Abstract
Folding and insertion of integral β-barrel proteins in the outer membrane is an essential process for Gram-negative bacteria that requires the β-barrel Assembly Machinery (BAM). Efficient OMP folding and insertion appears to require a consensus C-terminal signal in OMPs characterized by terminal F or W residues. The BAM complex is embedded in the outer membrane and, in E. coli, consists of the β-barrel BamA and four lipoproteins BamBCDE. BamA and BamD are broadly distributed across all species of Gram-negative bacteria whereas the other components are present in only a subset of species. BamA and BamD are also essential for viability suggesting that these two proteins constitute the functional core of the bacterial BAM complex. Here we present the crystal structure of BamD from the thermophilic bacteria Rhodothermus marinus refined to 2.15 Å resolution. The protein contains 5 tetratricopeptide repeats (TPRs) organized into two offset tandems, each capped by a terminal helix. The N-terminal domain contains three TPRs and displays remarkable structural similarity with proteins that recognize targeting signals in extended conformations. The C-terminal domain harbors the remaining 2 TPRs and previously described mutations that impair binding to other BAM components map to this domain. Therefore, the structure suggests a model where the C-terminal domain provides a scaffold for interaction with BAM components, while the N-terminal domain participates in interaction with the substrates, either recognizing the C-terminal consensus sequence or binding unfolded OMP intermediates.
Introduction
The outer membrane of Gram-negative bacteria is unique and essential for its survival. This semi-permeable membrane is typically asymmetric composed of phospholipids on the inner leaflet and primarily lipopolysaccharide (LPS) on the outer leaflet 1. Proteins associated with the outer membrane can be divided into three groups based on their architecture: (i) lipoproteins, which do not contain transmembrane domains and are attached to the OM by lipid modification of their N-terminal cysteine residue 2; (ii) Outer Membrane Proteins (OMPs), characterized by a transmembrane β-barrel architecture 3; and (iii) periplasm-spanning export channels, whose trans-outer-membrane components can have β-barrel structures as in TolC 4, or a novel α-barrel architecture typified by Wza 5; 6.
Transmembrane β-barrel OMPs carry out a variety of functions in bacteria including nutrient and waste product exchange with the environment, cell adhesion and fulfillment of structural and enzymatic roles 3. Folding and insertion of OMPs in the outer membrane is an essential process that requires a multiprotein complex in the outer membrane known as β-barrel Assembly Machinery (BAM) 7; 8. In E. coli, this complex is composed of BamA, a β-barrel OMP also known as Omp85/YaeT, and four lipoproteins, BamBCDE (previously known as YfgL, NlpB, YfiO and SmpA respectively) 8; 9. Periplasmic chaperones such as SurA assist in shuttling nascent OMPs to the BAM complex 10. The process was recently reconstituted in vitro indicating that SurA and BAM can mediate OMP folding and insertion into membranes 11. In vivo, the efficiency of the process appears to depend on a consensus sequence at the C-terminus of β-barrel OMPs 12; 13. The last residue in OMPs is almost always F or W and, starting from the C-terminus, positions 3, 5, 7 and 9 are hydrophobic residues with Y frequently occupying position 3 13. Thus, it is likely that this signal is important for one or more mechanistic steps in OMP folding and insertion.
BamA is essential for viability 8; 14; 15 and homologs are retained in mitochondria and chloroplasts where they also mediate insertion of β-barrel proteins 16. BamD is the only BAM lipoprotein required for viability 8; 9; 17; 18. Both BamA and BamD are broadly distributed in Gram-negative bacteria 19 which, together with their requirement for viability, suggest that they constitute the core of the BAM complex. The phylogenetic distribution of BamB, C and E is more restricted 19, and individual deletion of their genes in E. coli result in membrane permeability defects but not loss of viability 8; 9.
The structural information necessary to understand the molecular mechanisms of OMP folding and insertion mediated by BAM is beginning to emerge. BamA contains a large periplasmic domain with five POlypeptide-TRanslocation-Associated (POTRA) repeats in addition to its transmembrane β-barrel. Crystallographic and solution scattering data revealed a superhelical arrangement of the POTRA domains 20; 21; 22 thought to interact with nascent OMPs and perhaps nucleate folding of their β-strands 20; 21; 23. Structures of the non-essential lipoproteins BamB and BamE have recently been reported 24; 25; 26; 27. However, structural data on the essential lipoprotein BamD has remained elusive, although sequence analysis suggested that the protein contains tetratricopeptide repeats 19. Here we report the crystal structure of BamD from Rhodothermus marinus refined to 2.15 Å resolution and discuss the implications for the role of BamD in the BAM-mediated mechanism of OMP folding and insertion.
