Abstract
An important challenge in translational bioinformatics is to understand how genetic variation gives rise to molecular changes at the protein level that can precipitate both monogenic and complex disease. To this end, we compiled datasets of human disease-associated amino acid substitutions (AAS) in the contexts of inherited monogenic disease, complex disease, functional polymorphisms with no known disease association, and somatic mutations in cancer, and compared them with respect to predicted functional sites in proteins. Using the sequence homology-based tool SIFT to estimate the proportion of deleterious AAS in each dataset, only complex disease AAS were found to be indistinguishable from neutral polymorphic AAS. Investigation of monogenic disease AAS predicted to be non-deleterious by SIFT were characterized by a significant enrichment for inherited AAS within solvent accessible residues, regions of intrinsic protein disorder, and an association with the loss or gain of various post-translational modifications. Sites of structural and/or functional interest were therefore surmised to constitute useful additional features with which to identify the molecular disruptions caused by deleterious AAS. A range of bioinformatic tools, designed to predict structural and functional sites in protein sequences, were then employed to demonstrate that intrinsic biases exist in terms of the distribution of different types of human AAS with respect to specific structural, functional and pathological features. Our web tool, designed to potentiate the functional profiling of novel AAS, has been made available at http://profile.mutdb.org/.
Keywords: amino acid substitutions, missense mutations, translational bioinformatics, disease mechanism, association study, SNP
Introduction
Understanding the molecular consequences of the mutations that cause human genetic disease remains an important research challenge [Karchin, 2009; Mooney, 2005; Ng & Henikoff, 2006; Steward et al., 2003]. There are now several resources available which employ annotations describing biochemical features that are potentially useful for identifying function-altering and/or disease-associated amino acid substitutions (AAS), including SNPs3D [Yue et al., 2006], the SNP Function Portal [Wang et al., 2006], PolyDoms [Jegga et al., 2007], LS-SNP [Karchin et al., 2005a] and MutDB [Singh et al., 2007] among others. However, these resources typically use only sequence and structural features, such as evolutionary conservation in the vicinity of the site of mutation, and make no attempt to quantify the relative contributions made by specific molecular functions (features) that have either been introduced or disrupted by the mutations in question. Additionally, various tools have been developed to predict dysfunctional and/or disease-causing AAS. These include SIFT [Ng & Henikoff, 2003], PolyPhen [Ramensky et al., 2002], PMUT [Ferrer-Costa et al., 2005], PANTHER [Mi et al., 2007], LS-SNP [Karchin et al., 2005a], RCOL profiles [Terp et al., 2002], SNAP [Bromberg & Rost, 2007] and the SVM at SNPs3D [Yue et al., 2006] among others. All these tools operate using approximately the same principles, i.e. they are all supervised and employ features based on protein sequence, sequence conservation and/or protein structure. For example, approaches to classification of mutation sites have used linear regression [Chasman & Adams, 2001], neural networks [Bromberg & Rost, 2007], support vector machines [Krishnan & Westhead, 2003] and decision trees [Karchin et al., 2005a; Saunders & Baker, 2002]. These tools differ, however, in terms of their choice of training data, which can be datasets of human disease alleles as in the case of PolyPhen [Ramensky et al., 2002], evolutionary mutations that differentiate closely related species [Arbiza et al. 2006; Capriotti et al., 2008b], or experimentally induced mutations such as those originally studied with SIFT [Ng & Henikoff, 2003]. Other groups have used additional novel features such as physicochemical properties [Jiang et al., 2006], structural information [Tavtigian et al., 2008], information theory [Karchin et al., 2005b] and Gene Ontology terms [Calibrese et al., 2009]. These features have served to improve predictive accuracy. Despite differences in dataset construction and statistical inference models, the tools listed above yield remarkably similar predictions, an unsurprising finding since they were designed with a similar goal in mind: viz. to predict functional vs. non-functional mutations or disease vs. non-disease mutations.
However, since the features to be examined are nearly all exclusively based on protein sequence and structure, the currently available tools are inherently incapable of shedding light on the molecular causes of disease beyond simple disruptions of protein structure or sequence conservation. We have therefore set out to extend this area of enquiry by attempting to quantify the relative contributions made by different protein features when disrupted by mutation. To this end, we have evaluated the presence (and mutation-induced disruption) of a range of structural and functional features predicted by several different bioinformatics tools. Our approach was conceptually straightforward in that we utilized statistical inference methods to predict amino acid functions and then estimated how these predictions were altered by AAS.
Machine learning methods that predict structural and functional sites in amino acid sequences are well established and facilitate the prediction of secondary structure [Faraggi et al., 2009], solvent accessibility [Faraggi et al., 2009], post-translational modification [Iakoucheva et al., 2004] and enzyme catalysis [Youn et al., 2007]. These tools typically employ both sequence- and structure-based features, and have been trained on datasets of well characterized functional sites. For example, residues involved in enzyme catalysis can be predicted using machine learning methods trained on a database of catalytic sites [Porter et al., 2004]. Here, we have assessed the relative contributions of a range of functional site features to protein disruption in several disease-associated mutation datasets as well as a dataset of mutations which (in all likelihood) lack functional significance. We used several methods to predict structural features, post-translational modification and catalytic residues in the analysis of five test datasets containing different types of human amino acid substitutions: (i) mutations causing inherited disease, (ii) somatic cancer-associated mutations identified in breast and colorectal tumours, (iii) somatic cancer-associated amino acid substitutions identified in protein kinase genes from diverse human tumours, (iv) functional polymorphisms (with no known disease association), (v) putatively functional polymorphisms associated with human inherited disease, and a control dataset of putatively neutral polymorphisms. The results of our study indicate a significant difference between disease and non-disease associated variants in terms of both the structural and functional features disrupted.
Data Sources and Methods
Datasets
Five distinct sets of amino acid substitutions (AAS) with different disease annotations were collected for the purposes of this analysis. Firstly, heritable AAS from the Human Gene Mutation Database (HGMD, August 2007; http://www.hgmd.org; Stenson et al., 2009) were grouped into three different categories:
Germline disease-causing amino acid substitutions underlying human inherited disease (29,485 AAS from 1,877 different human nuclear genes) which will henceforth be referred to as the ‘Inherited disease’ dataset.
Disease-associated polymorphisms of putative functional significance (761 AAS from 496 different human nuclear genes), henceforth referred to as the ‘Disease-associated polymorphism’ dataset). To be included in this category, variants had to have occurred at polymorphic frequency (i.e. a minor allele frequency of ≥1% in the population under study). In addition, not only must a statistically significant (P < 0.05) association between the polymorphism and a clinical phenotype have been reported, but also other information (e.g. in vitro or in vivo expression/functional data, replicated association studies, epidemiological information, evolutionary conservation data etc.) must also have been presented to support the contention that the polymorphism in question was of bona fide functional significance.
Polymorphisms shown experimentally (e.g. by in vitro reporter gene expression assays) to be of functional significance albeit with no known disease association (893 AAS from 177 different human nuclear genes; henceforth referred to as the ‘Functional polymorphism’ dataset). All AAS from HGMD with reported consequences for the mRNA splicing phenotype were excluded from this analysis.
