in triplicate with overnight incubation at 35 or 37°C.
TABLE 1.
Bacterial isolates evaluated by real-time PCR
| Strain | Sourcea | PCR resultb |
|---|---|---|
| V. vulnificus | ||
| MO6-24/O | Clinical, Calif. | + |
| MO6-24/T | MO6-24/O phase variant | + |
| MO6-24/31T | MO6-24/O acapsular mutant | + |
| CVD752 | MO6-24/O acapsular mutant | + |
| CVD737 | MO6-24/O acapsular mutant | + |
| E4125/O | Clinical | + |
| NJMSA/O | Clinical, N.J. | + |
| BO6312/O | Clinical, Md. | + |
| 52785/O | Clinical, N.Y. | + |
| 6353/O | Clinical, Md. | + |
| 1015H | Clinical, La. | + |
| EDL174/O | Clinical, Ga. | + |
| LC4/O | Clinical | + |
| LC4/T | LC4/O phase variant | + |
| 5C1326/O | Clinical, Md. | + |
| 85A667/O | Clinical, Calif. | + |
| 2400112 | Clinical, Fla. | + |
| LL728 | Clinical, Fla. | + |
| VV1009 | Environmental, Fla. | + |
| 345/O | Environmental, La. | + |
| 345/T | 345/O phase variant | + |
| UNCC 913 | Environmental, N.C. | + |
| UNCC1015 | Environmental, N.C. | + |
| MLT 365 | Environmental, Fla. | + |
| MLT 367 | Environmental, Fla. | + |
| MLT 403 | Environmental, Fla. | + |
| 409/O | Unknown | + |
| 80363/O | Unknown | + |
| V. cholerae | ||
| NRT36S | Clinical, Japan | − |
| JVB52 | NRT36S phase variant | − |
| A 5 | Environmental | − |
| JVB25 | Unknown | − |
| 2076 | Unknown | − |
| YVY 210 | Unknown | − |
| YVY 212 | Unknown | − |
| V. parahaemolyticus | ||
| ATCO 1782 | Clinical, Japan | − |
| NY 3547 | Clinical, N.Y. | − |
| TX2103 | Clinical, Tex. | − |
| NY3483 | Clinical, N.Y. | − |
| 10290 | Clinical, Wash. | − |
| ATCC 43996 | Clinical, England | − |
| Salmonella enterica | ||
| Cholerasius 10708 | Clinical | − |
| Enteridis 13076 | Clinical | − |
| L. monocytogenes | ||
| ATCC 7644 | Clinical | − |
| ATCC 49594 | Clinical | − |
| E. coli | ||
| O157:H7 | Clinical | − |
| JM109 | K-12 | − |
| ATCC 25922 | Clinical, Wash. | − |
| P. aeruginosa ATCC 15442 | Environmental | − |
| Shigella flexneri ATCC 12022 | Clinical | − |
Suppliers of bacterial strains are provided in the text.
Real-time PCR assay results are shown as either positive ΔRn (+) or no observed amplification (−) as described in Materials and Methods.
DNA extractions.
Bacterial cultures (1.0 ml) and whole-oyster homogenates (250 μl; see details below) were centrifuged at 5,000 × g for 10 min, and pellets were extracted with a QIAamp DNA minikit (Qiagen, Valencia, Calif.). DNA yield and purity were determined spectrophotometrically by measuring 260-nm/280-nm absorbance ratios (SPECTRAmax Plus 384; Molecular Devices, Sunnyvale, Calif.). DNA was concentrated by ethanol precipitation, followed by centrifugal vacuum evaporation (DNA Speed Vac; Savant Instruments Inc., Holbrook, N.Y.). Pellets were suspended in 50 or 100 μl of Tris-EDTA buffer (pH 8.0; 10 mM Tris, 1 mM EDTA) and incubated at 65°C for 10 min to solubilize DNA. Concentrated DNA was stored at −20°C.
V. vulnificus real-time PCR assay.
Oligonucleotide sequences were derived from the V. vulnificus structural gene for cytolysin, vvhA (GenBank accession number M34670). The probe is localized to a region of the gene that previously had been used as a species-specific genetic probe (36). Primer Express software (version 1.5; PE Applied Biosystems) was used to design the TaqMan probe (5′CCG TTA ACC GAA CCA CCC GCA A3′) and the forward (5′TGT TTA TGG TGA GAA CGG TGA CA3′) and reverse (5′TTC TTT ATC TAG GCC CCA AAC TTG3′) PCR primer set. Probe and primers were assessed for species specificity by a BLAST (National Center for Biotechnology Information) search to determine homology to known sequences.
