Abstract
Background
Sulphadoxine and pyrimethamine are anti-folate drugs that show synergistic anti-malarial effect. Point mutations in dihydrofolate reductase (dhfr) and dihydropteorate synthatase (dhps) cause anti-folate drug resistance phenotype in human malaria parasites. This study presents pattern of point mutations in dhfr/dhps genes among Indian sub-continent.
Methods
Microscopically diagnosed one hundred Plasmodium vivax field isolates were collected from five widely separated geographical regions of India. Dhfr and dhps genes were PCR amplified and sequenced. Previously published mutations data were collected and analyzed using Chi square test to identify geographical cluster of mutant/wild type genotypes.
Results
Sequence analysis revealed single (S58R), double (S58R/S117N) and quadruple (F57L/S58R/T61M/S117T/) point mutations at dhfr and single (A383G) to double (A383G/A553G) mutations at dhps in P. vivax field isolates. In addition, three new mutations were also observed at dhfr. Both, dhfr and dhps genes revealed tandem repeat variations in field isolates. Dhps revealed very low mutation frequency (14.0%) compared to dhfr (50.70%). Comparative analysis revealed a progressive increase in frequency of quadruple mutant dhfr genotype (p < 0.001) within five years in north-eastern state (Kamrup, Assam). Frequency of dhfr genotypes revealed three distinct geographical clusters of wild (northern India), double mutant (southern India), and quadruple mutant (north-eastern and island regions of India) on the Indian sub-continent.
Conclusion
Study suggests that SP may be susceptible to P. vivax in India, except Andaman and north-eastern state. The distinction of geographical regions with sensitive and resistant parasite phenotypes would be highly useful for designing and administering national anti-malarial drug policy.
Background
Malaria is a life-threatening ancient parasitic disease and causes 250-500 million clinical episodes and nearly one million deaths annually [1]. Among the five human malaria species, Plasmodium falciparum is the most severe form, causing malignant malaria globally, while Plasmodium vivax is the most widespread species outside Africa, causing huge morbidity and can be severe and fatal [2-7].
The worldwide spread of chloroquine (CQ) resistant strains of P. falciparum has led to use of sulphadoxine-pyrimethamine (SP) as the first-line anti-malarial drug in Southeast Asian countries. Sulphadoxine and pyrimethamine sequentially inhibits dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR) enzymes respectively in the folate biosynthesis pathway resulting synergistic anti-malarial effect [8]. Parasite has overcome the effect of SP by evolving point mutations in the respective genes encoding enzymes involved in the folate biosynthesis pathway. The mutated DHPS and DHFR enzymes have reduced binding affinity with SP drug and thus parasite survives in the presence of drug [9,10].
In India, resistance to CQ was reported for the first time in 1973 in P. falciparum from north-eastern states [11] and later it spread throughout the country [12]. To overcome CQ resistance problem, in 1982, SP was employed as first-line anti-malarial therapy for treatment of falciparum malaria in areas with >25% CQ resistant level, complicated malaria case, and higher malaria endemicity [12]. Currently, according to the national anti-malarial drug policy, artimisinin-based combination therapy (ACT) (Artesunate+SP) is being used in most of the malaria endemic regions [12].
Reduced susceptibility to CQ in P. vivax was for the first time observed in 1989 in Australian soldiers returned from Papua New Guinea [13]. Later, several cases from Papua New Guinea, Indonesia, New Guinea, Brazil and India were documented [14-19]. Recent studies from P. vivax ex-vivo maturation experiment showed reduction in the susceptibility to CQ in Southeast Asian counties [20,21]. This information indicates gradual increase in CQ resistance cases of P. vivax.
India contributes more than 78% of total malaria cases of Southeast Asia and P. vivax accounts for more than 50% of annual malaria cases [22]. Point mutations in P. vivax dhfr had been documented from different parts of India [23-25], however mutation data of dhps is yet to identified in order to understand the molecular epidemiology of anti-folate resistance. Therefore, identifying information about presence of anti-folate drug resistance related point mutations (dhfr/dhps) in P. vivax from Indian sub continent would be highly helpful to understand the global pattern of anti-folate drug resistance. This study aims to identify point mutations in dhfr/dhps and the spatio temporal pattern of anti-folate drug resistance in Indian sub-continent.