Secondly, two additional datasets of somatic cancer-associated AAS were obtained from recent cancer resequencing studies [Sjöblom et al., 2006; Greenman et al., 2007]. The first of these datasets comprised those AAS identified in exons derived from 20,857 transcripts from 11 breast and colorectal tumours. This breast and colorectal cancer dataset (henceforth referred to as the ‘Cancer’ dataset) represents 1099 somatic substitutions from 847 different human genes [Sjöblom et al., 2006]. The second dataset of cancer-associated AAS comprises 695 somatic substitutions identified in the exons of 518 protein kinase genes from 312 diverse human tumours, henceforth referred to as the ‘Kinase’ dataset [Greenman et al., 2007].
Finally, a set of AAS (annotated as ‘polymorphism’), downloaded from the UniProtKB/Swiss-Prot database [Boeckmann et al., 2003; ftp://ftp.ebi.ac.uk/pub/databases/swissprot/release/docs/humsavar.txt], comprised the neutral AAS used in this study. This dataset represents one of the most extensive sources of putatively neutral polymorphism data available but is nevertheless unlikely to represent a truly neutral dataset since at least some of the component AAS could yet prove to have an association with disease [Care et al., 2007]. To further improve the neutral credentials of this dataset, any AAS that were concurrently annotated in HGMD as being disease-causing or of potential functional significance were removed (N.B. 1,589 AAS were excluded in this way). In addition, since rare missense alleles are inherently more likely to be deleterious than common missense alleles [Kryukov et al., 2007], only those AAS in the UniProtKB/Swiss-Prot dataset that occurred at polymorphic frequencies (≥1% in a population of European descent; Hap-Map-CEU) were retained. This putatively neutral set of AAS therefore contained 8,509 human polymorphisms (taken from a total of 4,864 different genes) and shall henceforth be known as the ‘Swiss-Prot neutral’ dataset. Once again, it should be noted that we cannot wholly exclude the possibility that a subset of these supposedly neutral polymorphisms could be of functional importance or that they might have a role either in complex disease or as modifiers of disease susceptibility. Table 1 summarizes the above mutation datasets.
Table 1.
Summary of mutation datasets (comprising exclusively human missense mutations) used in the analysis. See text for full descriptions and sources of datasets.
| Name of dataset | Description | Number of amino acid substitutions (AAS) |
Number of distinct genes |
|---|---|---|---|
|
Inherited disease |
Heritable disease-causing mutations from HGMD |
29,485 | 1,485 |
|
Disease- associated polymorphisms |
Heritable disease-associated polymorphisms from HGMD, of putative functional significance |
761 | 496 |
|
Functional polymorphisms |
Heritable polymorphisms of functional significance from HGMD but with (as yet) no reported disease phenotype |
893 | 177 |
| Kinase | Somatic mutations in kinase genes identified in an analysis of >200 individual tumours |
695 | 312 |
| Cancer | Somatic mutations from breast and colorectal cancer tumours |
1,099 | 847 |
|
Swiss-Prot neutral |
Validated polymorphic AAS from Swiss-Prot annotated as ‘polymorphism’. |
8,509 | 4,864 |
Inherited disease-causing mutations in oncogenes and tumour suppressor (TS) genes
Using gene ontology (GO) terms (http://www.geneontology.org), lists of inherited disease genes matching the GO terms for oncogene (GO:0008151) or ‘tumour suppressor’ and ‘anti-oncogene’ (GO:0045786) were compiled. The first subset of AAS in tumour suppressor genes comprised 1,227 AAS from 33 genes. The second subset of AAS in oncogenes contained 288 AAS from 26 genes.
Profiling tools
The tools employed in this analysis were sequence-based and included measures of structure, function and post-translational modification. The tools chosen were of sufficiently high accuracy to be useful in testing biological hypotheses. Secondary structure (80% accuracy) and solvent accessibility (79% accuracy) were predicted using SPINE [Dor & Zhou, 2005]. Protein structure stability was assessed using Imutant [Capriotti et al., 2008a] (77% accuracy). Regions of intrinsic protein disorder were predicted using VSL2B predictor [Peng et al., 2006] (>85% accuracy). Short structured or loosely structured helical regions within long disordered regions (so-called Molecular Recognition Fragments, MoRFs) were identified using a predictor of calmodulin-binding targets, CaMBTP [Radivojac et al., 2006] (81% accuracy). Post-translational modification sites were identified using DisPhos to identify phosphorylation sites [Iakoucheva et al., 2004] (75% accuracy claimed for serine, threonine and tyrosine residues), OGlycoPred to identify O-linked glycosylation sites (77% accuracy claimed for serine, threonine, proline and lysine; Radivojac, unpublished work), UbPred to predict sites of ubiquitination [Radivojac et al., 2009] (72% accuracy claimed) and MethylPred to predict sites of protein methylation [Daily et al., 2005] (71% accuracy claimed for arginine and lysine residues). Catalytic sites were ascertained with a catalytic residue predictor termed CRP [Youn et al., 2007] (65% accuracy claimed over all residues). Finally, SIFT [Ng & Henikoff, 2003] was used to predict whether or not the AAS were deleterious. The tools described above were employed in order to interrogate both the wild-type and the mutant sequences; any change in prediction scores between the wild-type and mutant sequences was recorded. Conservative cutoffs were employed to minimize the false discovery rates. For the tools that generated predictions as probabilities (i.e. disorder, calmodulin-binding sites, phosphorylation, O-linked glycosylation, ubiquitination, methylation and catalytic residues), only ‘high confidence sites’ were considered, defined here as sites with a false positive prediction rate of ~0.1 (estimated during model evaluation).
Evaluation of in silico predictions
The large size of the combined datasets (41,442 AAS in total) from this study makes it impractical for each individual AAS to be functionally investigated in vitro. However, it should be possible, at least in principle, to validate a subset of our in silico predictions against a series of amino acid residues of known structural/functional importance. To this end, a test sample of 6,073 AAS (from 1209 distinct proteins) was selected from the total (41,442) AAS under study. These AAS represented all AAS from the proteins for which functional data on stability, secondary structure, solvent accessibility, disordered regions, calmodulin-binding sites, catalytic site residues and post-translational modification (methylation, phosphorylation, O-linked glycosylation and ubiquitination) could be obtained. This test sample of AAS were then assessed in order to establish whether the original structural/functional predictions were true positives (TP), false positives (FP), true negatives (TN) or false negatives (FN).
The in vitro data on the 6,073 AAS test sample, required for validating our original in silico predictions, were obtained from publically available databases augmented by searches of the scientific literature. In vitro data on the consequences of AASs (in 16 human proteins) for protein stability were obtained from Allali-Hassani et al. (2009). The program DSSP (Kabsch et al. 1983) was used to extract secondary structure and solvent accessibility information from 12 human proteins with known X-ray crystallographic structures. The locations of disordered regions within 61 human proteins were obtained from DisProt v. 4.9 (Sickmeier et al. 2007). The locations of known calmodulin-binding sites in 10 human proteins were obtained from the Calmodulin Target Database (Yap et al. 2000). The locations of catalytic site residues in 65 human proteins were obtained from the Catalytic Site atlas (v. 2.2.11; Porter et al., 2004). Finally, the UniProt Knowledgebase (Release 15.7) and Human Protein Reference database (HPRD; Keshava Prasad et al. 2009) together yielded data on post-translational modifications for 1140 human proteins.