Real-time PCR assays used the TaqMan (PE Applied Biosystems) reagents and technology. This assay is based on the fluorescence emitted from the cleavage of a reporter dye during PCR; fluorescence of the reporter dye is suppressed in the intact probe due to the close proximity of a quencher dye. PCR amplification reaction mixtures (50 μl) contained various concentrations of the DNA sample (3.0 μl), TaqMan buffer A (5 mM MgCl2; 200 μM dATP, dGTP, dCTP, and 400 μM dUTP), a TaqMan fluorogenic probe (0.25 μM), primers (0.90 μM each), and AmpliTaq Gold DNA polymerase (2.5 U). Reactions were performed in triplicate for each PCR sample in capped 0.2-ml thin-walled reaction tubes (Bio-Rad Laboratories, Hercules, Calif.). The PCR protocol consisted of holding samples at 50°C for 2 min, followed by denaturation at 95°C for 10 min and then by 40 cycles of 95°C for 15 s and 60°C for 1 min. Reactions were performed with the GeneAmp 5700 sequence detection system, and data were analyzed with GeneAmp 5700 sequence detection system software (PE Applied Biosystems). The amplified PCR product was detected by monitoring the increase in fluorescence signal generated from the 6-carboxyfluorescein-labeled probe. Quencher dye was TAMARA (6-carboxy-N,N,N′,N′-tetramethylrhodamine). Amplified PCR products (100 bp) were verified by electrophoresis with a 4% low-melting-temperature agarose (NuSieve GTG; BioWhittaker Molecular Applications, Rockland, Maine) with ethidium bromide staining as described by Sambrook et al. (28).
Fluorescence intensity, as an indicator of amplicon concentration, was calculated from the equation ΔRn = (Rn+) − (Rn−) (reporter signal fluorescence minus normalized background). The signal amplification (ΔRn) was then plotted against PCR cycles to generate cycle threshold (Ct) values. The Ct is the initial cycle in which amplification is detected as exceeding an arbitrary threshold. Standard curves for quantitation were plotted from triplicate samples by using Ct values of 10-fold dilutions of template extracted from 2 × 109 CFU of V. vulnificus MO6-24/O ml−1. The Ct values for experimental samples were also calculated from the means of triplicate PCRs.
Sensitivity and specificity of V. vulnificus real-time PCR.
Specificity of real-time PCR was determined by comparing PCR products derived from V. vulnificus (n = 28) to non-V. vulnificus (n = 22) DNA templates. For sensitivity assays, DNA was extracted from pure cultures of V. vulnificus MO6-24/O (2 × 109 CFU ml−1) and serially diluted in Tris-EDTA buffer to generate a standard curve used for enumeration of unknown samples. Enumeration of V. vulnificus by real-time PCR was compared to plate count values. Cells were incubated at 30°C with agitation in LB until early stationary phase. Samples were serially diluted in ASW to determine CFU per milliliter on LA plates. DNA from diluted cells was extracted and precipitated with ethanol as described above. These experiments were repeated with or without the addition of V. parahaemolyticus strain NY 3547 (3.2 × 106 CFU ml−1) cells prior to DNA extractions in order to determine the influence of background DNA.
DNA probing of colony blot hybridizations.
V. vulnificus from oyster homogenates was enumerated using the species-specific VVAP oligonucleotide probe and colony blot hybridization procedure previously described by Wright et al. (36). Briefly, colonies from plate counts on LA were transferred in triplicate to Whatman (Maidstone, England) 541 filters and hybridized with a species-specific alkaline phosphatase-labeled probe (DNA Technologies A/S Denmark). Probe-positive colonies were detected by measuring alkaline phosphatase enzymatic activity with an appropriate chromogenic substrate and were enumerated to determine concentrations of V. vulnificus.
Detection of V. vulnificus in artificially inoculated oysters.
Oyster (Crassostrea virginica) meats (ca. 30 g) were diluted (1:10) in ASW and homogenized (Lab-Blender 400 stomacher; Tekmar Company, Cincinnati, Ohio) for 90 s. Oyster homogenates (10 ml) were seeded with early-stationary-phase V. vulnificus MO6-24/O cells diluted in ASW to concentrations of 102 to 107 CFU ml−1. Bacterial inocula were determined by colony blot hybridization, as described above. Aliquots (1.0 ml) of seeded homogenates were centrifuged (5,000 × g for 10 min), and DNA was extracted and precipitated with ethanol for real-time PCR quantification, as described above.
Detection of indigenous V. vulnificus in oysters.
Oysters were purchased from Florida gulf coast wholesalers, stored at 4 ± 2.0°C, and assayed within 2 days of harvest date. To increase background levels of V. vulnificus, oysters were incubated, with agitation, in a 30°C water bath (Gyrotory; New Brunswick Scientific, Edison, N.J.) for 12 to 24 h in ASW. Oyster sample preparation included scrubbing with a brush under running tap water and shucking using an aseptic technique. Thirty to 50 g of oyster meat (approximately three to five oysters) was homogenized with the stomacher or a conventional blender (Waring Commercial, Torrington, Colo.) for 90 s in an equal weight of ASW. Homogenates (250 μl) were centrifuged (5,000 × g for 10 min), and DNA was extracted and precipitated with ethanol as described above. Magnetic bead DNA purification (Dynal AS, Oslo, Norway) was also performed on selected samples as previously described (18). V. vulnificus cells in dilutions of oyster homogenates were enumerated by both real-time PCR and colony blot hybridization as described above. Statistical analysis was based on the Student t test using paired and equal variance of the log CFU per gram.
Enumeration of VBNC cells.