Methods
Study sites and sample collection
Blood samples were collected from five widely separated geographical regions of the Indian subcontinent namely Delhi (2005); Chennai, Tamil Nadu (2005); Kamrup, Assam (2007); Nadiad, Gujarat (2005) and Panna, Madhya Pradesh (2006). Details of epidemiological and geographical information about study sites are reported elsewhere [26]. Finger prick blood was spotted on autoclaved Whatman filter paper strips (Number 3) from the symptomatic patients in active case detection surveys as well as from patient attending clinics. A total of 100 microscopically-diagnosed P. vivax positive blood samples were spotted and dried blood spots were stored at 4.0°C. Only P. vivax infected samples were included in this study. This study was approved by the ethics committee of the National Institute of Malaria Research, New Delhi. All blood spots were collected only after obtaining consent of the patients.
DNA extraction, PCR, and DNA sequencing
Genomic DNA was extracted from blood spots using QIAamp mini DNA kit (Qiagen, Germany) according to manufacturer instructions. Genomic DNA was eluted in 120.0 μl triple sterile water and store in -20°C until use. One step modified PCR strategy was employed for amplification of dhfr/dhps using nested primers reported earlier [27,28]. The modified PCR conditions for dhfr was- initial denaturation at 95°C/5.0 minute, denaturation at 95°C/30 second, annealing at 64°C/30 second, and extension at 72°C/60 second for 40 cycle, and a final extension at 72°C/5.0 minute. The modified PCR amplification for dhps was same as dhfr except annealing step, which is 55°C/30 second. PCR products were purified with Exonuclease I/Shrip alkaline phosphates treatment according to manufacturer instruction (Fermentas, USA). Purified PCR products were outsourced to Macrogen Inc, Korea for DNA sequencing [29]. Each sample was sequenced with both forward and reverse primers. DNA sequences were edited and aligned (ClustalW method) with EditSeq and MegAlign module of DNA Lasergene software version 7.0 (Madison, USA). Samples that show novel point mutations were re-sequenced from another independent PCR product. All sequences have been submitted to the GenBank (EU149665-EU149764, EU145878-EU145947).
Collection of dhfr mutations data and statistical analysis
Dhfr point mutations data from Panna, Madhya Pradesh (N = 30); Nadiad, Gujarat (N = 19); Mukherji Nagar, Delhi (N = 68); Cuttack, Orissa (N = 16); Mohan Nagar (N = 49) Aligarh (N = 38) and Mirzapur (N = 8), Uttar Pradesh; Chennai, Tamil Nadu (N = 67) Car Nicobar, Andaman Nicobar (N = 38); Panjim, Goa (N = 76); Kamrup, Assam (N = 24) and Navi Mumbai, Maharashtra (N = 24) were collected from the earlier studies [23-25,30]. On the basis of mutations in dhfr and geographical location, grouping between different geographical regions was done. Geographical regions were grouped according to the higher prevalence of wild type, double mutant, and quadruple mutant dhfr genotypes. Chi square test was done to check the significance of prevalence of dhfr mutant genotypes in geographical locations. Confidence interval at 95% was done to check the proportion dhfr genotypes in each group.
Results
Mutations analysis in dhfr and dhps
For dhfr, a total of 71 P. vivax isolates were successfully PCR amplified and sequenced. PCR amplified dhfr was 711 bp in length and covers complete coding region. All sequences were compared with wild type reference sequence (GenBank accession no. X98123) to detect point mutations in dhfr. Point mutations observed at dhfr were given in table 1. Approximately 50% (35/71) sequences at dhfr were wild type. DNA sequence alignment revealed point mutations at 57(F→L), 58(S→R), 61(T→M), 64(V→L), 117(S→T), and 173(I→F). Point mutations at codon 64 (V→L) and 173 (I→F) were first time observed in the Indian isolates. One isolate showed amino acid substitution (57F→L) by changing nucleotide TTG rather than TTA (TTC wild type). Dhfr sequence analysis revealed two synonymous mutations at codon 69Y (TAT→TAC) and 134 V (GTC→GTT). Synonymous mutation 134 V (GTC→GTT) is novel and observed only in the wild type dhfr genotype. The majority of the mutated codons were S58R (47.88%), S117N/T (46.47%), F57L (21.12%), and T61M (19.71%). Study isolates revealed single (S58R), double (S58R/S117N) and quadruple (F57L/S58R/T61M/S117T/) mutations.
Table 1.
Distribution of Plasmodium vivax dhfr/dhps point mutations in field isolates
| DHFR | DHPS | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Regions | Sample size | Wild type | F57L | S58R | T61M | V64L | S117N/T | I173F | T69C | C134T | Sample size | Wild type | A383G | A553G |
| Nadiad | 3 | 2 | 1 | 1 | 20 | 20 | ||||||||
| Delhi | 11 | 10 | 1 | 2 | 20 | 17 | 3 | 3 | ||||||
| Panna | 17 | 13 | 4 | 3 | 8 | 2 | 20 | 20 | ||||||
| Chennai | 19 | 3 | 1 | 16 | 1 | 16 | 1 | 20 | 9 | 11 | 8 | |||
| Kamrup | 21 | 7 | 14 | 13 | 13 | 13 | 1 | 20 | 20 | |||||
| Total | 71 | 35 | 15 | 34 | 14 | 1 | 33 | 1 | 11 | 2 | 100 | 86 | 14 | 11 |
Boldface: synonymous substitution,
Values indicate number of isolates.