Disease annotations
For the Inherited disease AAS, the disease terms recorded in the original publications were mapped to the Unified Medical Language System (UMLS) using a simple word permutation-based method developed and tested by Shah et al. [Shah et al., 2006; Shah et al., 2007]. The disease names were mapped to UMLS concept identifiers (CUI) using the open source UMLS-Query module [Shah & Musen, 2008]. UMLS-Query provides a function called maptoId, which accepts a phrase (up to 10 words) and maps it to a CUI (and can be restricted by a vocabulary if so desired). The function first looks for an exact match for the phrase; if none is found, it will generate all possible permutations and attempt an exact match for each one. The function also performs right truncation to look for partial matches. For example, calling the function to find a CUI belonging to the SNOMED-CT for ‘intraductal carcinoma of prostate’ will match concepts “intraductal” (C0007124) as well as “carcinoma of prostate” (C0600139). Permutation generation along with right truncation is conceptually similar to using skip n-grams for matching concepts. Skip bigrams have been shown to perform at or above state-of-the-art measures with less complexity, for the purpose of identifying matching concepts [Reeve & Han, 2007]. Some 23,594 (~80% of the total) disease terms relating to the Inherited disease AAS were mapped to the UMLS with high confidence. The hierarchy of disease terms from the SNOMED-CT ontology was used to explore the relationships between the disease states and the underlying molecular phenotypes.
Enrichment analysis
Using Swiss-Prot neutral as a control dataset, we compared the distribution of the structural and functional sites for each dataset (Inherited disease, Disease-associated polymorphism, Functional polymorphism, Kinase and Cancer) against the Swiss-Prot neutral distribution. To allow for multiple testing, the significance of any difference noted was then assessed by means of Fisher's exact test with Bonferroni correction. Only P values < 0.00172 (0.05/29) were considered significant.
Results & Discussion
In silico profiling of amino acid substitutions of potential pathological importance
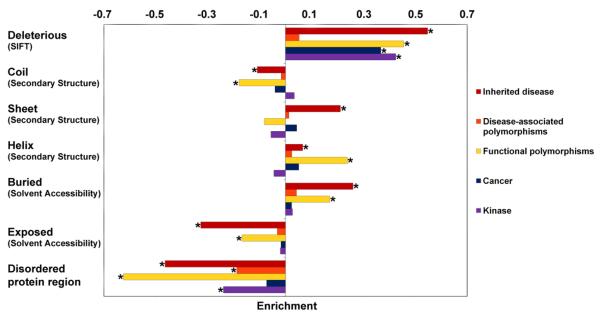
Identifying the biological functions disrupted by specific amino acid substitutions (AAS) is an important challenge that has relevance both for understanding the underlying molecular mechanism(s) of a given disease and for identifying functional polymorphic sites that may impact upon both complex disease and disease susceptibility. The enrichment of AAS at residues of structural or functional importance in each dataset was compared and contrasted, as depicted in Figures 1 and 2.
Figure 1.
Relative enrichment of structural amino acid residues by comparison with the dataset of putatively neutral polymorphism (Swiss-Prot neutral). For each feature, the bars represent the relative proportion [(Dataset − Swiss-Prot neutral) / (Dataset + Swiss-Prot neutral)] of entries in the dataset associated with that feature as compared to the Swiss-Prot neutral polymorphism dataset. For SIFT, the bars represent the proportion of entries in each dataset that are predicted to be deleterious. *indicates a significant p-value with Fisher's exact test after Bonferroni correction for multiple comparisons (P < 1.72.10−3).
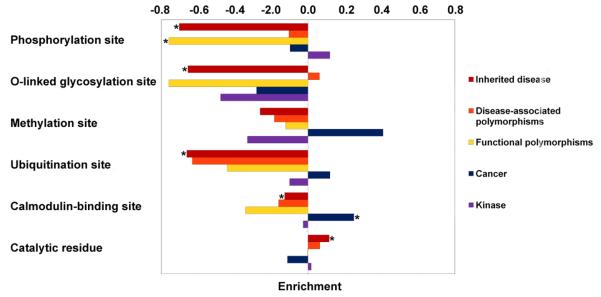
Figure 2.
Relative enrichment of functional sites by comparison with a dataset of putatively neutral polymorphisms (Swiss-Prot neutral). For each feature, the bars represent the relative proportion [(Dataset − Swiss-Prot neutral) / (Dataset + Swiss-Prot neutral)] of entries in the dataset associated with that feature as compared to the Swiss-Prot neutral polymorphism dataset. *indicates significant p-value with Fisher's exact test after Bonferroni correction for multiple comparisons (P < 1.72.10−3).
Prediction of deleterious amino acid substitutions
SIFT was used to predict the proportion of deleterious AAS in each missense mutation dataset. The Inherited disease, Functional polymorphism, Cancer and Kinase datasets were all characterized by a significant enrichment in the proportion of substitutions predicted to be deleterious when compared to the putatively neutral Swiss-Prot neutral dataset (see Figure 1; SIFT). For the Inherited disease dataset, ~76% of AAS were predicted to be deleterious (average SIFT score 0.072), a value very similar to the proportion (69%) previously predicted to be deleterious using disease-causing AAS from UniProtKB/Swiss-Prot [Ng & Henikoff, 2003]. For the Functional polymorphisms, 59% of AAS were predicted to be deleterious using SIFT (average SIFT score 0.162).
By contrast, SIFT predicted that only ~25% of Disease-associated polymorphisms were deleterious (average SIFT score 0.38), a proportion almost identical to the 22% noted for the Swiss-Prot neutral control dataset. Since the ±20% accuracy range of the SIFT method [Ng & Henikoff, 2003] renders reliable discrimination of these datasets impossible, we must conclude that there is no evidence for a significant difference between the two datasets. There are two plausible explanations to account for the marked similarity between the Swiss-Prot neutral and Disease-associated polymorphism datasets in terms of their SIFT scores. Firstly, the contribution of disease-associated polymorphisms to disease may well be additive via the net effect of multiple subtle modifications to function [Schork et al., 2009]. In agreement with this assertion, we found that the Disease-associated polymorphisms were located mainly in exposed residues (55.2%) or within disordered regions (19.2%). Such residues tend be less highly conserved evolutionarily than those which are buried within the protein structure. Hence, polymorphic variants in these locations may exert a subtle influence on protein function rather than a drastic one. Since SIFT employs evolutionary conservation as a proxy to predict function, it may be beneficial to retrain the method with these AAS when using SIFT to make predictions regarding polymorphic AAS located within exposed residues or disordered regions. For example, disordered protein regions have been shown to exhibit different rates of evolution (Brown et al., 2002) and different amino acid substitution patterns (Radivojac et al., 2002) than ordered regions. The alternative possibility is that a large proportion of disease-associated polymorphisms (considered by the original authors reporting them to be directly causative of the disease association) are not in reality the variants directly responsible for the disease association. Instead, they may simply be closely linked to (and/or in strong linkage disequilibrium with) those additional, and hitherto undetected, functional variants actually responsible for the observed disease associations.