To evaluate real-time PCR detection of VBNC cells, V. vulnificus MO6-24/O was incubated at 30°C in LB until the early-stationary-growth phase (109 CFU ml−1). Cells (1.0 ml) were centrifuged (5,000 × g for 10 min), washed three times in 1.0 ml of ASW to remove residual nutrients from the growth media, and resuspended in ASW. Microcosm flasks (n = 2) containing ASW (99 ml) were inoculated with washed cultures (1.0 ml) to achieve an inoculum concentration of ca. 107 CFU ml−1. Microcosms were stored at 4 ± 2.0°C and monitored to assess VBNC induction during a 43-day period by real-time PCR assay, plate counts, and growth in alkaline peptone water (APW), as previously described (10). Cultures were considered nonculturable when plate counts were below detectable levels (<100 CFU ml−1) and no visible turbidity was observed in APW enrichment tubes. The viability of cells was assessed with nucleic acid staining dyes (LIVE/DEAD BacLight bacterial viability kit; Molecular Probes, Eugene, Oreg.). This two-color fluorescence assay determines bacterial viability based on cell membrane integrity. Cell membranes of all bacteria cells stained with SYTO 9 have a green fluorescence. Propidium iodide stain penetrates bacteria with damaged cell membranes and generates a red fluorescence. Therefore, viable bacterial cells with intact membranes stain green, while nonviable cells are red. Acridine orange direct counts (AODC) of samples were estimated with 0.01% acridine orange staining (J. T. Baker Chemical Co., Phillipsburg, N.J.). Fluorescent cells in microcosm samples were visualized and enumerated with a fluorescence microscope (Nikon; Labphot) in a counting chamber (Bright-Line; Hausser Scientific, Horsham, Pa.). To further confirm the viability of VBNC cells, cultures were resuscitated following a temperature upshift of the microcosm as described by Whitesides and Oliver (33). Samples (10 ml) of VBNC microcosms were incubated at room temperature for 24 h, and culturability was determined by plate counts and growth in APW enrichment broth.
RESULTS
Real-time PCR assay.
PCR primers and a TaqMan probe were designed to target and overlap the nucleotide region of the species-specific DNA probe (VVAP) described previously (19, 36). The 100-bp amplicon of the hemolysin showed no homology to the published sequence in the GenBank database. Species specificity was confirmed by positive signal amplification (ΔRn) of all 28 isolates of V. vulnificus and lack of product from all 22 non-V. vulnificus strains (Table 1). Amplified PCR product size was verified by gel electrophoresis (not shown).
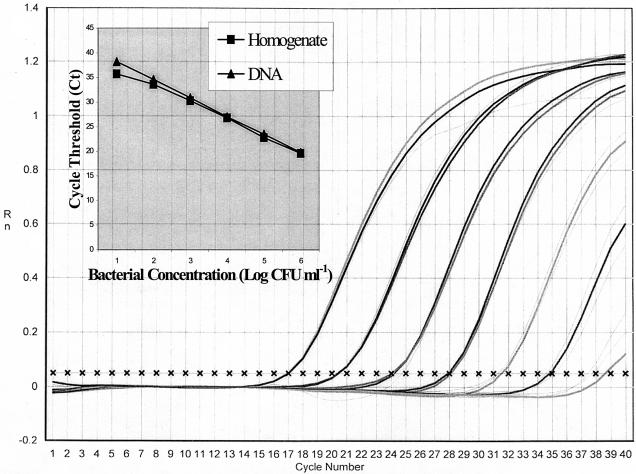
Limits of sensitivity of V. vulnificus real-time PCR were determined from end point titration of DNA extracted from pure culture. Linear values for PCR amplification were achieved for dilutions of purified DNA concentrations ranging from 7.2 × 10−5 to 72.0 ng μl−1 (Fig. 1). Real-time PCR amplification of DNA obtained from serial dilutions of pure culture in ASW was detected at <250 CFU ml−1 but showed a linear range of detection from 2.3 × 102 to 2.3 × 108 CFU ml−1 (log 2.4 to 8.4 CFU ml−1) based on plate counts (Table 2). The limit of linear detection in the PCR assay with a total reaction volume of 50 μl using 3.0 μl of genomic DNA, assuming 100% extraction efficiency, was extrapolated to be six cells per PCR. Comparison of bacterial concentrations, as determined by plate counts on LA and real-time PCR results, showed excellent correlation (r2 = 0.99). Linear detection of amplification product also was observed in the presence of exogenous cells, with 3.2 × 106 CFU of V. parahaemolyticus NY 3547 ml−1 added to V. vulnificus cultures (Table 2) and showed correlation to real-time PCR enumeration without added V. parahaemolyticus (r2 = 0.99) and to plate counts (r2 = 0.99).
FIG. 1.
Standard curves for V. vulnificus real-time PCR. Standard curves were plotted for the log cell number of bacteria versus the number of cycles required to reach Ct and were based on the means of triplicate samples. Samples were derived from dilutions of DNA extracted from cells either in pure culture or in oyster homogenates. Equations of lines for pure culture and homogenates were y = 3.50x + 14.5 and y = 3.33x + 30.7, respectively.
TABLE 2.
