Abstract
Ralstonia solanacearum is the causative agent of bacterial wilt in many important crops. A specific and sensitive PCR detection method that uses primers targeting the gene coding for the flagella subunit, fliC, was established. Based on the first fliC gene sequence of R. solanacearum strain K60 available at GenBank, the Ral_fliC PCR primer system was designed; this system yielded a single 724-bp product with the DNAs of all of the R. solanacearum strains tested. However, R. pickettii and four environmental Ralstonia isolates also yielded amplicons. The Ral_fliC PCR products obtained with 12 strains (R. solanacearum, R. pickettii, and environmental isolates) were sequenced. By sequence alignment, Rsol_fliC primers specific for R. solanacearum were designed. With this primer system, a specific 400-bp PCR product was obtained from all 82 strains of R. solanacearum tested. Six strains of R. pickettii and several closely related environmental isolates yielded no PCR product; however, a product was obtained with one Pseudomonas syzygii strain. A GC-clamped 400-bp fliC product could be separated in denaturing gradient gels and allowed us to distinguish P. syzygii from R. solanacearum. The Rsol_fliC PCR system was applied to detect R. solanacearum in soil. PCR amplification, followed by Southern blot hybridization, allowed us to detect about one target DNA molecule per PCR, which is equivalent to 103 CFU g of bulk soil−1. The system was applied to survey soils from different geographic origins for the presence of R. solanacearum.
Ralstonia solanacearum is the causal agent of bacterial wilt in solanaceous crops but has also been recorded to infect a large range of more than 200 species representing over 50 families of plants (17). Traditionally, the pathogen has been classified in five biovars according to carbon source utilization (16, 18) and in six races based on host range (8, 26). R. solanacearum is supposed to be a soil-borne bacterium originating from the tropics, subtropics, and warm temperate regions (15), but strains causing brown rot of potato in geographic regions with a temperate climate are possibly adapted to lower temperatures (20, 24). In recent years, the increasing number of sites infested with potentially cold-adapted strains of R. solanacearum in several places in Europe dramatically enhanced the threat posed to European potato crops (20, 24, 43). Thus, reliable methods to detect the pathogen not only in tubers but also in soil or soil-related habitats are required. Several PCR-based methods for the detection of R. solanacearum have been described in the literature. These approaches are usually based on the amplification of ribosomal gene sequences (i.e., 16S or 16S-23S intergenic spacer region of the ribosomal DNA [rDNA]) (5, 12, 25, 35, 42, 44). However, due to the high degree of conservation of the ribosomal genes within the genus Ralstonia, 16S rDNA sequence similarities between species can be as high as 98% (30, 35, 38). This can lead to positive signals with related species, such as R. picketti, and thus rRNA-based methods have drawbacks. In other studies, the low level of resolution of 16S rDNA-based sequence analysis has been circumvented by using primers targeting functional genes such as endoglucanase and hrpB (28) or a random fragment that was claimed to be R. solanacearum specific (21).
In the present study, the fliC gene coding for the flagellar subunit protein flagellin was used to develop a highly specific and sensitive PCR-based detection system for R. solanacearum. The suitability of flagellin fliC genes for taxonomic applications has been shown in a number of studies for a large variety of bacterial species of several major bacterial groups: for α-proteobacteria (36); for β-proteobacteria, to which R. solanacearum belongs (11, 14); for low-G+C gram-positive bacteria (40); for the genus Pseudomonas (4, 23); and most notably for most enterobacterial species (3, 22).Flagellin genes have been used for detection, studies of population genetics, and epidemiological analyses (46). Due to their structure, which is conserved in the terminal regions that flank a variable, central region, flagellin genes are regarded as good candidates for PCR-based detection (46).
The main goal of the present study was to develop specific primers for amplification of a flagellin gene fragment that target all subgroups of the R. solanacearum species complex and to investigate their application to the detection of R. solanacearum in soil. Therefore, special attention was paid to achieving a sensitive amplifiability of the fliC gene fragments from DNA directly extracted from soil. The sensitivity of the detection system was enhanced by Southern blot hybridization. The novel method allowed the specific and sensitive detection of this major bacterial pathogen in soils and related habitats. Sequence heterogeneities of fliC DNA fragments amplified from different R. solanacearum strains were detected by using denaturing gradient gel electrophoresis (DGGE).
MATERIALS AND METHODS
Bacterial strains.
A list of all bacterial strains used to amplify fliC sequences is given in Table 1. Four environmental isolates from agricultural field soils in Germany (OV203, OV225, and Q3-8/14) or Brazil (L3L) (group F, Table 1) were included in the present study. These environmental isolates were identified by fatty acid methyl ester analysis (FAME) either as R. pickettii (Q3-8/14) or as R. solanacearum (OV203 and OV225) or by 16S rDNA sequencing (Escherichia coli positions 968 to 1401) as R. solanacearum (L3L).
TABLE 1.