For dhps, a total of 100 P. vivax isolates were successfully PCR amplified (950 bp) which covers hotspot region for drug resistance related point mutations (codon 350 to 652). All sequences were compared with wild type reference sequence (GenBank accession no. AY186730) to detect point mutations in dhps. Sequence analysis revealed point mutation at codon A383G and A583G (Table 1). Majority of the sequences were wild type (86%). Only single (A383G) and double (A383G/A583G) point mutations were observed at dhps in the study isolates (Table 1).
Geographical distribution of dhfr and dhps genotypes
Four distinct genotypes of dhfr were observed and these are wild type, single mutant, double mutant and quadruple mutant. In total, among mutant genotypes, frequency of double mutant (28.16%) and quadruple mutant (19.71%) was higher compared with single mutant (2.81%). Isolates of Panna, Delhi, and Nadiad were dominated by wild type genotype (Table 2). Frequency of mutant dhfr genotypes was highest in isolates of Chennai (84.24%) and Kamrup (66.67%) regions. In contrast, isolates from Delhi (9.0%), Panna (23.52%), and Nadiad (33.34%) showed relatively low proportion of mutant genotypes. Geographical distribution of various genotypes observed at dhfr is given in Table 2. Double mutant genotype was observed to be predominated in Chennai isolates (78.94%) whereas quadruple mutant genotype was dominated in Kamrup region (61.90%). The silent point mutation T69C was observed in wild type dhfr sequences only. The point mutation T69C was found at a high frequency in isolates of Panna (47.05%) but in lesser extent in Delhi (18.18%) and Chennai (5.26%) isolates. Another silent point mutation C134T was limited to isolates of Panna (11.76%).
Table 2.
Region wise distribution of Plasmodium vivax dhfr and dhps genotypes
| DHFR (%) | DHPS (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Region | Sample size | WT | SM | DM | QM | Sample size | WT | SM | DM |
| Nadiad | 3 | 66.67 | 33.33 | 20 | 100 | ||||
| Delhi | 11 | 90.9 | 9.1 | 20 | 85 | 15 | |||
| Panna | 17 | 76.47 | 5.88 | 17.65 | 20 | 100 | |||
| Chennai | 19 | 15.79 | 78.95 | 5.26 | 20 | 45 | 15 | 40 | |
| Kamrup | 21 | 33.34 | 4.76 | 61.9 | 20 | 100 | |||
| Total | 71 | 49.29 | 2.81 | 28.16 | 19.71 | 100 | 86 | 3 | 11 |
Three distinct dhps genotypes were observed in the field isolates: wild type, single mutant and double mutant. The wild type genotype was observed in high frequency in field isolates of Delhi (85%) and Chennai (55%) whereas isolates from Panna, Nadiad, and Kamrup were exclusively wild type. The mutant genotypes (single and double) were only observed in isolates from Delhi and Chennai (Table 2).
Tandem repeat variation, distribution, and association with mutations
At dhfr, four tandem repeat variants were observed and designated as Type 1-4, on the basis of six amino acid deletion/insertion (Figure 1). In total, tandem repeat Type 2 (84.50%) was the major repeat polymorph observed in field isolates (Table 3). Type 2 tandem repeat was observed in high frequency in all study sites except for Delhi where Type 1 predominates. Distribution of tandem repeat variants in different study sites is given in Table 3. Tandem repeat variant was monomorphic (Type-2) in field isolates of Nadiad, Chennai and Kamrup, whereas it was polymorphic in field isolates of Delhi and Panna. This suggests that monomorphic nature of tandem repeat was only observed in a region where mutant dhfr genotypes predominate. In study isolates, all mutant dhfr genotypes were observed in isolates having Tandem repeat Type-2.
Figure 1.
Tandem repeat variation in Plasmodium vivax DHFR.
Table 3.