Under the assumption that all the AAS in the Inherited disease dataset do indeed represent causative variants underlying the various genetic diseases as claimed by the original reporting authors, it can be seen that only 76% of them are predicted by SIFT to disrupt protein function. If we break down the SIFT predictions on a gene-wise basis for inherited disease, we see that SIFT prediction accuracy (i.e. the proportion of inherited disease-causing AAS predicted to disrupt protein function) ranged from 31-100% (Supp. Table S1 & S2). Analyzing a subset of 6,457 Inherited disease AAS which SIFT had predicted not to be of functional significance (i.e. ‘tolerant’, denoting tolerated), revealed that ~50% (3,210 AAS) were located in surface exposed regions, representing a significant enrichment over the Inherited disease dataset as a whole (+20%; P = 4.15·10−199, Fisher's exact test). The predicted ‘tolerant’ Inherited disease subset was also significantly depleted, as compared to the entire Inherited Disease dataset, with respect to AAS giving rise to a decrease in protein stability of ≥1 kcal/mol (−5.4%; P = 2.14·10−7, Fisher's exact test) and enriched for AAS located in disordered regions (+1.5%; P = 3.2·10−4, Fisher's exact test). The ‘tolerant’ Inherited disease subset also exhibited a significant enrichment for AAS predicted to result in the loss of phosphorylation sites (+0.4%; P = 2.55·10−5, Fisher's exact test) and AAS giving rise to a gain of ubiquitination sites (+0.3%; P = 4.43·10−4, Fisher's exact test).
The ‘tolerant’ Inherited disease subset exhibited similarities to both the Disease-associated polymorphism and Cancer datasets e.g. in terms of the distribution of mutations in both surface-exposed residues (>50%) and within disordered regions (~20%). It may nevertheless be important, when evaluating AAS in exposed or disordered regions, to attribute a lower confidence level to the ‘tolerant’ label assigned by SIFT; this may hold true especially when evaluating polymorphic AAS which by their very nature tend to be located in evolutionarily less highly conserved regions.
Evaluation of in silico predictions
The 6,073 AAS of the test sample represented all AAS from the proteins for which functional data on stability, secondary structure, solvent accessibility, disordered regions, calmodulin-binding sites, catalytic site residues and post-translational modification (methylation, phosphorylation, O-linked glycosylation and ubiquitination) could be obtained. Nine examples of these AAS, representing nine of the features being considered here, and known to be associated with a human inherited disease, are listed in Supp. Table S3.
The standard benchmarking statistics used to evaluate the structural/functional predictions made on the test sample of 6,073 AAS were the false positive rate (FPR); sensitivity; specificity; Matthews Correlation Coefficient (MCC; Matthews, 1975) and the accuracy (the mean of sensitivity and specificity scores). The Matthews Correlation Coefficient (MCC) was employed since it represents one of the best available measures of prediction quality. It returns a value between −1 and +1; a coefficient of −1 represents the worst possible prediction, 0 a random prediction and +1 a perfect prediction. The validation of the original in silico predictions for the test sample of 6,073 AAS is summarized in Supp. Table S4. MCC values for the various predictors were in the range of 0.125-0.701. Our combined algorithm would therefore appear to have performed best on the features; secondary structure, solvent accessibility, calmodulin-binding, O-linked glycosylation and ubiquitination (MCC>0.50). The in silico predictions for phosphorylation sites had by far the lowest MCC score (0.125) but it should be appreciated that even in quite well characterized proteins, it is highly likely that not all bona fide phosphorylation sites will have been identified experimentally. In summary, using our combined algorithm on our test sample of 6,073 AAS, we were able to achieve a sensitivity of 0.63 and a specificity of 0.93 with respect to identifying sites of known structural/functional importance. Despite the relatively small size of the test sample, this validation serves to confirm that our original in silico predictions are, at the very least, of sufficient quality for the generation and testing of biological hypotheses. Furthermore, since virtually all the prediction models we employed were trained on data derived from a number of different species, the chance of ‘overfitting’ to human data is minimized.
Structural properties
The structural properties of the sites altered by amino acid substitution in the different datasets are also summarized in Figure 1. Buried residues located within the core of a protein have long been known to be important for protein folding and stability [Sandberg et al., 1995] whereas residues located at or close to the surface of a protein are more likely to be involved in protein-protein interactions [Ye et al., 2006]. When solvent accessibility was considered, a significant enrichment of AAS at buried sites was noted for both the Inherited disease and Functional polymorphism datasets: 70% (P < 2.2·10−16; Fisher's exact test) and 58% (P < 2.2·10−16; Fisher's exact test) respectively as compared to 41% of Swiss-Prot neutral (Figure 1, solvent accessibility, ‘buried’). By contrast, the Kinase (solvent accessibility, 43% AAS buried), Cancer (solvent accessibility, 43% AAS buried) and Disease-associated polymorphism datasets (solvent accessibility, 44% AAS buried) were indistinguishable from the Swiss-Prot neutral dataset in terms of their solvent accessibility, indicating that the pathogenic effects of their AAS may not be biased towards the disruption of intrinsic structural properties in the same way that Inherited Disease and Functional polymorphisms are.
Since our prediction algorithms are sequence-based, we can readily evaluate (without structural modeling) changes in prediction for specific AAS by running the bioinformatics tools on both the wild-type and mutant sequences. It should be noted that large predicted structural changes are based primarily upon local sequence features and do not take the entire protein sequence into account. However, although these features may not indicate actual mutation-induced conformational changes (e.g. the conversion of an alpha-helix into a beta-sheet), by performing this experiment we are testing whether these features represent good indicators of a disruptive mutation. To test whether these findings were significant by comparison with the putatively neutral AAS, we calculated the enrichment (or depletion) with respect to the Swiss-Prot neutral dataset, determining significance using Fisher's exact test with a Bonferroni correction. When we examined the change in protein stability consequent to a given amino acid substitution, we observed a significant enrichment for AAS that give rise to a decrease in stability of ≥1kcal/mol but only for the Inherited disease dataset (+9.7%; P = 3.9·10−24; Fisher's exact test). In terms of the change of predicted solvent accessibility due to amino acid substitution (Exposed>Buried & Buried>Exposed, Figure 3), only the Cancer and Kinase datasets exhibited a significant enrichment for AAS predicted to be located at surface exposed residues in the wild-type protein but buried as a consequence of the amino acid substitution (Exposed>Buried; Cancer = +2.9%; P = 1.3·10−3; Kinase = +3.9%; P = 6.2·10−4; Fisher's exact test; Figure 3). Such Exposed>Buried transitions are likely to exert a dramatic effect upon protein function.
Figure 3.
Relative enrichment of changes at structural amino acid sites brought about by amino acid substitution. For each mutation, if the prediction method altered its prediction in a given context upon substitution of the wild-type residue, then the nature of that alteration is indicated. Enrichment was then compared with a dataset of putatively neutral polymorphisms (Swiss-Prot neutral). *indicates significant p-value with Fisher's exact test after Bonferroni correction for multiple comparisons (P < 1.72.10−3).