Sensitivity of V. vulnificus real-time PCR assay
| Plate countsa (log CFU ml−1 ± SD) | Real-time PCR resultb (log CFU ml−1 ± SD) (Ct ± SD) with V. parahaemolyticus:
|
|
|---|---|---|
| Absent | Present | |
| 8.4 ± 0.07 | 7.9 ± 0.02 (17.7 ± 0.08) | 7.9 ± 0.01 (17.8 ± 0.04) |
| 7.5 ± 0.11 | 7.2 ± 0.03 (20.1 ± 0.10) | 7.1 ± 0.04 (20.5 ± 0.14) |
| 6.5 ± 0.02 | 6.4 ± 0.05 (23.0 ± 0.17) | 6.3 ± 0.02 (23.4 ± 0.06) |
| 5.6 ± 0.03 | 5.4 ± 0.07 (26.6 ± 0.25) | 5.3 ± 0.04 (26.8 ± 0.16) |
| 4.5 ± 0.08 | 4.2 ± 0.03 (30.8 ± 0.11) | 4.3 ± 0.04 (30.5 ± 0.14) |
| 3.4 ± 0.04 | 3.3 ± 0.22 (34.1 ± 0.78) | 3.4 ± 0.17 (33.6 ± 0.60) |
| 2.4 ± 0.07 | 2.6 ± 0.08 (36.5 ± 0.27) | 2.5 ± 0.17 (36.6 ± 0.60) |
| <250 | 2.8 ± 0.04 (37.2 ± 0.16) | 2.7 ± 0.10 (37.5 ± 0.35) |
V. vulnificus MO6-24/O levels were determined by plate counts on LA spread plates as described in the text.
Real-time PCR determination of V. vulnificus concentrations used a standard curve for the mean of triplicate Ct values, for 10-fold dilutions of DNA template extracted with or without V. parahaemolyticus cells at a concentration of 3.2 × 106 CFU ml−1. Data represent means of two individual experiments based on triplicate samples for each.
Enumeration of V. vulnificus in artificially inoculated oyster homogenate.
Oyster homogenates were seeded with pure cultures of V. vulnificus ranging from 102 to 107 CFU ml−1. The background of indigenous V. vulnificus cells from uninoculated homogenate was determined to be 2.1 × 103 CFU ml−1 by real-time PCR. As shown in Table 3, the presence of oyster tissue did not affect real-time PCR detection of V. vulnificus, and results of colony blot hybridization enumeration using the VVAP gene probe showed good correlation (r2 = 0.97) between the two assays. Comparison of standard curves, based on either dilutions of DNA template extracted from pure culture or DNA extracted from serial dilutions of bacteria in seeded oyster homogenates, also showed excellent agreement (Fig. 1). Thus, although the addition of homogenate did not appear to interfere with the real-time PCR assay, sensitivity in these experiments was only assessable above the threshold of V. vulnificus background in uninoculated oysters. Therefore, we used standard curves derived from pure cultures for subsequent studies to eliminate problems with background contamination.
TABLE 3.
Enumeration of V. vulnificus from inoculated oysters
| Inoculum (log CFU ml−1) | Result for:
|
|
|---|---|---|
| DNA probea (log CFU ml−1 ± SD) | Real-time PCRb (log CFU ml−1 ± SD) (Ct ± SD) | |
| 7.3 | 7.0 ± 0.01 | 7.7 ± 0.02 (19.5 ± 0.08) |
| 6.3 | 5.8 ± 0.14 | 6.8 ± 0.05 (22.7 ± 0.17) |
| 5.3 | 4.9 ± 0.11 | 5.7 ± 0.02 (26.9 ± 0.06) |
| 4.3 | 4.0 ± 0.07 | 4.8 ± 0.01 (30.3 ± 0.06) |
| 3.3 | 3.7 ± 0.06 | 4.0 ± 0.03 (33.3 ± 0.10) |
| 2.3 | 3.6 ± 0.17 | 3.6 ± 0.04 (34.8 ± 0.15) |
V. vulnificus MO6-24/O levels in inoculated oyster homogenate were determined by colony blot hybridization using the VVAP alkaline phosphatase probe (36) as described in the text.
Real-time PCR determination of V. vulnificus concentrations was based on the mean of triplicate samples. Concentrations were derived from a standard curve by using the mean of triplicate Ct values for serial 10-fold dilutions of DNA extracted from known concentrations of bacteria as described in text.
Enumeration of indigenous V. vulnificus in oysters.
Ten lots of oysters, freshly harvested from Florida gulf coast waters, were used for real-time PCR analysis to enumerate naturally occurring V. vulnificus. In some cases mild temperature abuse was employed to increase the number of bacteria in oyster samples. Consistent PCR signals were not obtained from indigenous V. vulnificus in oysters under conditions described for experiments with seeded oyster homogenates; therefore, the protocol was further optimized for these experiments. The use of magnetic bead DNA purification, which has been shown to produce real-time PCR sensitivities of 6 to 8 CFU g−1 with V. cholerae in inoculated oysters (18), did not achieve comparable results with V. vulnificus in unseeded oysters (data not shown). We found that decreasing the homogenate volume from 1.0 ml to 250 μl for Qiagen DNA extraction and increasing the PCR volume from 25 to 50 μl with 3.0 μl of template yielded the greatest sensitivity and reproducibility for detection by real-time PCR. These conditions were incorporated into the protocol for subsequent assays. As shown in Table 4, the numbers of indigenous V. vulnificus in oysters, as determined by a gene probe of colony blot hybridizations, ranged from nondetectable levels to 3.5 log CFU g−1 (4.5 × 103 CFU g−1). No significant difference (t test; P = 0.43) between the overall average values obtained with real-time PCR enumeration and those obtained in the VVAP probe assay was observed. Significant differences were observed for individual lots 2, 7, and 8 (P < 0.004), and V. vulnificus probe-positive colonies were not detected by colony hybridization in lot 3, which was shown to contain 2.0 log CFU g−1 (1.1 × 102 CFU g−1) by real-time PCR analysis. Thus, the detection limits of the V. vulnificus TaqMan assay approached 102 CFU g−1 for native bacteria in oyster tissues.