Strains used in this studya
| Group | Species | Strain | No. of strains | Biovar(s) | Host plants (no. of strains) | Geographic origin (no. of strains) | Source or reference |
|---|---|---|---|---|---|---|---|
| A | R. solanacearum | 39 | 2 | Solanaceae: potato (28), tomato (4), egg plant (1), and bittersweet (1); soil (1); ND (4) | Europe (39) | 41 | |
| B | NDc | 7 | ND | Solanaceae: potato (6) and nightshade (1) | Europe: Friesland (6) and Aquitaine (1) | 41 | |
| C | R. solanacearum | 27 | 2 | Solanaceae: potato (21), tomato (2), pepper (2), egg plant (1), and soil (1) | Africa (12), South America (14), Indonesia (1) | 41 | |
| D | R. solanacearum | 9 | 1, 3, 4 | Musaceae (3), Solanaceae (4), Eupatorium odoratum (1), and ginger (1) | America (7), Australia (1), Cyprus (1) | 41 | |
| E | R. solanacearum | DSM1993 | 1 | 3 | Common bean | Mauritius | DSMZ |
| R. solanacearum | DSM9544 | 1 | 1 | Tomato | United States | DSMZ | |
| R. solanacearum | DSM50905 | 1 | ? | Banana | Costa Rica | DSMZ | |
| R. solanacearum | JS740 | 1 | 1 | Potato | Colombia | 27 | |
| R. solanacearum | JS778 | 1 | 3 | Potato | Réunion Island | 27 | |
| R. solanacearum | JS783 | 1 | 1 | Tomato | United States | 27 | |
| R. solanacearum | JS841 | 1 | 4 | Tomato | Sri Lanka | 27 | |
| F | P. syzygii | JV1010 | 1 | Clove | Indonesia | 27 | |
| R. pickettii | JR660 | 1 | 27 | ||||
| R. pickettii | DSM6297 | 1 | DSMZ | ||||
| R. pickettii | LMG5942 | 1 | LMG | ||||
| R. pickettii | LMG6871 | 1 | LMG | ||||
| R. pickettii | LMG7001 | 1 | LMG | ||||
| R. mannitolytica | LMG6866T | 1 | LMG | ||||
| Environmental isolates | Q3-8/14, OV203, OV225, and L3L | 4 | Potato rhizosphere | Germany and Brazil (L3L) | This study | ||
| R. eutropha | DSM531 | 1 | DSMZ | ||||
| R. basilensis | DSM | 1 | DSMZ |
Genomic DNA of Ralstonia isolates, which was also used in the study by Timms-Wilson et al. (41), was applied in order to test the specificity of the primer systems R sol fliC_for/rev; group E contains additional R. solanacearum strains that were not tested by Timms-Wilson et al. (41), and group F contains strains belonging to the Ralstonia genus but not to the R. solanacearum species complex.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; LMG, Laboratorium voor Mikrobiologie Gent, Universiteit Gent.
ND, not determined.
Soil used for testing the sensitivity of the detection system.
Samples of bulk soil used to test the sensitivity of the method originated from either (i) an experimental field site in The Netherlands, where R. solanacearum was initially added to a final level of ca. 106 cells/g of soil, or (ii) a microcosm experiment performed at Plant Research International, Wageningen, The Netherlands. From the latter experiment, immunofluorescence colony-staining counts (IFC) (42) of R. solanacearum are available for all sampling time points (34).
Survey of soil samples originating from different geographic areas.
To assess the prevalence of R. solanacearum cells in soils, a set of soil samples was tested. The samples originated from Brazil (six composite samples from a maize field), Thailand (six composite samples from a tomato field with infected plants), Cuba (four composite samples each from a sugarcane and a tobacco field and from forest soil), The Netherlands (six composite samples from a potato field with R. solanacearum infections), Spain (six composite samples from a potentially infested potato field), and Germany (six composite samples from a noninfested potato field from which strains OV203 and OV225 were isolated).
DNA extraction.
Total DNA of all soil samples was either extracted following the method of Smalla et al. (37) or by using the UltraClean soil DNA kit (MoBio Laboratories, Solana Beach, Calif.). Both procedures include a combined enzymatic and bead-beating step (cell homogenizer; Braun, Melsungen, Germany) for cell lysis.
Genomic DNA of strains was obtained by sodium dodecyl sulfate and proteinase K cell lysis, selective precipitation of cell debris and polysaccharides with CTAB (cetyltrimethylammonium bromide), and isopropanol precipitation of DNA according to the protocol of Wilson et al. (45).
The amplifiability of DNA was checked by PCR amplification of the eubacterial 16S rDNA fragment between positions 968 and 1401 published in Heuer et al. (19).
Primer development.
Based on the first fliC gene sequence of R. solanacearum strain K60 (available at GenBank under accession number AF283285 [submitted in 2000 and published in 2001]) (39), the Ral_fliC primer system (forward [5′-CCTCAGCCTCAATASCAACATC-3′] and reverse [5′-CATGTTCGACGTTTCMGAWGC-3′]), resulting in an amplicon size of 724 bp, was derived and optimized by using the Oligo program (version 4.0.). The sequences of amplicons obtained with R. solanacearum, R. pickettii, or the four environmental isolates (Table 1) were used together with all of the fliC sequences available in the database to design the R. solanacearum-specific primers Rsol_fliC (forward [5′-GAACGCCAACGGTGCGAACT-3′] and reverse [5′-GGCGGCCTTCAGGGAGGTC-3′]) giving an amplicon size of 400 bp. For DGGE separation of the Rsol_fliC amplicons, the GC-rich sequence described by Heuer et al. (19) was attached to the 5′ end of the reverse primer to prevent complete melting. Both primer sets were analyzed by basic local alignment search tool (BLAST) (2).
PCR amplifications.
For PCR amplification of R. solanacearum fliC gene fragments, the reaction mixture contained Stoffel buffer (10 mM KCl, 10 mM Tris-HCl [pH 8.3]), 0.2 mM deoxynucleoside triphosphates, 3.75 mM MgCl2, 4% [wt/vol] acetamide, 100 nM concentrations of each forward and reverse primer, and 2 U of AmpliTaq Stoffel fragment/25 μl. For amplification of environmental DNA extracted from soils, bovine serum albumin (1.25 μg/25 μl) was used to prevent inhibition. The PCR was carried out as follows. After an initial denaturation step for 5 min at 94°C, amplification was performed by using 25 cycles for DNA from isolates and 35 cycles for environmental DNA. Cycles consisted of a 30-s denaturation at 94°C, 2 min of primer annealing at 60°C for the Ral_fliC system and at 63°C for Rsol_fliC system, and a 1-min primer extension at 72°C, followed by a final step at 72°C (10 min) and cooling to 4°C. Products were analyzed by electrophoresis in 1% (wt/vol) agarose gels and ethidium bromide staining (32).