When secondary structure was explored, the Inherited disease and Functional polymorphism datasets were both found to be significantly enriched in AAS within alpha-helical regions (Inherited disease = +4.3%; P = 8.9·10−14; Functional polymorphism = +19.2%; P = 1.7·10−29; Fisher's exact test) but significantly reduced in AAS located in coiled regions; as compared to the Swiss-Prot neutral dataset (Inherited disease = −11.2%; P = 6.4·10−73; Functional polymorphism = −17.3%; P = 9.8·10−23; Fisher's exact test; Figure 1). The increased number of AAS in the alpha-helical regions for both the Inherited disease and Functional polymorphism datasets may be attributed to the fact that helices constitute one of the most common recognition motifs in proteins [Che et al., 2007]. It follows that modifying these regions may alter the biological activities of the protein involved. An example of a protein with an enrichment of disease-causing AAS within alpha-helical regions is keratin 12 (KRT12) in which AAS often only occur in the highly conserved alpha-helical regions essential for keratin filament assembly (alpha-helix-initiation motif of rod domain 1A or alpha-helix-termination motif of rod domain 2B) [Nishida et al., 1997]. The depletion of AAS within coiled regions for Inherited disease mutations and Functional polymorphisms may be related to the lack of a specific three dimensional structure (barring a few exceptions) in coiled regions.
The Inherited disease dataset was also significantly enriched for AAS in beta-sheet regions (+6.8%; P = 1.62·10−49, Fisher's exact test) and for changes of predicted secondary structure due to amino acid substitution from a beta-sheet region to a coiled or alpha-helical region (Sheet>Helix,Coil; +0.9%; P = 3.4·10−7; Fisher's exact test; Figure 3). The conversion of a beta-sheet into an alpha-helical region may lead to new and deleterious interactions in the disease state since helical regions are the most common recognition motifs of proteins [Che et al., 2007]. The strong bias of Inherited disease mutations and (especially) Functional polymorphisms towards alpha-helical regions suggests that secondary structure may represent a particularly informative feature for machine learning and the computational classification of deleterious AAS.
Role of intrinsically disordered proteins in both complex and somatic disease
Intrinsically disordered (ID) protein regions lack a unique 3-D structure and exist in a dynamic ensemble of different conformations [Dunker et al., 2001]. Their functional roles are well documented and they tend to be enriched in regulation and signaling via protein–protein and protein-nucleic acid interactions [Dyson & Wright, 2005; Radivojac, 2007]. The number of AAS from the Inherited disease (3034 AAS; −17.83%; P = 2.2·10−16; Fisher's exact test), Disease-associated polymorphism (146 AAS; −8.89%; P = 6.2·10−8; Fisher's exact test), Functional polymorphism (58 AAS; −21.63%; P = 8.83·10−56; Fisher's exact test) and Kinase (120 AAS; −10.86%; P = 1.44·10−10; Fisher's exact test) datasets occurring within ID regions was significantly reduced by comparison with the Swiss-Prot neutral dataset (Figure 1). Although ID regions were significantly depleted in the vicinity of Disease-associated polymorphisms, AAS within ID regions still account for ~19% of this dataset, as compared to 10% of the Inherited Disease dataset.
Disease-associated polymorphisms in ID regions may play an additive role in complex disease [e.g. p.G460W (ADD1, MIM# 102680) which is associated with hypertension; Cusi et al., 1997] or may in some cases act as disease modifiers for disease-causing mutations [as in the case of p.H558R (SCN5A, MIM# 600163) which modifies the effects of the disease-causing p.T512I on Na+ channel function [Viswanathan et al., 2003] or p.A115S in xylosyltransferase I (XYLT1, MIM# 608124) which is associated with higher serum XT activity and acts as a disease modifier in pseudoxanthoma elasticum (PXE, MIM# 264800); Schön et al., 2006].
It was also noted that 26% of those entries in the Disease-associated polymorphism dataset which were located within ID regions, were associated with cancer susceptibility. This supports previous work which has highlighted the importance of intrinsic disorder in cell signaling and cancer-associated proteins [Iakoucheva et al., 2002]. One example is the missense polymorphism p.A538T (HIF1A, MIM# 603348) which is located within an ID region and is associated with renal carcinoma [Ollerenshaw et al., 2004].
Functional sites
The distributions of AAS predicted at functional amino acid residues are summarized in Figure 2 for the different datasets. The Inherited disease dataset was characterized by a significant enrichment of AAS located at catalytic residues (+2.32%; P = 7.54·10−10; Fisher's exact test) but displayed a significant paucity of AAS at calmodulin-binding sites (−0.62%; P = 1.12·10−3; Fisher's exact test) and at three different sites of post-translational modification including O-linked glycosylation (−0.64%; P = 7.29·10−17; Fisher's exact test), ubiquitination (−0.45%; P = 1.12·10−12; Fisher's exact test) and phosphorylation (−2.01%; P = 1.47·10−54; Fisher's exact test) (Figure 2). The Functional polymorphism dataset also displayed a paucity of AAS at phosphorylation sites (−2.1%; P = 2.17·10−6; Fisher's exact test). The Cancer dataset was significantly enriched for AAS at calmodulin-binding sites (+1.82%; P = 1.62·10−3; Fisher's exact test; Figure 2, Calmodulin-binding sites). When we examined the change in functional site (gain or loss) consequent to a given amino acid substitution, the Functional polymorphisms dataset was significantly enriched for gains of catalytic residues (+2.63%; P = 3.76·10−4; Fisher's exact test; Figure 4) consequent to AAS. The Inherited disease and Functional polymorphism datasets both displayed a paucity of AAS giving rise to losses or gains of phosphorylation whilst the Inherited disease dataset also exhibited a paucity of AAS resulting in the loss of O-linked glycosylation sites and the gain or loss of ubiquitination sites.
Figure 4.
Relative enrichment of change at functional amino acid sites brought about by amino acid substitution. For each mutation, if the prediction method altered its prediction in a given context upon substitution of the wild-type residue, then the nature of that alteration is specified. *indicates significant p-value using Fisher's exact test after Bonferroni correction for multiple comparisons (P < 1.72.10−3).
In terms of functional sites, the Inherited disease dataset was found to be enriched in AAS at catalytic residues but depleted in AAS at three types of post-translational modification tested. The Functional polymorphisms dataset was also enriched in AAS giving rise to the gain of catalytic sites whereas the Cancer dataset was enriched for AAS at calmodulin-binding sites.
Germline AAS in oncogenes and tumour suppressor genes
Two subsets of missense mutations were derived from the Inherited disease dataset viz. germline AAS from tumour suppressor genes (1,227 AAS; 33 genes) and germline AAS from oncogenes (288 AAS; 26 genes). Disease-causing missense substitutions in oncogenes are usually dominant gain-of-function mutations whereas their counterparts in tumour suppressor genes tend to be recessive loss-of-function mutations. Overall, disease-causing AAS in oncogenes and tumour suppressor genes exhibited significant differences in terms of both their SIFT-predicted deleteriousness and the distribution of AAS within regions of intrinsic protein disorder (Table 2). Some 69.2% of tumour suppressor AAS and 82.3% of oncogene AAS were predicted by SIFT to be deleterious (‘intolerant’) (P = 8.89·10−3; Fisher's exact test). With respect to the distribution of AAS within protein regions of intrinsic disorder, these two subsets exhibited significant differences, with 15.1% of tumour suppressor AAS and 4.3% of oncogene AAS located within disordered regions (P = 1.8·10−7; Fisher's exact test).
Table 2.