Results and Discussion
Crystal Structure of BamD
A lipid-free, soluble form of BamD from the thermophilic bacteria Rhodothermus marinus (rmBamD) was expressed in E. coli from a plasmid encoding the mature protein (amino acids 23–280) with the N-terminal cysteine mutated to alanine. The structure was determined using single anomalous dispersion methods and seleno-methionine substituted protein. Data collected from native crystals was then used to refine the structure to 2.15Å resolution (Table 1). The final model contains residues 30–280 and is thus only missing seven amino acids from the N-terminus due to conformational flexibility.
Table 1.
Data Collection and Refinement Statistics.
| Native BamD | SeMet BamD | |
|---|---|---|
| Data Collection | ||
| Space Group | R3a | R3a |
| Unit Cell: a=b (Å) | 114.37 | 114.22 |
| c (Å) | 77.63 | 77.62 |
| Wavelength (Å) | 1.0000 | 0.9792 |
| Resolution (Å)b | 70.0–2.15 (2.23–2.15) | 50.0–2.7 (2.80–2.70) |
| Rsymc (%) | 7.3 (33.7) | 8.5 (22.8) |
| I / s | 20.3 (2.7) | 15.2 (3.7) |
| Data Completeness (%) | 99.7 (99.0) | 97.7 (87.7) |
| Redundancy | 2.8 (2.6) | 3.6 (2.7) |
|
| ||
| Phasing | ||
| FOM before DMd | 0.36 | |
| FOM after DM | 0.69 | |
|
| ||
| Refinement Statistics | ||
| Resolution (Å) | 57.2 - 2.15 (2.26 – 2.15) | |
| Reflections in work set | 19492 | |
| Reflections in test set | 1032 | |
| Protein atoms | 2117 | |
| Water molecules | 152 | |
| Rwork (%) | 16.9 (20.7) | |
| Rfreee (%) | 21.0 (27.9) | |
| Mean B-value protein (Å2) | 37.1 | |
| Mean B-value water (Å2) | 41.5 | |
| RMS dev. bonds (Å) | 0.007 | |
| RMS dev. angles (°) | 1.007 | |
The hexagonal axis setting was used.
Values in parentheses are for the highest-resolution shell.
Rsym=ΣhΣi |(Ii(h)-< I(h)>|/ ΣhΣI Ii(h), where Ii(h) is the I-th measurement of reflection h, and <I(h)> is the weighted mean of all measurements of h.
Figure Of Merit before Density Modification
Rwork = Σ|Fobs−Fcalc|/ΣFobs where Fobs = observed structure factor amplitude and Fcalc = structure factor calculated from model. Rfree is computed in the same as Rwork, but using the test set of reflections.
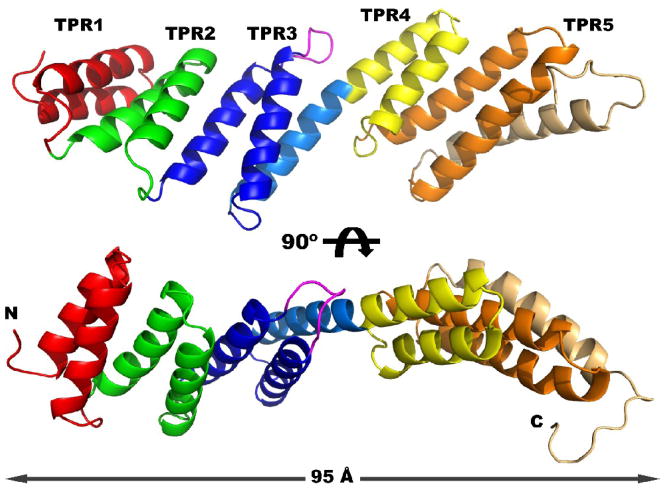
The structure of rmBamD consists of five tetratricopeptide repeats (TPR) arranged in two offset tandem arrays (Figure 1). TPRs are 34 amino acid degenerate sequences that fold into a helix-turn-helix motif 28; 29. TPRs are typically arranged in tandem arrays such that the two antiparallel helices (A and B) from one repeat pack against the A helix of the next motif. The tandem is frequently terminated by a capping helix. Packing several TPRs in this manner results in a right-handed superhelical structure 30. In rmBamD however, three N-terminal TPRs (TPR1–3, red, green and blue in Figure 1) are capped by a helix that is continuous with the A helix of TPR 4 (light blue and yellow). As a consequence, the two C-terminal TPRs (TPR4–5 yellow and orange) are offset from the N-terminal three, breaking the superhelical arrangement. rmBamD is therefore a very elongated molecule measuring more than 95 Å between the N-terminus and the capping helix of TPR5 (light orange). In general, the turns connecting the A and B helices are short and fairly typical. TPR3 however is notable by the extended loop that connects the two helices (magenta in Figure 1).