Southern blot hybridization.
Probes were generated from the Rsol_fliC PCR product obtained with either strain R. solanacearum 1609 (race 3/biovar 2), strain DSM9544 (race 1/biovar 1), or strain DSM1993 (race 1/biovar 3) by labeling the PCR product, which was excised from the agarose gel after electrophoresis with digoxigenin. The probes were used either separately or as a mix of the three probes. Southern blotting was done according to the method of Sambrook et al. (32). Hybridization was performed under conditions of medium stringency following the protocol given in Fulthorpe et al. (13). Hybridization of Southern blotted fliC PCR products obtained from soil DNA was performed with a mix of the three separately prepared probes.
DGGE.
DGGE systems (D-Code System; Bio-Rad, Inc., Hercules, Calif.) were used according to the protocols previously published by Heuer et al. (19). Polyacrylamide gels were composed of 0.17% (vol/vol) TEMED (N,N,N′,N′-tetramethylethylenediamine), 0.047% (wt/vol) ammonium persulfate, a 60:1 ratio of acrylamide/N,N′-methylene bisacrylamide, 1× buffer, and 2% glycerol. Denaturing gradients of 30 to 70% of denaturant substances (100% denaturant corresponds to 7 M urea plus 40% [vol/vol] deionized formamide) were used. Gradients were poured by using a gradient former and a peristaltic pump, and gels were allowed to polymerize for at least 2 h. Electrophoresis was performed in 0.5× Tris-acetate-EDTA buffer at 58°C at a constant voltage of 220 V for 5.5 h. Samples of the PCR mixture (varying between 2 and 8 μl to adjust differences in DNA concentration to ∼40 ng) were applied to the polyacrylamide gels.
Silver staining.
An acid silver-staining protocol was used for detection of DNA in DGGE gels (29). Gels were dried and digitalized with a translucent scanning device (Epson Deutschland GmbH, Düsseldorf, Germany).
Cloning and sequencing of fliC fragments.
For cloning of Ral_fliC fragments, 12 bacterial strains of groups A, D, E, and F (Table 1) were chosen. The 724-bp PCR product was ligated into the pGEM-T vector (Promega, Madison, Wis.) and introduced into competent E. coli JM109 cells (Promega) via transformation. Sequencing was done with the standard primers SP6 and T7 (IIT GmbH, Bielefeld, Germany).
Sequence analysis.
Analysis of cloned sequences was performed by using the software package BioEdit available from the internet (www.mbio.ncsu.edu/BioEdit). Multiple alignments were done applying the CLUSTALW tool delivered by the package. Maximum-likelihood phylogenetic trees were calculated by using the fastDNAml algorithm, an accessory application within BioEdit.
Sequence accession numbers.
All fliC sequences generated in this study were deposited in the GenBank database under accession numbers AY192716 to AY192727. The 16S rDNA sequences generated in the present study were deposited in the GenBank database under accession numbers AY216797 (OV225), AY216796 (R. solanacearum 1609), and AY216798 (Q3-8/14).
RESULTS AND DISCUSSION
Primer development.
The Ral_fliC primer gave a single product of 724 bp not only with DNA of R. solanacearum but also with R. pickettii and with four environmental isolates identified by FAME as R. pickettii or R. solanacearum (Table 1). The PCR products obtained from 12 strains belonging to group A (R. solanacearum strains [biovar 2] 1609, 737, and 267), group E (R. solanacearum strain [biovar 1] DSMZ9544), and group F (R. pickettii strains LMG5942, LMG6871, LMG7001, and DSM6297 and environmental isolates OV203, OV225, Q3-8/14, and L3L) were cloned and sequenced.
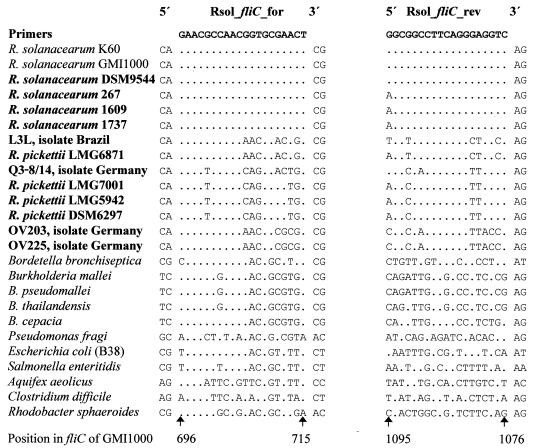
A comparison of the cloned Ral_fliC sequences with 36 homologous fliC sequences obtained from the GenBank database, two of them being published R. solanacearum fliC sequences (31, 39), revealed four regions that were potentially specific for all R. solanacearum sequences analyzed. Based on these regions, a primer system was designed that gave single product of defined length (400 bp) after PCR amplification with R. solanacearum strains DSM9544 and 1609. This primer system, denoted Rsol_fliC, was further used in the study. Comparison of aligned target sequences for primer annealing revealed that a product of 400 bp would theoretically be obtained with all R. solanacearum strains, since both primers matched the corresponding target sequences in the first 17 nucleotides counted from the 3′ end. In contrast, none of the other sequences, most notably the R. pickettii strains and the four environmental isolates, were expected to give a signal by PCR with this primer system. At least three of five bases at the 3′ end of the forward primer Rsol_fliC showed mismatches with the sequences of all nontarget bacteria for which fliC sequences are available in GenBank (Aquifex, low-G+C gram-positive bacteria, α-proteobacteria, β-proteobacteria, Pseudomonas spp., and enterobacteria). In respect to the Rsol_fliC reverse primer, all R. pickettii sequences share a mismatch at position 5 from the 3′ end of the primer. Sequences of the other bacterial taxa analyzed (see above) had additional mismatches at the 3′ end. Thus, the forward primer was predicted to be the most important for the specificity of the primer system (Fig. 1).