Comparison of Inherited disease amino acid substitutions (AAS) subsets from tumour suppressor genes (1227 AAS from 33 genes) and oncogenes (288 AAS from 26 genes),
| Structural/functional features | Germline AAS in oncogenes (% of dataset) |
Germline AAS in tumour suppressor genes (% of dataset) |
p-value |
|---|---|---|---|
| Deleterious (SIFT)* | 82.33 | 69.24 | 1.17E-05 |
| Coil (Secondary structure) at wild-type residue | 41.52 | 43.77 | 5.02E-01 |
| Sheet (Secondary structure) at wild-type residue |
25.27 | 18.11 | 8.89E-03 |
| Helix (Secondary structure) at wild-type residue |
33.21 | 38.12 | 1.30E-01 |
| Buried (Solvent accessibility) at wild-type residue |
64.98 | 63.46 | 6.78E-01 |
| Exposed (Solvent accessibility) at wild-type residue |
35.02 | 36.54 | 6.78E-01 |
|
Disordered protein region at wild-type residue* |
4.33 | 15.12 | 1.83E-07 |
| Phosphoryation site at wild-type residue | 0.36 | 0.83 | 7.00E-01 |
| O-linked glycosylation site at wild-type residue |
0.36 | 0.25 | 5.64E-01 |
| Methylation site at wild-type residue | 0.00 | 0.50 | 6.01E-01 |
| Ubiquitination site at wild-type residue | 0.00 | 0.58 | 3.59E-01 |
| Calmodulin-binding site at wild-type residue | 1.81 | 2.24 | 8.20E-01 |
| Catalytic residue site at wild-type residue | 14.80 | 8.97 | 5.40E-03 |
| Decrease in stability >1 kcal/mol | 66.10 | 67.79 | 7.44E-01 |
| Coil -> Helix,Sheet (Secondary structure) | 1.44 | 2.91 | 2.13E-01 |
| Helix -> Coil,Sheet (Secondary structure) | 1.81 | 3.41 | 2.47E-01 |
| Sheet -> Helix,Coil (Secondary structure) | 2.17 | 3.07 | 5.52E-01 |
| Buried residue to exposed (solvent accessibility) |
3.61 | 6.64 | 6.85E-02 |
| Exposed residue to buried (solvent accessibility) |
7.94 | 8.39 | 9.04E-01 |
| Loss of phosphoryation site | 0.00 | 0.75 | 2.23E-01 |
| Gain of phosphoryation site | 1.81 | 0.42 | 2.45E-02 |
| Loss of O-linked glycosylation site | 0.36 | 0.17 | 4.63E-01 |
| Gain of O-linked glycosylation site | 0.72 | 0.00 | 3.49E-02 |
| Loss of methylation site | 0.00 | 0.42 | 5.91E-01 |
| Gain of methylation site | 0.36 | 0.25 | 5.64E-01 |
| Loss of ubiquitination site | 0.00 | 0.58 | 3.60E-01 |
| Gain of ubiquitination site | 0.00 | 0.42 | 5.91E-01 |
| Loss of catalytic residue | 8.66 | 4.73 | 1.26E-02 |
| Gain of catalytic residue | 3.97 | 3.99 | 1.00E+00 |
indicates significant p-value with Fisher's exact test after Bonferroni correction (P < 1.72·10−3).
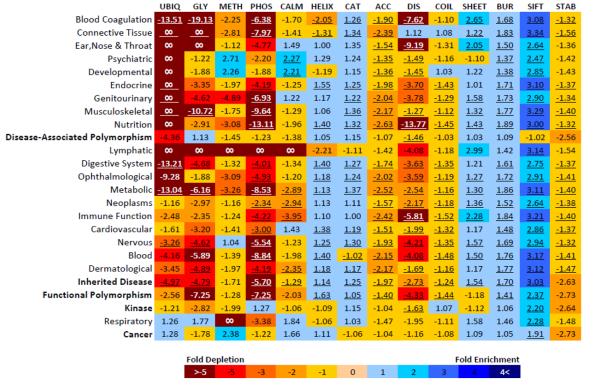
Enrichment of specific features by disease
Inspection of a ‘heat map’ depicting the enrichment or depletion of all AAS by disease category (Figure 5) reveals that only a few of the inherited disease AAS classes exhibit statistically significant differences in terms of the underlying molecular function disrupted. Blood coagulation disorders were found to exhibit a significant depletion in terms of post-translational modification sites, including a 19-fold depletion in AAS at O-linked glycosylation sites (P = 1.2·10−6; Fisher's exact test; Figure 5) and a 13-fold depletion at ubiquitination sites (P = 1.7·10−4; Fisher's exact test; Figure 5). Genitourinary disorders exhibited a 6-fold depletion in AAS at phosphorylation sites (P = 2.4·10−10; Fisher's exact test; Figure 5). Nutritional diseases exhibited a 13-fold depletion for AAS located both within disordered regions and at phosphorylation sites (P = 6.7·10−80 and P = 1.6·10−8 respectively; Fisher's exact test; Figure 5). Developmental and psychiatric disorders both showed a 2-fold enrichment for AAS located in calmodulin-binding sites (P = 3.6·10−4 and P = 2.9·10−4 respectively; Fisher's exact test; Figure 5). Overall, however, the predicted enrichment or depletion of specific protein features was not found to be an inherited disease-specific phenomenon.
Figure 5.
Fold-enrichment for the distribution of wild-type structural and functional sites for different disease concepts from the UMLS (Unified Medical Language System). Fold - enrichment for each disease concept is compared against a dataset of putatively neutral polymorphisms (Swiss-Prot neutral). Datasets are shown in bold. Statistically significant results are underlined (Bonferroni correction, P < 1.72.10−3). Colour coding indicates the magnitude of the fold-enrichment (see below). Datasets are UBIQ = Ubiquitination; GLY = O-linked glycosylation; METH = Protein methylation; PHOS = Phosphorylation; CALM = Calmodulin-binding site; HELIX = Secondary structure helix; CAT = Catalytic site; ACC = Solvent accessibility; DIS = Protein disorder; COIL = Secondary structur e, coil; SHEET = Secondary structure, beta sheet; BUR = Buried; SIFT = SIFT score; STAB = Protein stability.
Inherited disease, disease-associated polymorphisms and functional polymorphisms
The Inherited disease dataset displayed significant differences with respect to the Swiss-Prot neutral dataset in terms of SIFT predictions, structural features (protein stability, secondary structure, solvent accessibility, protein disorder) and functional sites (catalytic residues, sites of phosphorylation, ubiquitination & O-linked glycosylation). When the Disease-associated polymorphism dataset was compared against the neutral Swiss-Prot neutral dataset, the only significant differences identified were those involving the depletion of AAS in intrinsically disordered regions of proteins. Functional polymorphisms were found to be intermediate between the Inherited disease mutations and the Disease-associated polymorphisms, and differed from the neutral Swiss-Prot neutral dataset in terms of SIFT prediction, solvent accessibility, secondary structure, disordered regions, phosphorylation sites and gain of catalytic residues. Disease-associated polymorphisms are often associated with complex traits and it is therefore very likely that they exert subtle effects which, either singly or in combination with other genetic or environmental factors, give rise to a disease state/susceptibility. This contrasts with monogenic disease in which we show [and others (Wang and Moult, 2001; Yue et al., 2005) have previously shown] that disruption of protein stability is the main underlying causative factor. We further postulate that disease-associated polymorphisms are biased towards exerting their influence via the subtle modification of functional sites at exposed residues (~55%) or by modifying functional sites within disordered regions (~20%).