TABLE 4.
Enumeration of indigenous V. vulnificus in oysters
| Sample lot | Result for:
|
|
|---|---|---|
| Gene probea (log CFU g−1 ± SD) | Real-time PCRb (Log CFU g−1 ± SD) (Ct ± SD) | |
| 1 | 1.8 ± 0.67c | 2.0 ± 0.17 (37.0 ± 0.59) |
| 2d | 3.4 ± 0.15 | 2.3 ± 0.15 (35.7 ± 0.66) |
| 3d | <1.0 ± 0c | 2.0 ± 0.12 (36.6 ± 0.45) |
| 4 | 2.3 ± 0.13 | 2.3 ± 0.16 (35.0 ± 0.16) |
| 5 | 3.4 ± 0.21 | 3.1 ± 0.12 (33.0 ± 0.43) |
| 6 | 3.5 ± 0.35 | 3.6 ± 0.06 (30.6 ± 0.29) |
| 7d | 3.1 ± 0.00 | 2.6 ± 0.06 (32.4 ± 0.17) |
| 8d | 3.1 ± 0.06 | 3.6 ± 0.12 (28.9 ± 0.58) |
| 9 | 2.0 ± 0.20 | 2.4 ± 1.02 (33.1 ± 3.90) |
| 10 | 2.7 ± 0.30 | 2.1 ± 1.04 (33.9 ± 3.80) |
V. vulnificus MO6-24/O levels in uninoculated oyster homogenates were determined by DNA probe colony blot hybridization as described in the text.
Real-time PCR determination of V. vulnificus concentrations was based on the means of triplicate samples, and levels were derived from a standard curve by using the mean of triplicate Ct values for serial 10-fold dilutions of DNA extracted from a known concentration of bacteria as described in the text.
For statistical purposes, samples with probe-negative colony blot hybridizations were assigned a value corresponding to the lowest possible number detectable by the assay (1.0 log CFU g−1).
Lot for which there was a significant difference (P < 0.004) between bacterial concentrations from the gene probe and those from real-time PCR.
Detection of VBNC V. vulnificus.
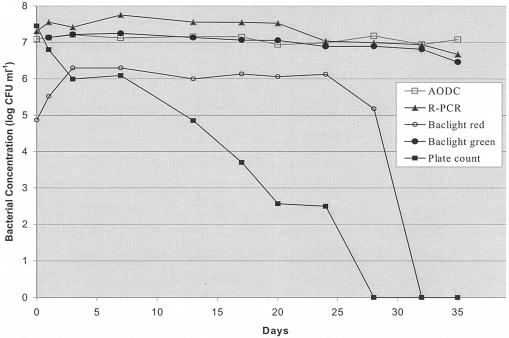
V. vulnificus MO6-24/O became VBNC after incubation in ASW at low temperature. Figure 2 shows that cells (initial level, 2.8 × 107 CFU ml−1) steadily declined in culturabilityon LA plates and were not culturable by day 28 of the study, as confirmed by lack of growth in APW enrichment medium (data not shown). However, the BacLight assay indicated that large numbers of cells maintained viability throughout cold storage, as shown by the number of cells with intact cell membranes (BacLight green). Real-time PCR enumeration was consistent with the viability assay and detected the same or slightly elevated levels of DNA throughout the study. Interestingly, the number of cells with red fluorescence generated by propidium iodide staining with the BacLight assay (indicative of loss of cell membrane integrity and hence viability) initially increased, but leveled off at day 3 and subsequently dropped to nondetectable levels by day 32. VBNC cells were resuscitated by temperature shift on day 43 to levels of ca. 106 CFU ml−1 on LA after incubation at room temperature for 24 h.
FIG. 2.
Enumeration of VBNC V. vulnificus cells from microcosms. Levels of V. vulnificus MO6-24/O (CFU per milliliter) were determined by real-time PCR (R-PCR) and plate count colony hybridizations, as described in the text. Bacterial viability was based on whether cells stained green (viable) or red (nonviable) in the LIVE/DEAD BacLight assay as observed by fluorescence microscopy. Total direct counts of cells per milliliter (AODC) were made by enumeration of cells stained with acridine orange. Standard deviations of the values obtained from these assays ranged from 0 to 0.5 log CFU ml−1 for duplicate microcosms.