FIG. 1.
Sequence comparison of the primers Rsol_fliC_for (forward) and Rsol_fliC_rev(reverse) with the corresponding sequences of target and other species. The alignment was based on amino acid sequences. Strain names in boldface indicate that the sequence was generated in the present study. Bases identical to those in the primer sequences are shown as dots.
A BLAST search performed with both primers on 25 September 2002 revealed that the only significant matches (100% identity) produced were with the two available R. solanacearum fliC sequences of strains GMI1000 (AL646078) and K60 (AF283285). Surprisingly, the nucleotide sequence similarities of fliC sequences of all species of the related genus Burkholderia included in the alignments to the R. solanacearum fliC sequences were very low, ranging from 30.2 to 34% sequence identity. In consequence, we assumed that the fliC gene offered a good phylogenetic resolution at the species level.
Testing the primer system with isolates.
All 82 strains tested that belong to the species complex of R. solanacearum (groups A, C, D, and E, Table 1) gave an amplification product of 400 bp in PCR with the primer system Rsol_fliC. Furthermore, a 400-bp PCR product was obtained from the Pseudomonas syzygii strain JV1010 (one strain), whereas no PCR products were observed with the strains of R. pickettii (five strains), R. eutropha (one strain), R. basilensis (one strain), and R. mannitolytica (one strain). Hence, the PCR-based approach was most likely specific for R. solanacearum. False-positive reactions with bacterial strains outside the genus Ralstonia are not likely to occur since, for instance, the sequence similarity within the fliC fragment to the proximate genus Burkholderia was surprisingly low. In particular, the corresponding priming sites did not match the primer sequence of Rsol_fliC.
The finding that a PCR product of the expected size was amplified from P. syzygii strain JV1010 genomic DNA confirms the assumption that P. syzygii is part of the R. solanacearum species complex (12). Until now, there have been no PCR-based detection systems available to differentiate between P. syzygii and R. solanacearum in a fast and simple way.
Use of the Rsol_fliC primer system to check the identification of presumptive R. solanacearum isolates.
Colonies isolated on R2A medium from field soils have been from time to time identified by fatty acid methyl ester analysis as R. solanacearum or R. pickettii. Thus, strains highly related to R. solanacearum can be isolated from uninfested soils. Since R. solanacearum is a quarantine organism, its isolation from field soils would be alarming and of enormous economic consequences for farmers. Strains OV225 and Q3-8/14 both gave positive signals with the primer system described by Seal et al. (35). The complete 16S rDNA sequence (positions 8 to 1513 based on E. coli numbering [6])was determined for strain OV225 (which has identical BOX fingerprints as strain OV203) and strain Q3-8/14. The 16S rDNA sequence of strain OV225 showed the highest sequence similarity to the 16S rDNA sequence of R. solanacearum 1609 (97.34%), whereas the 16S rDNA sequence of strain Q3-8/14 had 99.45% similarity to R. pickettii MSP3. Although all four environmental Ralstonia isolates yielded PCR products with the Ral_fliC primer system, no PCR products were obtained with the Rsol_fliC primer system. Thus, the latter PCR system allows to differentiate these environmental isolates from R. solanacearum.
Sensitivity of Rsol_fliC PCR applied to DNA extracted from soil samples.
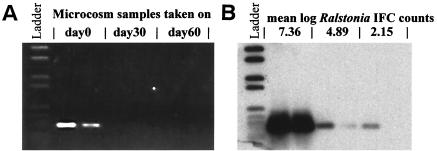
Special emphasis was placed on the applicability of Rsol_fliC PCR on soil DNA. The PCR amplifiability of all DNA samples directly extracted from soil was confirmed by the amplification of 16S rDNA fragments (positions 968 to 1401 [E. coli numbering]). The sensitivity of Rsol_fliC PCR detection of R. solanacearum in soils was evaluated with DNA extracted from soils from a microcosm experiment performed in The Netherlands; this microcosm had been inoculated with R. solanacearum at an initial concentration of 108 cells g of soil−1. IFC were available for five sampling time points at days 0, 33, and 54 after inoculation (Fig. 2; selected IFC data from Schönfeld et al. [34]).The detection limit of the direct Rsol_fliC PCR in soil was ca. 105 cells g of soil−1 when the PCR products were detected in ethidium bromide-stained agarose gels (Fig. 2A). Hybridization of Southern-blotted PCR products obtained with the Rsol_fliC primers by using a probe generated from a Rsol_fliC PCR of the introduced R. solanacearum strain 1609 revealed that cell densities of R. solanacearum down to ca. 103 cells g of soil−1 could be detected (Fig. 2B).
FIG. 2.
Agarose gel (A) and corresponding Southern blot (B) of Rsol_fliC PCR products obtained from PCR amplification of microcosm soils with IFC available for each time point of sampling. Positive and negative controls are notshown.