Distribution of tandem repeat variants at DHFR and DHPS in field isolates
| DHFR | Regions (%) | Total N (%) | ||||
|---|---|---|---|---|---|---|
| Repeat Type | Delhi | Nadiad | Panna | Chennai | Kamrup | |
| Type 1 | 54.54 | 0 | 11.76 | 0 | 0 | 8 (11.26) |
| Type 2 | 36.34 | 100 | 76.47 | 100 | 100 | 60 (84.5) |
| Type 3 | 9.09 | 0 | 0 | 0 | 0 | 1 (1.4) |
| Type 4 | 0 | 0 | 11.76 | 0 | 0 | 2 (2.81) |
| Total (N) | 11 | 3 | 17 | 19 | 21 | 71 |
| DHPS | ||||||
| Type A | 10 | 10 | 0 | 5 | 21.05 | 9.27 |
| Type B | 90 | 85 | 83.34 | 90 | 26.31 | 75.25 |
| Type C | 0 | 0 | 5.55 | 5 | 0 | |
| Type D | 0 | 5 | 5.55 | 0 | 15.79 | 5.15 |
| Type E | 0 | 0 | 0 | 0 | 10.52 | 2.06 |
| Type F | 0 | 0 | 0 | 0 | 10.52 | 2.06 |
| Type G | 0 | 0 | 5.55 | 0 | 10.52 | 3.09 |
| Type H | 0 | 0 | 0 | 0 | 5.26 | 1.03 |
| Total (N) | 20 | 20 | 18* | 20 | 19* | 97 |
*: DNA sequences of tandem repeat region was not good in some isolates and therefore not included in analysis of tandem repeats
At dhps, eight tandem repeats variants were observed and designated as Type A-H on the basis of deletion/insertion of seven amino acid repeat whereas (Figure 2). Tandem repeat Type-B was the major variant in the field isolates (75.25%) and predominate in all study sites (Table 3). Distribution of tandem repeat variants in different study sites is given in Table 3. Seven tandem repeat variants were found in field isolates of Kamrup, three in Panna, Chennai, and Nadiad and only two in Delhi. Point mutations were only observed in field isolates having tandem repeat Type B.
Figure 2.
Tandem repeat variation in Plasmodium vivax DHPS.
Progressive increase in frequency of dhfr quadruple mutant
In Kamrup region, dhfr point mutation data from 2002-2003 revealed higher frequency of double mutant (54.16%) and low prevalence of wild (29.16%) and quadruple mutant (16.67%) genotypes [23]. Present study conducted in 2006-2007 revealed quite surprising result with respect to the data of dhfr point mutations studied in 2002-03. In the present study a high prevalence of the quadruple mutant genotype (61.90%) and an almost absence of the double mutant genotype (4.16%) were observed. However, the lower prevalence of wild type remains the same. The interesting finding was the drastic increase in the frequency of quadruple mutant dhfr genotype in Kamrup region (p = <0.0001) within a five year (2002 to 2007) span. This suggests that there is progressive increase in the frequency of quadruple mutant dhfr genotype.
Geographical clustering of dhfr genotypes among Indian continent
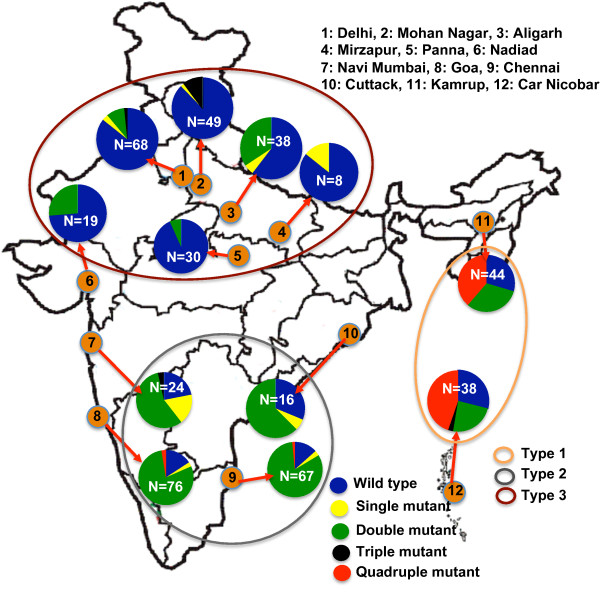
Dhfr point mutations data of earlier published work (n = 477) revealed single to quadruple mutant genotypes with their varied prevalence in different geographical regions of India (Figure 3). Comprehensive analysis of dhfr point mutations revealed three distinct pattern which are- 1) high prevalence of wild type, 2) high prevalence of double mutant type, and 3) high prevalence of quadruple mutant type. On the basis of above categorization, study sites were grouped together to see any trend of significance between geographical location and prevalence of dhfr genotypes. Three distinct geographical clusters were appeared at significant level (p < 0.0001). These geographical clusters are- Type-1) covers geographical areas of North India and majority of isolates were wild type (81.64%, 95% CI = ± 4.9) and very low proportion of single and or double mutant. Type-2) covers geographical areas of South India and majority of isolates were double mutant type (80.54%, 95% CI = ± 6.22) and very low proportion of wild and single mutant types. Type-3) covers geographical areas of north-eastern state and Island region where quadruple mutant (42.68%, 95% CI = ± 10.72) predominate over wild (29.26%) and double mutant type (28.04%). Details of geographical regions falling in specific cluster are given in figure 3. Comparison of Type-1 cluster with Type-2 or mutant versus wild types (p < 0.0001), and Type-2 cluster with Type-3 or double mutant versus quadruple mutant (p < 0.0001) strongly supports grouping or clustering of wild, double mutant, and quadruple mutant dhfr genotypes in three distinct geographical regions of the country.