Somatic disease (Cancer & Kinase datasets)
Both the somatic datasets are likely to contain a proportion of ‘passenger’ as opposed to ‘driver’ mutations. Consistent with this expectation, SIFT predicted 47% of Cancer mutations and 54% of Kinase mutations to be deleterious. Although one might intuitively expect there to be fewer passenger mutations in the more focused Kinase dataset [Torkamani & Schork, 2008], in practice the slight excess of deleterious mutations in this dataset was not statistically significant.
The Kinase dataset was significantly depleted in disordered regions reflecting the idiosyncrasies of the structure of the proteins in the Kinase dataset. Both the Cancer and Kinase datasets exhibited significant enrichment for radical changes to protein structure via changes of solvent accessibility from buried to exposed, consequent to AAS. The Cancer dataset was found to be significantly enriched for AAS at calmodulin-binding sites which are short or loosely structured helical segments within otherwise disordered regions and can be seen as being analogous to Molecular Recognition Fragments (MoRFs) [Mohan et al., 2006]. Since MoRFs exhibit molecular recognition and binding functions [Mohan et al., 2006], the Cancer AAS in these regions are likely to disrupt a wide range of functions in the cell including signaling and protein-protein interaction sites. We speculate that the ‘drivers’ in the Cancer and Kinase datasets act via radical changes to protein structure, indicated by the significant enrichment of AAS predicted to alter solvent accessibility (Buried>Exposed), whilst ‘drivers’ in Cancer are also likely to exert their effects via the disruption of molecular recognition sites (e.g. protein-protein interaction sites).
Limitations of the study
The limitations of our study revolve around both the datasets and the prediction tools employed. For the Inherited disease and Disease-associated polymorphism datasets, multiple lines of evidence were used to assign an AAS as being causative of a disease phenotype. Despite the best efforts of the reporting authors and database curators, there are however likely to be a proportion of AAS in each dataset that are not actually causative of the associated disease even although they have been reported as being so. This is especially true for the Disease-associated polymorphism dataset where the majority of AAS have been reported as being causative despite there often being no direct evidence for this assertion (e.g. from functional studies etc). Therefore, a proportion of the AAS in the Disease-associated polymorphism dataset may simply be in linkage disequilibrium with the actual causative variant(s) rather than being the causative variant(s) themselves. The future use of data derived from emerging functional assays holds out the promise of generating improved disease mutation datasets that can be used to train computational classifiers [Couch et al., 2008].
Both the somatic datasets (Cancer and Kinase) are also problematic in that they are expected not only to contain mutations that lead to neoplastic progression (‘drivers’) but also neutral mutations that have arisen as a consequence of the greatly increased mutation rates in tumour cells but do not directly influence the process of tumorigenesis in any way (‘passengers’) [Greenman et al., 2007].
The in silico tools selected for this study were validated by assessing the accuracy of predictions made for known functional sites (Suppl. Table S4). The accuracy of the predictions used in subsequent analyses should therefore have been high enough to be useful both in generating and in testing the various biological hypotheses put forward.
Conclusions
Our prediction that 70% of Inherited disease AAS and 58% of Functional polymorphisms are located within buried residues is consistent with the view that disruption of protein stability is a key feature of mutations that cause inherited disease. By contrast, Inherited disease AAS predicted to be non-deleterious (‘tolerant’) by SIFT were characterized by significant enrichment for AAS within solvent accessible residues, regions of intrinsic protein disorder, and in association with the loss or gain of various post-translational modifications. Although sequence conservation is a powerful feature for the prediction of deleterious AAS in the Inherited disease dataset, it lacks resolution especially when examining polymorphic AAS within exposed (solvent accessible) or disordered regions. It is important not to neglect the role that the disruption of functional residues undoubtedly plays in disease pathogenesis especially for complex disease. Therefore, the incorporation of structural and functional sites as additional features in machine learning algorithms is likely to improve our ability to identify computationally deleterious AAS especially in the case of polymorphic AAS.
Finally, we have constructed a web resource which can be used for in silico functional profiling. This Feature Server tool can be found at http://profile.mutdb.org. Using the CakePHP development framework, users can submit their own mutations for characterization. Once a protein sequence and amino acid substitution(s) has been submitted, a script is run that calculates the predicted gain and/or loss of all the bioinformatic features discussed here.
Supplementary Material
Acknowledgements
This research was supported by NSF awards DBI-0644017 (PI: Radivojac), K22LM009135 (PI: Mooney), R01LM009722 (PI: Mooney), a grant from IU Biomedical Research Council, Indiana University, the Showalter Trust and the Indiana Genomics Initiative. The Indiana Genomics Initiative (INGEN) is supported in part by the Lilly Endowment.
REFERENCES
- Allali-Hassani A, Wasney GA, Chau I, Hong BS, Senisterra G, Loppnau P, Shi Z, Moult J, Edwards AM, Arrowsmith CH, Park HW, Schapira M, Vedadi M. A survey of proteins encoded by non-synonymous single nucleotide polymorphisms reveals a significant fraction with altered stability and activity. Biochem J. 2009;424:15–26. doi: 10.1042/BJ20090723. [DOI] [PubMed] [Google Scholar]
- Arbiza L, Duchi S, Montaner D, Burguet J, Pantoja-Uceda D, Pineda-Lucena A, Dopazo J, Dopazo H. Selective pressures at a codon-level predict deleterious mutations in human disease genes. J Mol Biol. 2006;358:1390–1404. doi: 10.1016/j.jmb.2006.02.067. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Rossi I, Casadio R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinform. 2008a;9(Suppl. 2):S6. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Arbiza L, Casadio R, Dopazo J, Dopazo H, Marti-Renom MA. Use of estimated evolutionary strength at the codon level improves the prediction of disease-related protein mutations in humans. Hum Mutat. 2008b;29:198–204. doi: 10.1002/humu.20628. [DOI] [PubMed] [Google Scholar]
- Care MA, Needham CJ, Bulpitt AJ, Westhead DR. Deleterious SNP prediction: be mindful of your training data! Bioinformatics. 2007;23:664–672. doi: 10.1093/bioinformatics/btl649. [DOI] [PubMed] [Google Scholar]
- Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J Mol Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- Che Y, Brooks BR, Marshall GR. Protein recognition motifs: design of peptidomimetics of helix surfaces. Biopolymers. 2007;86:288–297. doi: 10.1002/bip.20744. [DOI] [PubMed] [Google Scholar]
- Couch FJ, Rasmussen LJ, Hofstra R, Monteiro AN, Greenblatt MS, de Wind N, IARC Unclassified Genetic Variants Working Group Assessment of functional effects of unclassified genetic variants. Hum Mutat. 2008;29:1314–1326. doi: 10.1002/humu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M, Glorioso N, Lanzani C, Manunta P, Righetti M, Rivera R, Stella P, Troffa C, Zagato L, Bianchi G. Polymorphisms of α-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349:1353–1357. doi: 10.1016/S0140-6736(97)01029-5. [DOI] [PubMed] [Google Scholar]