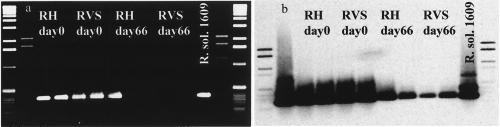
To demonstrate that Rsol_fliC PCR in combination with Southern hybridization is a useful tool for estimating unknown concentrations of R. solanacearum cells, a set of samples from a field experiment performed in The Netherlands were analyzed (T. Gorissen et al., unpublished data). The soil had been inoculated with 106 R. solanacearum cells g of soil−1 and was treated afterward by different measures (i.e., water-amended control and a combination of piggery manure amendment and solarization). DNA extracted from samples taken directly after inoculation (106 R. solanacearum cells g of soil−1) and 62 days after inoculation (unknown titer of R. solanacearum) was used to amplify the 400-bp R. solanacearum-specific fragment. A decline of R. solanacearum cells over time was suggested by agarose gel and in the corresponding blot, as the band intensity declined (Fig. 3a). However, no differences in band intensities could be observed between control and treated samples (i.e., a combination of piggery manure amendment and solarization). However, comparison of relative band intensities of samples at days 0 and 62 allowed an estimation of the presumptive cell densities of R. solanacearum in soil. In most day 62 samples very faint bands were detected in the agarose gel, indicating that the cell densities were near the detection limit of direct Rsol_fliC PCR (105 R. solanacearum cells g of soil−1) (Fig. 3a). Strong hybridization signals were observed at 105 cells g of soil−1, but differences in relative intensities were still distinguishable (Fig. 3b). No hybridization signal was detected in uninoculated soil samples (data not shown).
FIG. 3.
(a) Agarose gel of Rsol_fliC PCR products amplified from soils from a field experiment in The Netherlands obtained 0 or 66 days after inoculation with R. solanacearum strain 1609 at an initial concentration of 106 cells g of soil−1. Duplicates of samples are shown. Soil treatments are abbreviated as follows: RH, inoculated with strain 1609, untreated, water added; RVS, inoculated with strain 1609, manure amendment plus solarization. (b) Corresponding Southern blot of the agarose gel in panel a hybridized with an Rsol_fliC probe derived from fliC PCR of strain 1609.
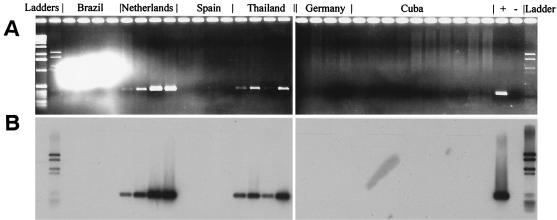
The Rsol_fliC PCR combined with Southern hybridization was applied to detect R. solanacearum of different biovars in a survey of soil samples from various geographic sites. Rsol_fliC PCRs and subsequent Southern hybridization with a mixed probe composed of probes for biovars 1, 2, and 3 were applied to DNA extracted directly from soils originating from three continents. All four soil samples originating from an infested field site in The Netherlands showed an amplification of the specific 400-bp fragment, as evidenced by agarose gel electrophoresis (Fig. 4). Symptoms of bacterial wilt were reported for this field. Positive signals were also found in DNA from soil samples from Thailand. These soil samples originated from a tomato field in which wilted tomato plants were observed (K. Wydra, unpublished data). In soil samples from the other regions, vestiges of R. solanacearum could not be detected, indicating that R. solanacearum was either not present or was present at concentrations of <103 cells g of soil−1.
FIG. 4.
Agarose gel (A) and corresponding Southern blot (B) of Rsol_fliC PCR products obtained from PCR amplification of soil samples originating from three continents. For hybridization, a mixed probe generated from three strains representing R. solanacearum biovars 1, 2, and 3 was used. The origins of the samples are indicated above the agarose gel image. A selection of the samples analyzed (four samples for each origin; 10 samples from Cuba) is presented.
Considering that the regulatory network R. solanacearum uses for virulence is activated only at cell densities of >107 ml−1 (7) and since it was possible to detect the 400-bp fliC fragment down to a level of 103 R. solanacearum cells g of soil−1, it can be assumed that the pathogen is detectable by Rsol_fliC PCR even if no symptoms of brown rot occur.
A promising application of the Rsol_fliC PCR in combination with Southern hybridization is in monitoring the fate of the pathogen over time. Shifts in the R. solanacearum population size can be detected by quantifying the Rsol_fliC PCR product, for example, by real-time PCR. Thus, one of the main advantages of the approach is the possibility to follow the fate of the pathogen without the necessity of introducing a special marker.
Another useful aspect of this approach is the possibility of detecting avirulent forms of the pathogen. Although flagellum-dependent motility was shown to play an important role in virulence during the early stages of disease manifestation, i.e., invasion and dissemination (39), the genes needed for flagellum constitution are not directly involved in virulence, i.e., they are not part of the virulence regulatory network (1, 9, 33). Therefore, the 400-bp Rsol_fliC fragment is detectable from strains which exhibit limited virulence in planta.
Variability within the 400-bp Rsol_fliC fragment.