Figure 3.
Frequencies of Plasmodium vivax dhfr genotypes and their geographical clustering among Indian subcontinent.
Discussion
Due to emergence and widespread of drug resistant strains of human Plasmodium species against chemotherapeutic intervention agents, clinical trial for monitoring drug efficacy over regular intervals as well as monitoring mutation data in relevant genes related with anti-malarial resistance are the crucial steps in deciding scope of a drug in field. This study uncovers 1) progressive increase in the frequency of quadruple mutant dhfr genotypes, 2) limited point mutations in dhps among Indian isolates, and 3) distinct pattern of anti-folate resistance in Indian sub-continent linked with geographical regions.
Plasmodium vivax is still susceptible to chloroquine in India [30,31] and anti-folate drugs are not used for treatment of vivax malaria, therefore, the appearance of point mutations in dhfr/dhps is surprising. This has been explained earlier because of the use of sulphadoxine-pyrimethamine to treat chloroquine-resistant P. falciparum is creating selection pressure in the P. vivax population. This has been reflected in the isolates of areas with sympatricity of P. falciparum and P. vivax [23,25], which presented a higher proportion of double and quadruple mutant dhfr genotypes than other regions. Cotrimoxazole (sulfamethoxazole and trimethoprim), an anti-microbial drug that target bacterial dhfr/dhps, is widely used in bacterial infection as prophylaxis, and has potential to select mutant dhfr/dhps of malaria parasites [32,33]. Cotrimoxazole is being used in India to treat bacterial infection in wide spectrum [34,35], which suggests that it could be a potential factor to select anti-folate point mutations in the Indian subcontinent. Other factors, i.e. presumptive treatment of malaria without diagnosis and use of S/P by private practitioners, cannot be ruled out.
The comprehensive analysis of point mutations in dhfr among Indian sub-continent revealed three distinct clusters/groups/types linked with geographical locations. North and south India revealed a prevalence of wild and double mutant dhfr, respectively, whereas north-eastern and island regions revealed a high prevalence of quadruple mutant dhfr genotype. Single to quintuple mutations have been reported in dhfr but only triple to quintuple mutant alleles are known to confer high level of resistance [36-38]. This strongly suggests that P. vivax isolates from northern and southern part of India could be sensitive to anti-folate drug, whereas isolates from north-eastern state and Island regions will be resistant to anti-folate drug.
A significant association between dhfr mutations and tandem repeat polymorphism was observed in studied isolates. The mutated dhfr genotype was exclusively associated with Type-2 tandem repeat and is strongly supported by previous study from India [23]. Further quadruple mutant dhfr alleles were exclusively associated with type 1 tandem repeat and are supported by earlier studies from India [23], Thailand [38] and Myanmar [39], however exception to this association was only showed by a single isolate from Myanmar [39]. These observations speculate dhfr allele with type 1 tandem repeat could be more prone for mutations and development of higher level of resistance conferring genotypes. Therefore, it could act as molecular marker to predict the risk of mutant/higher level resistance conferring mutant genotypes in any geographical area of Indian subcontinent.
Frequency of quadruple mutant dhfr genotype was progressively increased in Kamrup isolates from 2005 to 2007 [23]. The high proportion of quadruple mutant dhfr genotype in endemic area may be: 1) because of the characteristics of mutant parasites associated with higher gametocytogenesis [40-43] which favour transmission of mutant parasites compared to wild type dhfr, and 2) north-eastern states of India are highly endemic for malaria and international borders with neighbouring countries surround the region. The borders are very porous and illegal migrations of people across the borders are very common thus, importing drug resistant strains. The higher frequency of quadruple mutant dhfr genotypes in the isolates of Myanmar (71%) and Thailand (96%) [39,44], supports for inflow of quadruple mutant dhfr genotype in north-eastern states of India.