DISCUSSION
This is the first study to describe direct enumeration of indigenous vibrios in oysters by real-time PCR. In this report, we describe the development of a real-time PCR assay which uses the TaqMan system to quantify V. vulnificus in oysters and which targets the same locus of the vvhA hemolysin gene previously shown to be species specific by DNA oligonucleotide probe detection (19, 36, 37). The real-time format demonstrates both sensitivity and species specificity for V. vulnificus detection and provides quantitative analysis, which is not available with conventional PCR. The real-time PCR detection limit for V. vulnificus DNA derived from pure culture was 72 fg per μl of PCR mixture or the equivalent of 102 CFU ml−1 of culture, which would extrapolate to 6 CFU per PCR assay. Although PCR assay sensitivity is readily achieved from pure culture, complex food matrices can greatly inhibit these reactions and interfere with quantitative analysis. Limits of sensitivity for seeded oyster studies could not be determined beyond 103 CFU g−1 due to background V. vulnificus levels; however, detection of indigenous V. vulnificus was 102 CFU g−1 in unseeded samples.
Limiting factors for PCR sensitivity are generally sample purity and volume size. Greater sensitivity (6 to 8 CFU g−1) has been reported for real-time PCR analysis of V. cholerae (18) in seeded oyster homogenates using magnetic bead DNA purification, and similar levels have been enumerated in food with L. monocytogenes (1) or S. enterica (7). However, the use of magnetic beads did not increase the sensitivity of detection of V. vulnificus in oysters. In our studies, DNA template extraction methods were sufficient to remove impurities from seawater or complex oyster matrices that might interfere with the assay, and concentration of DNA templates by simple vacuum evaporation or ethanol precipitation increased the level of sensitivity. Initially, we observed decreased detection sensitivity in naturally infected oysters compared to that in artificially inoculated samples. We found that slight modifications in the Qiagen DNA extraction (decreasing the sample volume) and PCR (increasing reaction volume) protocols resulted in about a 10-fold increase in sensitivity. The reason for less-sensitive PCR detection by the initial protocol is not clear; however, decreased sample volumes may increase cell lysis and DNA extraction efficiency and/or reduce concentrations of PCR inhibitors. Also, native vibrios are more resistant to depuration protocols than those in artificially inoculated oysters (31), suggesting greater affinity or compartmentalization in oyster tissues (12). Natural bacterial populations may also exhibit stress responses or other unknown factors that make them more refractory to lysis than those in seeded samples. These data suggest that evaluation of treatments for the reduction of V. vulnificus in oysters should be based on examination of natural populations, as results may differ from those obtained through artificial inoculation
Numbers obtained by V. vulnificus real-time PCR correlated well with plate counts based on colony blot hybridization enumeration, supporting the use of real-time PCR for quantitative analysis. For one lot, the real-time PCR assay was able to enumerate V. vulnificus cells that were not detected by the gene probe, suggesting greater sensitivity for the PCR assay. Although the colony blot assay will theoretically detect 10 CFU g−1, this sensitivity is difficult to attain in high levels of background colonies commonly seen in oyster homogenates. On the other hand, PCR may amplify dead cells that are not detected by viable plate counts, and amplification could represent a false-positive result (14). Comparison of individual lots indicated significant differences in the numbers obtained by the two methods for 3 of 10 lots; however, there was no significant difference between methods when comparisons were based on the means of all the samples. For two lots showing significant differences in comparisons between methods, numbers were actually higher for colony hybridizations than for PCR, suggesting that the source of variation was not dead cells but rather inherent assay variability. This issue is further complicated by the observation that VBNC vibrios may retain viability but lose the ability to grow on solid medium.
Oysters commonly harbor 103 to 105 CFU of aerobic, heterotrophic bacteria g−1 in their tissues during summer months in temperate climates (30, 37). V. vulnificus may comprise 10 to 50% of these culturable populations and is easily enumerated from spread plates by colony hybridization to a DNA probe (37). However, in colder environments, numbers decline rapidly, and the organism becomes nondetectable by standard culture methods. In vitro microcosm studies have indicated that starvation and/or temperature downshifts induce a dormant state that is not culturable on standard media; however, culturability may be resuscitated with a temperature upshift (33). Other studies have argued that viability is not recovered upon resuscitation by the culture as a whole, but, rather, small selected numbers of cells remain culturable and are able to multiply under these conditions (4). Conditions that induce or resuscitate growth of VBNC V. vulnificus in oysters or seawater are unclear but may play a role in assessing virulence potential. The decline in reported V. vulnificus cases in colder months strongly suggests that under these environmental conditions, which could induce the VBNC state, cells are not virulent. However, experimental VBNC Vibrio spp. cells have demonstrated virulence in animal (13, 26, 27) and human (8) infections. Unfortunately, evaluation of VBNC is hampered by the fact that independent indicators of viability (i.e., RNA expression and electron transport) may also shut down to nondetectable levels (9, 17, 32).