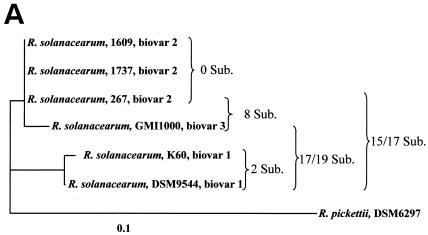
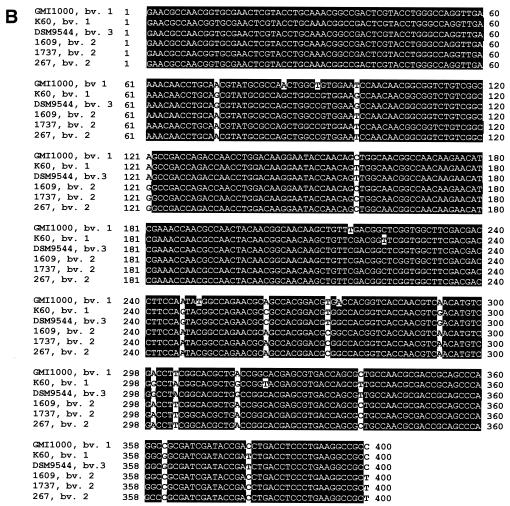
From sequence alignments, it appeared that the 400-bp fliC fragment of all R. solanacearum strains sequenced in the present study corresponds to the region between positions 696 and 1095 of the fliC sequence of R. solanacearum GMI1000. As representatives of three different biovars, fliC sequences of the six R. solanacearum strains were analyzed: DSM9544 and K60 (biovar 1); 1609, 1737, and 267 (biovar 2); and GMI1000 (biovar 3). Considerable sequence variability was found among these R. solanacearum strains (Fig. 5A). Although biovar 2 strains 1609, 1737, and 267 had identical sequences over the whole 400 bp, biovar 1 strains DSM9544 and K60 differed in two positions. GMI1000 (biovar 3) and 1609 (biovar 2) showed differences in eight positions. The differences of the two biovar 1 strains DSM9544 and K60 to biovar 2 strain 1609 and biovar 3 strain GMI1000 were more pronounced: strain 1609 differed from DSM9544 in 15 positions and from K60 in 7 positions. GMI1000 differed from DSM9544 in 19 positions and from K60 in 17 positions (see also Fig. 5A). All differentiating positions, occurring as single sites were found in the region from positions 73 to 400 of the PCR product (Fig. 5B).
FIG. 5.
(A) Neighbor-joining tree based on a comparison of 400-bp fragments of the fliC gene. The number of nucleotide substitutions between sequences is given at the branches. Sub., substitutions. The tree was rooted with R. pickettii DSM6297 as an outgroup. (B) Sequence comparison of the 400-bp fliC fragments of strains belonging to different biovars.
The selected 400-bp fragment is located within the central region of the fliC gene, where the greatest sequence variability between strains can be expected because this region codes for a nonfunctional domain of the protein (flagellin) (46). Furthermore, the fliC gene is not a part of a pathogenicity island, nor was it found to be transmitted by horizontal gene transfer (46). Therefore, it may represent a good phylogenetic marker, such as the 16S rDNA, but with a different scale of resolution and, presumably, a better discriminatory power, when closely related species or subgroups within a species are compared. Regarding the distinct positional variability within the 400-bp fragment, the application of DGGE as a sequence-based separation technique seemed to be promising.
Separation of fliC fragments in DGGE.
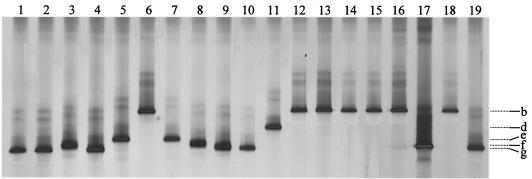
A single strong and sharp band was observed in DGGE of Rsol_fliC fragments of each of the 82 R. solanacearum strains and one P. syzygii strain analyzed. According to their melting behavior in DGGE, at least seven different band positions could be identified (Fig. 6). Of the 66 biovar 2 strains analyzed by Rsol_fliC PCR-DGGE, 63 had an electrophoretic mobility identical to that of strain 1609 (Fig. 6, b and g). The biovar 2 strains originating from potato plants (52 strains), tomato plants (6 strains), soil (2 strains), egg plants (2 strains), and bittersweet (1 strain) were obtained from various geographic regions. Only two isolates from pepper, originating from Brazil and Indonesia (bands with the electrophoretic mobility of bands a and c [results not shown in Fig. 6]), and one potato isolate had a different electrophoretic mobility. Although only eight biovar 1 isolates were included in the present study, four different electrophoretic mobilities corresponding to band g (UW20 [banana, Venezuela], UW28 [origin not available], and DSM9544 [3]), band f (UW25 [tomato, United States] and JS783 [tomato, United States]), band e (UW70 [plantain, Colombia] and JS740 [potato, Colombia]), and band b (UW136 [heliconia, Costa Rica]) were observed (Fig. 6). Thus, in contrast to biovar 2 strains, a higher genetic diversity can be supposed for biovar 1 isolates. This observation confirms the finding reported by Poussier et al. (27). All four biovar 3 strains (UW8 [Eupatorium odoratum, Costa Rica], UW255 [pepper, Australia], JS778 [potato, Réunion Island], and DSM1993) had the same electrophoretic mobility (band b) as for two of the three biovar 4 isolates (JS841 [potato, Sri Lanka] and UW27 [tobacco, United States]). The P. syzygii product had an electrophoretic mobility different from that of all R. solanacearum strains. Thus, Rsol_fliC PCR-DGGE offers an alternative method to the approach previously described by Poussier et al. (27) for differentiating this species from R. solanacearum. Although the limited number of isolates belonging to biovars 1, 3, and 4 that were included in the present study does not allow us to fully evaluate the discriminatory power of the Rsol_fliC PCR-DGGE approach, it might be particularly useful to analyze mixed infections with strains of different biovars of R. solanacearum in the plant or in the soil on the condition that sufficiently high numbers of R. solanacearum cells are present.
FIG. 6.