The development and spread of drug resistant parasite strains is a major obstacle to the malaria elimination programme. Therefore, it is essential to identify drug resistance areas/regions on the basis of point mutations in order to manage the national anti-malarial drug policy. In this regard, this study revealed a higher and increasing prevalence of quadruple mutant dhfr genotype in north-eastern and Island regions, therefore, additional caution may be taken for treatment of vivax malaria in these regions to stop the flourishing of quadruple mutant dhfr genotypes in remaining part of country. The study concluded that P. vivax isolates from northern and southern part of India could be susceptible to SP except for the isolates from Car Nicobar (Island) and Assam (north-eastern region). Geographical clustering of dhfr mutant genotypes would provide a rationale for a more appropriate national anti-malarial drug policy.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SKP: Experiment design, experimental work, data analysis, manuscript writing
HJ: Conceptual design of work, data analysis, manuscript writing,
VD: Sample collection, parasite identification, and experimental design
VKD: Conceptual design of work, Overall supervision of work, and manuscript writing
All authors read and approved the final manuscript.
Contributor Information
Surendra K Prajapati, Email: surendramrc@gmail.com.
Hema Joshi, Email: hema_joshi_mrc@yahoo.com.
Vas Dev, Email: mrcassam@hotmail.com.
Virendra K Dua, Email: vkdua51@gmail.com.
Acknowledgements
This work is supported by Department of Biotechnology, New Delhi. SKP is an ICMR Postdoctoral Fellow. The authors are thankful to the patients for their co-operation during the study. The authors are also thankful to scientist/staff of National Institute of Malaria Research, New Delhi as well as from field station for their support and help during the study. Authors expressed their deep gratitude to Dr. Hema Joshi, who passed away recently and she had contributed significantly to the present study.
References
- WHO. World Health Organization. Geneva; 2008. Annual Report. [Google Scholar]
- Andrade BB, Reis-Filho A, Souza-Neto SM, Clarencio J, Camargo LM, Barral A, Barral-Netto M. Severe Plasmodium vivax malaria exhibits marked inflammatory imbalance. Malar J. 2010;9:13. doi: 10.1186/1475-2875-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, Kochar A, Khatri MP, Gupta V. Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg. 2009;80:194–198. [PubMed] [Google Scholar]
- Kochar DK, Saxena V, Singh N, Kochar SK, Kumar SV, Das A. Plasmodium vivax malaria. Emerg Infect Dis. 2005;11:132–134. doi: 10.3201/eid1101.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genton B, D'Acremont V, Rare L, Baea K, Reeder JC, Alpers MP, Muller I. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008;5:e127. doi: 10.1371/journal.pmed.0050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson SJ, Carter R. Severe vivax malaria: newly recognised or rediscovered. PLoS Med. 2008;5:e136. doi: 10.1371/journal.pmed.0050136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjitra E, Anstey NM, Sugiarto P, Warikar N, Kenangalem E, Karyana M, Lampah DA, Price RN. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5:e128. doi: 10.1371/journal.pmed.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chulay JD, Watkins WM, Sixsmith DG. Synergistic antimalarial activity of pyrimethamine and sulfadoxine against Plasmodium falciparum in vitro. Am J Trop Med Hyg. 1984;33:325–330. doi: 10.4269/ajtmh.1984.33.325. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal PN, Sharma MID, I SS, Gogal S. Resistance to chloroquine in falciparum malaria in Assam state, India. J Commun Dis. 1973;5:175–180. [Google Scholar]
- National Vector Borne Disease Control Programme. http://www.nvbdcp.gov.in
- Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine? Lancet. 1989;2:1183–1184. doi: 10.1016/s0140-6736(89)91792-3. [DOI] [PubMed] [Google Scholar]
- Garavelli PL, Corti E. Chloroquine resistance in Plasmodium vivax: the first case in Brazil. Trans R Soc Trop Med Hyg. 1992;86:128. doi: 10.1016/0035-9203(92)90535-K. [DOI] [PubMed] [Google Scholar]