- Daily KM, Radivojac P, Dunker AK. Intrinsic disorder and protein modifications: building an SVM predictor for methylation. IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, CIBCB. 2005:475–481. [Google Scholar]
- Dor O, Zhou Y. Real-SPINE: an integrated system of neural networks for real-value prediction of protein structural properties. Proteins. 2005;68:76–81. doi: 10.1002/prot.21408. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Faraggi E, Xue B, Zhou Y. Improving the prediction accuracy of residue solvent accessibility and real-value backbone torsion angles of proteins by guided-learning through a two-layer neural network. Proteins. 2009;74:847–856. doi: 10.1002/prot.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Costa C, Gelpí JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegga AG, Gowrisankar S, Chen J, Aronow BJ. PolyDoms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Res. 2007;35:D700–706. doi: 10.1093/nar/gkl826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Yang H, Sun F, Chen T. Searching for interpretable rules for disease mutations: a simulated annealing bump hunting strategy. BMC Bioinform. 2006;7:417. doi: 10.1186/1471-2105-7-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005a;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- Karchin R, Kelly L, Sali A. Improving functional annotation of non-synonomous SNPs with information theory. Pac Symp Biocomput. 2005b:397–408. doi: 10.1142/9789812702456_0038. [DOI] [PubMed] [Google Scholar]
- Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinform. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan VG, Westhead DR. A comparative study of machine-learning methods to predict the effects of single nucleotide polymorphisms on protein function. Bioinformatics. 2003;19:2199–2209. doi: 10.1093/bioinformatics/btg297. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Nishida K, Honma Y, Dota A, Kawasaki S, Adachi W, Nakamura T, Quantock Isolation and chromosomal localization of a cornea-specific human keratin 12 gene and detection of four mutations in Meesmann corneal epithelial dystrophy. Am J Hum Genet. 1997;61:1268–1275. doi: 10.1086/301650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerenshaw M, Page T, Hammonds J, Demaine A. Polymorphisms in the hypoxia inducible factor-1α gene (HIF1A) are associated with the renal cell carcinoma phenotype. Cancer Genet Cytogenet. 2004;153:122–126. doi: 10.1016/j.cancergencyto.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–133. doi: 10.1093/nar/gkh028. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Obradovic Z, Brown CJ, Dunker AK. Improving sequence alignments for intrinsically disordered proteins. Pac Symp Biocomput. 2002:589–600. [PubMed] [Google Scholar]
- Radivojac P, Vucetic S, O'Connor TR, Uversky VN, Obradovic Z, Dunker AK. Calmodulin signaling: analysis and prediction of a disorder-dependent molecular recognition. Proteins. 2006;63:398–410. doi: 10.1002/prot.20873. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Baenziger PH, Kann MG, Mort ME, Hahn MW, Mooney SD. Gain and loss of phosphorylation sites in human cancer. Bioinformatics. 2008;24:i241–247. doi: 10.1093/bioinformatics/btn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2009 doi: 10.1002/prot.22555. In press [doi>10.1002/prot.22555] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve NH, Han H. CONANN: An Online Biomedical Concept Annotator. Lecture Notes in Computer Science. 2007:4544. [Google Scholar]
- Sandberg WS, Schlunk PM, Zabin HB, Terwilliger TC. Relationship between in vivo activity and in vitro measures of function and stability of a protein. Biochemistry. 1995;34:11970–11978. doi: 10.1021/bi00037a039. [DOI] [PubMed] [Google Scholar]
- Saunders CT, Baker D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J Mol Biol. 2002;322:891–901. doi: 10.1016/s0022-2836(02)00813-6. [DOI] [PubMed] [Google Scholar]
- Schön S, Schulz V, Prante C, Hendig D, Szliska C, Kuhn J, Kleesiek K, Götting C. Polymorphisms in the xylosyltransferase genes cause higher serum XT-I activity in patients with pseudoxanthoma elasticum (PXE) and are involved in a severe disease course. J Med Genet. 2006;43:745–749. doi: 10.1136/jmg.2006.040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Musen M. UMLS-Query: A Perl module for querying the UMLS. AMIA Annu Symp Proc. 2008:652–656. [PMC free article] [PubMed] [Google Scholar]
- Shah NH, Rubin DL, Supekar KS, Musen MA. Ontology-based annotation and query of tissue microarray data. AMIA Annu Symp Proc. 2006:709–713. [PMC free article] [PubMed] [Google Scholar]
- Shah NH, Rubin DL, Espinosa I, Montgomery K, Musen MA. Annotation and query of tissue microarray data using the NCI Thesaurus. BMC Bioinform. 2007;8:296. doi: 10.1186/1471-2105-8-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Singh A, Olowoyeye A, Baenziger PH, Dantzer J, Kann MG, Radivojac P, Heiland R, Mooney SD. MutDB: update on development of tools for the biochemical analysis of genetic variation. Nucleic Acids Res. 2007;36:D815–819. doi: 10.1093/nar/gkm659. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward RE, MacArthur MW, Laskowski RA, Thornton JM. Molecular basis of inherited diseases: a structural perspective. Trends Genet. 2003;19:505–513. doi: 10.1016/S0168-9525(03)00195-1. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB, IARC Unclassified Genetic Variants Working Group In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat. 2008;29:1327–1336. doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terp BN, Cooper DN, Christensen IT, Jørgensen FS, Bross P, Gregersen N, Krawczak M. Assessing the relative importance of the biophysical properties of amino acid substitutions associated with human genetic disease. Hum Mutat. 2002;20:98–109. doi: 10.1002/humu.10095. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Schork NJ. Prediction of cancer driver mutations in protein kinases. Cancer Res. 2008;68:1675–1682. doi: 10.1158/0008-5472.CAN-07-5283. [DOI] [PubMed] [Google Scholar]
- Viswanathan PC, Benson DW, Balser JR. A common SCN5A polymorphism modulates the biophysical effects of an SCN5A mutation. J Clin Invest. 2003;111:341–346. doi: 10.1172/JCI16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, Chapgier A, Yang K, Chuzhanova N, Feinberg J, Fieschi C, Boisson-Dupuis S, Alcais A, Filipe-Santos O, Bustamante J, de Beaucoudrey L, Al-Mohsen I, Al-Hajjar S, Al-Ghonaium A, Adimi P, Mirsaeidi M, Khalilzadeh S, Rosenzweig S, de la Calle Martin O, Bauer TR, Puck JM, Ochs HD, Furthner D, Engelhorn C, Belohradsky B, Mansouri D, Holland SM, Schreiber RD, Abel L, Cooper DN, Soudais C, Casanova JL. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wang P, Dai M, Xuan W, McEachin RC, Jackson AU, Scott LJ, Athey B, Watson SJ, Meng F. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–529. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. Calmodulin target database. J Struct Funct Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- Ye Y, Li Z, Godzik A. Modeling and analyzing three-dimensional structures of human disease proteins. Pacific Symp Biocomp. 2006;11:439–446. [PubMed] [Google Scholar]
- Youn E, Peters B, Radivojac P, Mooney SD. Evaluation of features for catalytic residue prediction in novel folds. Protein Sci. 2007;16:216–226. doi: 10.1110/ps.062523907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinform. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.