We employed quantitative real-time PCR to examine VBNC induction and found that the DNA concentration of nonculturable cells, as determined by real-time PCR, was sustained throughout these experiments. Viability was confirmed by the presence of a nonpermeable cell wall, as indicated by the BacLight assay. As expected, dead cells (i.e., without intact membranes) increased initially as the cultures aged; however, after extended incubation, dead cells also declined to nondetectable levels, presumably due to complete degradation of membranes and/or loss of nucleic acid integrity. Prior to the decline in dead cells, real-time PCR values slightly exceeded viable-cell counts by BacLight; however, once dead cells were no longer detected, concentrations of cells determined by real-time PCR closely paralleled concentrations of viable cells reported by BacLight. These data suggest that detectable DNA may persist in dead cell “ghosts,” initially contributing to real-time PCR amplification products, but was not a factor once cells were truly nonculturable in these microcosms. Whether or not DNA from dead cells remains stable in oyster homogenates is unclear. Endogenous DNase(s) may reduce the expected half-life of extracellular DNA (25), and experiments have shown only slight reduction of nucleic acid content following DNase treatment of cells prior to DNA extraction of VBNC cells (18).
Rapid, accurate enumeration methods are needed for monitoring shellfish harvesting areas and for evaluation of postharvest treatments to reduce V. vulnificus in oysters. Alternative detection strategies have combined real-time PCR and most-probable-number (MPN) enrichment protocols to enhance sensitivity and eliminate problems associated with DNA from dead cells (2). MPN enumeration is based on end-point titration of samples in enrichment medium, and real-time PCR provided improved detection for confirmation of V. vulnificus-positive growth. Recently, RT-PCR analysis of V. vulnificus demonstrated prolonged detection of hemolysin gene expression in VBNC cultures (9), and our assays are consistent with these results. These methods may be required for evaluation of treatments, such as freezing, that could preserve DNA from large numbers of dead cells and lead to false-positive amplification by PCR. Future studies will compare different applications for analysis of postharvest treatments, including enrichment methods, as well as use of RT-PCR for assessing more-transient RNA expression.
It should be noted that none of the available methods of V. vulnificus detection are able to discriminate virulent from avirulent strains of the species, as virulence determinants are not generally well defined. The genetic targets for the real-time PCR assay, as well as gene probes of most available detection methods, are based on the hemolysin gene. There is no in vivo evidence that expression of this gene contributes to virulence. Although the protein is a potent cytolysin and may be lethal in mice at nanogram-per-kilogram levels (15), it is expressed in both virulent and avirulent strains (20). Studies of isogenic mutants indicated that loss of vvhA gene function did not reduce virulence in mice (35). Conversely, expression of a capsular polysaccharide (CPS) has been clearly associated with disease in animal models (11, 29, 34, 40). Virulence and increased CPS expression of individual colonies are marked by opaque colony morphology, but a gene probe for detection of encapsulated V. vulnificus in food or environmental samples is not available. Our laboratory recently identified a V. vulnificus CPS operon (38), and current research is attempting to correlate genetic variation at this locus with the virulence phenotypes in order to identify a potential virulence gene targets.
Acknowledgments
We thank Robert Cousins and Raymond Blanchard for their invaluable advice on real-time PCR. G. E. Rodrick provided expertise for the oyster inoculation studies.
This study was funded in part by an NRI from USDA and by Florida Sea Grant.
REFERENCES
- 1.Bassler, H. A., S. J. A. Flood, K. J. Livak, J. Marmaro, R. Knorr, and C. A. Batt. 1995. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl. Environ. Microbiol. 61:3724-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackstone, G. M., J. L. Nordstrom, M. C. L. Vickery, M. D. Bownen, R. F. Meyer, and A. Depaola. 2003. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J. Microbiol. Methods 53:149-155. [DOI] [PubMed] [Google Scholar]
- 3.Blake, P. A., M. H. Merson, R. E. Weaver, D. G. Hollis, and P. C. Heublein. 1979. Disease caused by a marine vibrio: clinical characteristics and epidemiology. N. Engl. J. Med. 300:1-5. [DOI] [PubMed] [Google Scholar]
- 4.Bogosian, G., N. D. Aardema, E. V. Bourneuf, P. J. L. Morris, and J. P. O'Neil. 2000. Recovery of hydrogen peroxide-sensitive culturable cells of Vibrio vulnificus gives the appearance of resuscitation from a viable but nonculturable state. J. Bacteriol. 182:5070-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasher, C. W., A. Depaola, D. D. Jones, and A. K. Bej. 1998. Detection of microbial pathogens in shellfish with multiplex PCR. Curr. Microbiol. 37:101-107. [DOI] [PubMed] [Google Scholar]
- 6.Brauns, L. A., M. C. Hudson, and J. D. Oliver. 1991. Use of the polymerase chain reaction in detection of culturable and nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 57:2651-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S., A. Yee, M. Griffiths, C. Larkin, C. T. Yamashiro, R. Behari, C. Paszko-Kolva, K. Rahn, and S. A. De Grandis. 1997. The evaluation of a fluorgenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int. Food Microbiol. 35:239-250. [DOI] [PubMed] [Google Scholar]
- 8.Colwell, R. R., P. R. Brayton, D. Herrington, S. A. Huq, and M. M. Levine. 1996. Viable but non-culturable Vibrio cholera 01 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 12:28-31. [DOI] [PubMed] [Google Scholar]
- 9.Fischer-Le Saux, M., D. Hervio-Heath, S. Loaec, R. R. Colwell, and M. Pommepuy. 2002. Detection of cytotoxin-hemolysin mRNA in nonculturable populations of environmental and clinical Vibrio vulnificus strains in artificial seawater. Appl. Environ. Microbiol. 68:5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Food and Drug Administration. 1998. Bacteriological analytical manual, 8th ed. AOAC International. Arlington, Va.