- Garg M, Gopinathan N, Bodhe P, Kshirsagar NA. Vivax malaria resistant to chloroquine: case reports from Bombay. Trans R Soc Trop Med Hyg. 1995;89:656–657. doi: 10.1016/0035-9203(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Murphy GS, Basri H, Purnomo, Andersen EM, Bangs MJ, Mount DL, Gorden J, Lal AA, Purwokusumo AR, Harjosuwarno S. et al. Vivax malaria resistant to treatment and prophylaxis with chloroquine. Lancet. 1993;341:96–100. doi: 10.1016/0140-6736(93)92568-E. [DOI] [PubMed] [Google Scholar]
- Myat Phone K, Myint O, Myint L, Thaw Z, Kyin Hla A, Nwe Nwe Y. Emergence of chloroquine-resistant Plasmodium vivax in Myanmar (Burma) Trans R Soc Trop Med Hyg. 1993;87:687. doi: 10.1016/0035-9203(93)90294-Z. [DOI] [PubMed] [Google Scholar]
- Schuurkamp GJ, Spicer PE, Kereu RK, Bulungol PK, Rieckmann KH. Chloroquine-resistant Plasmodium vivax in Papua New Guinea. Trans R Soc Trop Med Hyg. 1992;86:121–122. doi: 10.1016/0035-9203(92)90531-G. [DOI] [PubMed] [Google Scholar]
- Schwartz IK, Lackritz EM, Patchen LC. Chloroquine-resistant Plasmodium vivax from Indonesia. N Engl J Med. 1991;324:927. doi: 10.1056/NEJM199103283241317. [DOI] [PubMed] [Google Scholar]
- Hasugian AR, Tjitra E, Ratcliff A, Siswantoro H, Kenangalem E, Wuwung RM, Purba HL, Piera KA, Chalfien F, Marfurt J, Penttinen PM, Russell B, Anstey NM, Price RN. In vivo and in vitro efficacy of amodiaquine monotherapy for treatment of infection by chloroquine-resistant Plasmodium vivax. Antimicrob Agents Chemother. 2009;53:1094–1099. doi: 10.1128/AAC.01511-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B, Chalfein F, Prasetyorini B, Kenangalem E, Piera K, Suwanarusk R, Brockman A, Prayoga P, Sugiarto P, Cheng Q, Tjitra E, Anstey NM, Price RN. Determinants of in vitro drug susceptibility testing of Plasmodium vivax. Antimicrob Agents Chemother. 2008;52:1040–1045. doi: 10.1128/AAC.01334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Valecha N, Jain T, Dash AP. Burden of malaria in India: retrospective and prospective view. Am J Trop Med Hyg. 2007;77:69–78. [PubMed] [Google Scholar]
- Alam MT, Bora H, Bharti PK, Saifi MA, Das MK, Dev V, Kumar A, Singh N, Dash AP, Das B, Wajihullah, Sharma YD. Similar trends of pyrimethamine resistance-associated mutations in Plasmodium vivax and P. falciparum. Antimicrob Agents Chemother. 2007;51:857–863. doi: 10.1128/AAC.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Prajapati SK, Kalyanaraman K, Mohmmed A, Joshi H, Chauhan VS. Plasmodium vivax dihydrofolate reductase point mutations from the Indian subcontinent. Acta Trop. 2006;97:174–180. doi: 10.1016/j.actatropica.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Prajapati SK, Joshi H, Valecha N, Reetha AM, Eapen A, Kumar A, Das MK, Yadav RS, Rizvi MA, Dash AP. Allelic polymorphism in the Plasmodium vivax dihydrofolate reductase gene among Indian field isolates. Clin Microbiol Infect. 2007;13:331–334. doi: 10.1111/j.1469-0691.2006.01639.x. [DOI] [PubMed] [Google Scholar]
- Prajapati SK, Verma A, Adak T, Yadav RS, Kumar A, Eapen A, Das MK, Singh N, Sharma SK, Rizvi MA, Dash AP, Joshi H. Allelic dimorphism of Plasmodium vivax gam-1 in the Indian subcontinent. Malar J. 2006;5:90. doi: 10.1186/1475-2875-5-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Pukrittakayamee S, Looareesuwan S, Pasvol G, Poirreiz J, White NJ, Snounou G. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob Agents Chemother. 2001;45:3122–3127. doi: 10.1128/AAC.45.11.3122-3127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegon M, Majori G, Severini C. Genetic variations of the Plasmodium vivax dihydropteroate synthase gene. Acta Trop. 2006;98:196–199. doi: 10.1016/j.actatropica.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Macrogen Inc. Korea. http://www.macrogen.com
- Valecha N, Joshi H, Eapen A, Ravinderan J, Kumar A, Prajapati SK, Ringwald P. Therapeutic efficacy of chloroquine in Plasmodium vivax from areas with different epidemiological patterns in India and their Pvdhfr gene mutation pattern. Trans R Soc Trop Med Hyg. 2006;100:831–837. doi: 10.1016/j.trstmh.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Nandy A, Addy M, Maji AK, Bandyopadhyay AK. Monitoring the chloroquine sensitivity of Plasmodium vivax from Calcutta and Orissa, India. Ann Trop Med Parasitol. 2003;97:215–220. doi: 10.1179/000349803235001868. [DOI] [PubMed] [Google Scholar]