Electrophoretic separation of GC-clamped Rsol_fliC PCR products of a selection of strains of R. solanacearum and P. syzygii strain JV1010 by 30 to 70% DGGE. Lanes (biovar/host plant/origin [NA = not available]): 1 and 19, strain 1609 (biovar 2/potato/The Netherlands); 2, strain UW20 (biovar 1/banana/Venezuela); 3, strain UW25 (biovar 1/tomato/United States); 4, strain UW28 (biovar 1/NA/NA); 5, UW70 (biovar 1/plantain/Colombia); 6, strain UW136 (biovar 1/heliconia/Costa Rica); 7, strain JS740 (biovar 1/potato/Colombia); 8, strain JS783 (biovar 1/tomato/United States); 9, strain DSM9544 (biovar 1/tomato/United States); 10, strain DSM50905 (biovar NA/banana/Costa Rica); 11, P. syzygii strain JV1010 (clove/Indonesia); 12, strain UW8 (biovar 3/E. odoratum/Costa Rica); 13, strain UW255 (biovar 3/pepper/Australia); 14, strain JS778 (biovar 3/potato/Réunion Island); 15, strain DSM1993 (biovar 3/Phaseolus vulgaris/Mauritius); 16, strain UW27 (biovar 4/tobacco/United States); 17, strain UW151 (biovar 4/ginger/Australia); 18, strain JS841 (biovar 4/potato/Sri Lanka). For further information on the above listed strains, see studies by Cook and Sequeira (10), Poussier et al. (27), and Timms-Wilson et al. (41).
Conclusion.
The Rsol_fliC primer system offers a specific and, in combination with Southern blot analysis, a very sensitive detection system for R. solanacearum. Although PCR products were obtained with R. solanacearum strains belonging to different biovars, no Rsol_fliC products were obtained with environmental soil isolates that seemed to be closely related to the R. solanacearum or R. pickettii. The PCR system can be used for cultivation-independent detection of the pathogen in DNA directly extracted from environmental samples. Since environmental stresses such as low temperature are known to induce the viable-but-nonculturable state in R. solanacearum (43), the use of cultivation-independent detection techniques such as the Rsol_fliC detection system, which allows a sensitive and specific detection of the pathogen, are crucial. Furthermore, this system could be a valuable tool for monitoring the expression of the fliC genes of R. solanacearum strains under different conditions in soil by Rsol_fliC PCR amplification of reverse-transcribed RNA that was directly extracted from soil.
Acknowledgments
We thank Leo van Overbeek and Ton Gorissen, Plant Research International, Wageningen, The Netherlands, for their collaboration; Tracy Timms-Wilson and Kirsty Howard, Centre for Ecology and Hydrology, Oxford, United Kingdom, for providing a collection of genomic DNA from Ralstonia strains; Patricia Ferreira, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil, for providing environmental isolates from Brazil; and Caitilyn Allen and Philippe Prior, INRA, Avignon, France, for constructive discussions.
This study was supported by EU projects FATE (FAIR 3632) and POTATOCONTROL (QLK3-2000-01598).
REFERENCES
- 1.Allen, C., J. Gay, and L. Simon-Buela. 1997. A regulatory locus, pehSR, controls polygalacturonase production and other virulence functions in Ralstonia solanacearum. Mol. Plant-Microbe Interact. 10:1054-1064. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belas, R., and D. Flaherty. 1994. Sequence and genetic analysis of multiple flagellin-encoding genes from Proteus mirabilis. Gene 148:33-41. [DOI] [PubMed] [Google Scholar]
- 4.Bellingham, N. F., J. A. W. Morgan, J. R. Saunders, and C. Winstanley. 2001. Flagellin gene sequence variation in the genus Pseudomonas. Syst. Appl. Microbiol. 24:157-165. [DOI] [PubMed] [Google Scholar]
- 5.Boudazin, G., A. C. Le Roux, K. Josi, P. Labarre, and B. Jouan. 1999. Design of division specific primers of Ralstonia solanacearum and application to the identification of European isolates. Eur. J. Plant Pathol. 105:373-380. [Google Scholar]
- 6.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organisation and primary structure of an rRNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 7.Brumbley, S. M., B. F. Carney, and T. P. Denny. 1993. Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of phcA, a putative lysR transcriptional activator. J. Bacteriol. 175:5477-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buddenhagen, I., and A. Kelman. 1964. Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 2:203-230. [Google Scholar]
- 9.Clough, S. J., A. B. Flavier, M. A. Schell, and T. P. Denny. 1997. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl. Environ. Microbiol. 63:844-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, D., and L. Sequeira. 1994. Strain differentiation of Pseudomonas solanacearum by molecular genetic methods, p. 77-93. In G. L. Hartman and A. C. Hayward (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 11.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fegan, M., M. Taghavi, L. I. Sly, and A. C. Hayward. 1998. Phylogeny, diversity and molecular diagnostics of Ralstonia solanacearum, p. 19-33. In P. Prior, C. Allen, and J. G. Elphinstone (ed.), Bacterial wilt disease—molecular and ecological aspects. Springer-Verlag, Berlin, Germany.
- 13.Fulthorpe, R. R., C. McGowan, O. V. Maltseva, W. E. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hales, B. A., J. A. W. Morgan, C. A. Hart, and C. Winstanley. 1998. Variation in flagellin genes and proteins of Burkholderia cepacia. J. Bacteriol. 180:1110-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayward, A. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 16.Hayward, A. 1964. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 27:265-277. [Google Scholar]
- 17.Hayward, A. 1994. Systematics and phylogeny of Pseudomonas solanacearum and related bacteria, p. 127-135. In G. L. Hartman and A. C. Hayward (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Oxford, England.
- 18.Hayward, A. C., H. M. El-Nashaar, U. Nydegger, and L. De Lindo. 1990. Variation in nitrate metabolism in biovars of Pseudomonas solanacearum. J. Appl. Bacteriol. 69:269-280. [Google Scholar]
- 19.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janse, J. D., F. A. X. Araluppan, J. Schans, M. Wenneker, and W. Westerhuis. 1998. Experiences with bacterial brown rot Ralstonia solanacearum biovar 2, race 3, in The Netherlands, p. 146-154. In P. Prior, C. Allen, and J. Elphinstone (ed.), Bacterial wilt disease—molecular and ecological aspects. Springer-Verlag, Berlin, Germany.