- 11.Gray, L. D., and A. S. Kreger. 1985. Purification and characterization of an extracellular cytolysin produced by Vibrio vulnificus. Infect. Immun. 48:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris-Young, L., M. L. Tamplin, J. W. Mason, H. C. Aldrich, and J. K. Jackson. 1995. Viability of Vibrio vulnificus in association with hemocytes of the American oyster (Crassostrea virginica). Appl. Environ. Microbiol. 61:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huq, A., I. N. G. Rivera, and R. R. Colwell. 2000. Epidemiological significance of viable but nonculturable microorganisms, p. 301-323. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 14.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreger, A. S., and D. Lockwood. 1981. Detection of extracellular toxin(s) produced by Vibrio vulnificus. Infect. Immun. 33:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linkos, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 17.Lleo, M. D., S. Pierobon, M. C. Tafi, C. Signoretto, and P. Canepari. 2000. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl. Environ. Microbiol. 66:4564-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon, W. J. 2001. TaqMan PCR for detection of Vibrio cholerae O1, O139, non-O1, and non-O139 in pure cultures, raw oysters, and synthetic seawater. Appl. Environ. Microbiol. 67:4685-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris, J. G., Jr., A. C. Wright, D. M. Roberts, P. K. Wood, L. M. Simpson, and J. D. Oliver. 1987. Identification of environmental Vibrio vulnificus isolates with a DNA probe for the cytotoxin-hemolysin gene. Appl. Environ. Microbiol. 53:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris, J. G., Jr., A. C. Wright, L. M. Simpson, P. K. Wood, D. E. Johnson, and J. D. Oliver. 1987. Virulence of Vibrio vulnificus: association with utilization of transferin-bound iron and lack of correlation with levels of cytotoxin or protease production. FEMS Microbiol. Lett. 40:55-59. [Google Scholar]
- 21.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norton, D. M., and C. A. Batt. 1999. Detection of viable Listeria monocytogenes with a 5′ nuclease PCR assay. Appl. Environ. Microbiol. 65:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuda, J., M. Ishibashi, S. L. Abbott, J. M. Janda, and M. Nishibuchi. 1997. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the west coast of the United States. J. Clin. Microbiol. 35:1965-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver, J. D., R. A. Warner, and D. R. Cleland. 1983. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 45:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver, J. D., J. E. Wear, M. B. Thomas, M. Warner, and K. Linder. 1986. Production of extracellular enzymes and cytotoxicity by Vibrio vulnificus. Diagn. Microbiol. Infect. Dis. 5:99-111. [DOI] [PubMed] [Google Scholar]
- 26.Oliver, J. D., and R. Bockian. 1995. In vivo resuscitation, and virulence toward mice, of viable but nonculturable cells of Vibrio vulnificus. Appl. Environ. Microbiol. 61:2620-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahman, I., M. Shahamat, M. A. R. Chowdhury, and R. R. Colwell. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 62:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Simpson, L. M., V. K. White, S. F. Zane, and J. D. Oliver. 1987. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect. Immun. 55:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamplin, M. L., G. E. Rodrick, N. J. Blake, and T. Cuba. 1982. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamplin, M. L., and G. M. Capers. 1992. Persistence of Vibrio vulnificus in tissues of gulf coast oysters, Crassostrea virginica, exposed to seawater disinfected with UV light. Appl. Environ. Microl. 58:1506-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weichart, D., D. McDougald, D. Jacobs, and S. Kjelleberg. 1997. In situ analysis of nucleic acids in cold-induced nonculturable Vibrio vulnificus. Appl. Environ. Microbiol. 63:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitesides, M. D., and J. D. Oliver. 1997. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl. Environ. Microbiol. 63:1002-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright, A. C., L. M. Simpson, J. D. Oliver, and J. G. Morris, Jr. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright, A. C., and J. G. Morris, Jr. 1991. The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect. Immun. 59:192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright, A. C., G. A. Miceli, W. L. Landry, J. B. Christy, W. D. Watkins, and J. G. Morris, Jr. 1993. Rapid identification of Vibrio vulnificus on nonselective media with an alkaline phosphatase-labeled oligonucleotide probe. Appl. Environ. Microbiol. 59:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright, A. C., R. T. Hill, J. A. Johnson, M. C. Roghman, R. R. Colwell, and J. G. Morris, Jr. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright, A. C., J. L. Powell, J. B. Kaper, and J. G. Morris, Jr. 2001. Identification of a group 1-like capsular polysaccharide operon for Vibrio vulnificus. Infect. Immun. 69:6893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, K., A. C. Wright, J. B. Kaper, and J. G. Morris, Jr. 1990. The cytolysin gene of Vibrio vulnificus: sequence and relationship to Vibrio cholerae El Tor hemolysin. Infect. Immun. 58:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, S. I., M. Ogawa, and Y. Mizuguchi. 1985. Relation of capsular materials and colony opacity to Vibrio vulnificus. Infect. Immun. 47:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]