- Jelinek T, Kilian AH, Curtis J, Duraisingh MT, Kabagambe G, von Sonnenburg F, Warhurst DC. Plasmodium falciparum: selection of serine 108 of dihydrofolate reductase during treatment of uncomplicated malaria with co-trimoxazole in Ugandan children. Am J Trop Med Hyg. 1999;61:125–130. doi: 10.4269/ajtmh.1999.61.125. [DOI] [PubMed] [Google Scholar]
- Kiara SM, Okombo J, Masseno V, Mwai L, Ochola I, Borrmann S, Nzila A. In vitro activity of antifolate and polymorphism in dihydrofolate reductase of Plasmodium falciparum isolates from the Kenyan coast: emergence of parasites with Ile-164-Leu mutation. Antimicrob Agents Chemother. 2009;53:3793–3798. doi: 10.1128/AAC.00308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada N, Chauhan LS, Babu BS, Thakur R, Khera A, Wares DF, Sahu S, Bachani D, Rewari BB, Dewan PK. Linking HIV-infected TB patients to cotrimoxazole prophylaxis and antiretroviral treatment in India. PLoS One. 2009;4:e5999. doi: 10.1371/journal.pone.0005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansa V, Jose V, Medhi B, Pandhi P. Decrease Use of Cotrimoxazole in Clinical Practice: Is it Justified? JK SCIENCE. 2010;12:214–215. [Google Scholar]
- Hastings MD, Maguire JD, Bangs MJ, Zimmerman PA, Reeder JC, Baird JK, Sibley CH. Novel Plasmodium vivax dhfr alleles from the Indonesian Archipelago and Papua New Guinea: association with pyrimethamine resistance determined by a Saccharomyces cerevisiae expression system. Antimicrob Agents Chemother. 2005;49:733–740. doi: 10.1128/AAC.49.2.733-740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins VN, Joshi H, Rungsihirunrat K, Na-Bangchang K, Sibley CH. Antifolates can have a role in the treatment of Plasmodium vivax. Trends Parasitol. 2007;23:213–222. doi: 10.1016/j.pt.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Imwong M, Pukrittayakamee S, Renia L, Letourneur F, Charlieu JP, Leartsakulpanich U, Looareesuwan S, White NJ, Snounou G. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 2003;47:1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na BK, Lee HW, Moon SU, In TS, Lin K, Maung M, Chung GT, Lee JK, Kim TS, Kong Y. Genetic variations of the dihydrofolate reductase gene of Plasmodium vivax in Mandalay Division, Myanmar. Parasitol Res. 2005;96:321–325. doi: 10.1007/s00436-005-1364-0. [DOI] [PubMed] [Google Scholar]
- Mockenhaupt FP, Teun Bousema J, Eggelte TA, Schreiber J, Ehrhardt S, Wassilew N, Otchwemah RN, Sauerwein RW, Bienzle U. Plasmodium falciparum dhfr but not dhps mutations associated with sulphadoxine-pyrimethamine treatment failure and gametocyte carriage in northern Ghana. Trop Med Int Health. 2005;10:901–908. doi: 10.1111/j.1365-3156.2005.01471.x. [DOI] [PubMed] [Google Scholar]
- Adedeji AA, Fehintola FA, Fateye BA, Happi TC, Amoo AO, Gbotosho GO, Sowunmi A. Plasmodium falciparum malaria in Nigerian children during high and low transmission seasons: gametocyte carriage and response to oral chloroquine. J Trop Pediatr. 2005;51:288–294. doi: 10.1093/tropej/fmi016. [DOI] [PubMed] [Google Scholar]
- Sowunmi A, Fateye BA, Adedeji AA, Fehintola FA, Gbotosho GO, Happi TC, Oduola AM. Predictors of the failure of treatment with chloroquine plus chlorpheniramine, in children with acute, uncomplicated, Plasmodium falciparum malaria. Ann Trop Med Parasitol. 2005;99:331–338. doi: 10.1179/136485905X36226. [DOI] [PubMed] [Google Scholar]
- Tjitra E, Suprianto S, Anstey NM. Higher gametocyte prevalence following failure of treatment of Plasmodium falciparum malaria with sulfadoxine-pyrimethamine and the combination of chloroquine plus sulfadoxine-pyrimethamine: implications for progression of anti-folate resistance. Trans R Soc Trop Med Hyg. 2002;96:434–437. doi: 10.1016/S0035-9203(02)90385-8. [DOI] [PubMed] [Google Scholar]
- Lu F, Lim CS, Nam DH, Kim K, Lin K, Kim TS, Lee HW, Chen JH, Wang Y, Sattabongkot J, Han ET. Mutations in the antifolate-resistance-associated genes dihydrofolate reductase and dihydropteroate synthase in Plasmodium vivax isolates from malaria-endemic countries. Am J Trop Med Hyg. 2010;83:474–479. doi: 10.4269/ajtmh.2010.10-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]