- 21.Lee, Y. A., and C. C. Wang. 2000. The design of specific primers for the detection of Ralstonia solanacearum in soil samples by polymerase chain reaction. Bot. Bull. Acad. Sin. 41:121-128. [Google Scholar]
- 22.Machado, J., F. Grimont, and P. A. D. Grimont. 2000. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res. Microbiol. 151:535-546. [DOI] [PubMed] [Google Scholar]
- 23.Morgan, J. A. W., N. F. Bellingham, C. Winstanley, M. A. Ousley, C. A. Hart, and J. R. Saunders. 1999. Comparison of flagellin genes from clinical and environmental Pseudomonas aeruginosa isolates. Appl. Environ. Microbiol. 65:1175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsson, K. 1976. Experience of brown rot caused by Pseudomonas solanacearum (Smith) in Sweden. OEPP/EPPO Bull. 6:199-207. [Google Scholar]
- 25.Pastrik, K. H., and E. Maiss. 2000. Detection of Ralstonia solanacearum in potato tubers by polymerase chain reaction. J. Phytopathol. 148:619-626. [Google Scholar]
- 26.Pegg, K., and M. Moffett. 1971. Host range of the ginger strain of Pseudomonas solanacearum in Queensland. Aust. J. Exp. Agric. Anim. Husb. 11:696-698. [Google Scholar]
- 27.Poussier, S., D. Trigalet-Demery, P. Vandewalle, B. Goffinet, J. Luisetti, and A. Trigalet. 2000. Genetic diversity of Ralstonia solanacearum as assessed by PCR-RFLP of the hrp gene region, AFLP and 16S rRNA sequence analysis, and identification of an African subdivision. Microbiology 146:1679-1692. [DOI] [PubMed] [Google Scholar]
- 28.Poussier, S., P. Prior, J. Luisetti, C. Hayward, and M. Fegan. 2000. Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 23:479-486. [DOI] [PubMed] [Google Scholar]
- 29.Riesner, D., G. Steger, R. Zimmat, R. A. Owens, M. Wagenhöfer, W. Hillen, S. Vollbach, and K. Henco. 1989. Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis 10:377-389. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, S. J., S. J. Eden-Green, P. Jones, and D. J. Ambler. 1990. Pseudomonas syzygii sp. nov, the cause of Sumatra disease of cloves. Syst. Appl. Microbiol. 13:34-43. [Google Scholar]
- 31.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schell, M. 2000. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38:263-292. [DOI] [PubMed] [Google Scholar]
- 34.Schönfeld, J., A. Gelsomino, L. S. van Overbeek, A. Gorissen, K. Smalla, and J. D. van Elsas. 2003. Effects of compost addition and simulated solarisation on the fate of Ralstonia solanacearum biovar 2 and indigenous bacteria in soil FEMS Microbiol. Ecol. 43:63-74. [DOI] [PubMed] [Google Scholar]
- 35.Seal, S. E., L. A. Jackson, J. P. Young, and M. J. Daniels. 1993. Differentiation of Pseudomonas solanacearum, Pseudomonas syzygii, Pseudomonas pickettii, and the blood disease bacterium by partial 16S rRNA sequencing: construction of oligonucleotide primers for sensitive detection by polymerase chain reaction. J. Gen. Microbiol. 139:1587-1594. [DOI] [PubMed] [Google Scholar]
- 36.Shah, D. S. H., T. Perehinec, S. M. Stevens, S. Aizawa, and R. E. Sockett. 2000. The flagellar filament of Rhodobacter sphaeroides: pH-induced polymorphic transitions and analysis of the fliC gene. J. Bacteriol. 182:5218-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalla, K., N. Cresswell, L. C. Mendonca-Hagler, A. Wolters, and J. D. van Elsas. 1993. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74:78-85. [Google Scholar]
- 38.Taghavi, M., C. Hayward, L. I. Sly, and M. Fegan. 1996. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 46:10-15. [DOI] [PubMed] [Google Scholar]
- 39.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tasteyre, A., T. Karjalainen, V. Avesani, M. Delmee, A. Collignon, P. Bourlioux, and M. C. Barc. 2000. Phenotypic and genotypic diversity of the flagellin gene (fliC) among Clostridium difficile isolates from different serogroups. J. Clin. Microbiol. 38:3179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timms-Wilson, T. M., K. Bryant, and M. J. Bailey. 2001. Strain characterization and 16S-23S probe development for differentiating geographically dispersed isolates of the phytopathogen Ralstonia solanacearum. Environ. Microbiol. 3:785-797. [DOI] [PubMed] [Google Scholar]
- 42.van der Wolf, J. M., S. G. C. Vriend, P. Kastelein, E. H. Nijhuis, P. J. van Bekkum, and J. W. L. van Vuurde. 2000. Immunofluorescence colony-staining (IFC) for detection and quantification of Ralstonia (Pseudomonas) solanacearum biovar 2 (race 3) in soil and verification of positive results by PCR and dilution plating. Eur. J. Plant Pathol. 106:123-133. [Google Scholar]
- 43.Van Elsas, J. D., P. Kastelein, P. van Bekkum, J. M. van der Wolf, P. M. de Vries, and L. S. van Overbeek. 2000. Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology 90:1358-1366. [DOI] [PubMed] [Google Scholar]
- 44.Weller, S. A., J. G. Elphinstone, N. C. Smith, N. Boonham, and D. E. Stead. 2000. Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66:2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 2.10-2.12. In F. M. Ausubel, R. Bent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.), Current protocols in molecular biology. Greene and Wiley, New York, N.Y. [DOI] [PubMed]
- 46.Winstanley, C., and J. A. W. Morgan. 1997. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology 143:3071-3084. [DOI] [PubMed] [Google Scholar]