Figure 1. Crystal structure of rmBamD.
Two views of rmBamD related by a 90º rotation. The N-terminal domain contains TPRs 1–3 (red, green and blue respectively) and is capped by a helix (light blue). The C-terminal domain contains TPRs 4 and 5 (yellow and orange) and the final capping helix (light orange). An extended loop in TPR3 is labeled magenta.
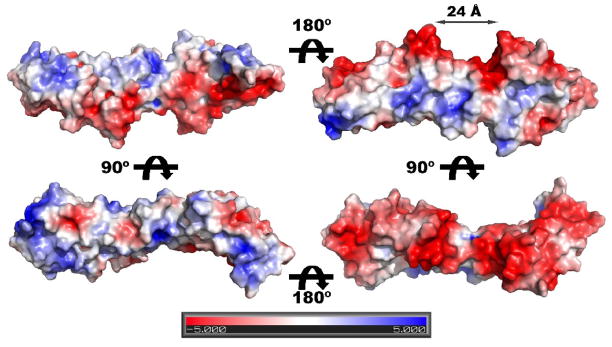
Mapping the electrostatic surface potential on the structure of rmBamD reveals a striking distribution of charge (Figure 2). One side of the molecule, harboring the turns linking the TPRs helices, is positively charged whereas the opposite side displays a strong electronegative character. Interestingly, the offset between the N and C-terminal domains creates a negatively charged semicircular groove at the bottom of rmBamD, approximately 24 Å in diameter that may be important for interaction with other BAM components.
Figure 2. Electrostatic surface potential of rmBamD.
The distribution of positive (blue) and negative (red) surface potential is mapped to a surface representation of rmBamD. For reference, the orientation of the molecule in the left panels is the same as in Figure 1. A negatively charged, semicircular groove between the N and C-terminal domains 24Å in diameter is labeled.
BamD across Gram-negative bacteria
BamD is essential in E. coli 8; 17; 18 as well as Neisseria meningitidis 31, and together with BamA is thought to constitute the core of the BAM machinery. Consistent with its essential role, BamD homologs are ubiquitous in Gram-negative bacteria 17; 19. We used hidden Markov models to iteratively search the KEGG database for BamD homologs using jackhmmer (part of the HMMER package 32). The alignment was manually edited in Jalview 33 to eliminate a few sequences that did not contain the invariant cysteine at the mature N-terminus that becomes lipidated in BamD 34. This resulted in 607 sequences with broad representation of Gram-negative bacteria.
The length of BamD is approximately 240–300 amino acids (from the translational start) in most bacteria, with rmBamD being 280 amino acids long. In E. coli BamD is 245 amino acids and the alignments suggest that the protein is missing the last helix capping TPR5 in rmBamD (Figure 3A). Conversely, BamD homologs in Pseudomonadales including Moraxella, Acinetobacter and Pseudomonas, have an extended C-terminus that makes them 340–380 amino acids long. This extension does not have clear sequence similarity to any other protein and may represent additional TPR repeats.
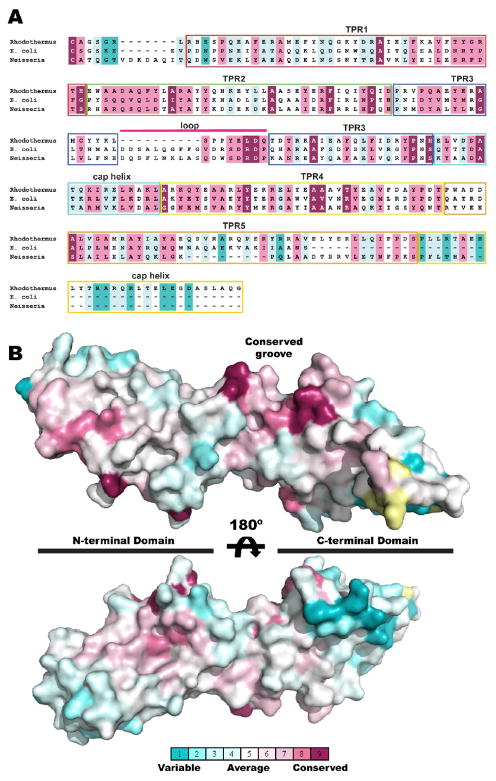
Figure 3. Sequence conservation in BamD.
A sequence alignment of BamD homologs (see text) was analyzed with the Consurf sever. Conservation scores are normalized so that the average score is zero and the standard deviation is 1. Thus rapidly evolving positions are negative (colored cyan) and slowly evolving positions are positive (colored purple) while positions with an average rate of evolution are colored white. (A) Aligned sequences of BamD from Rhodothermus marinus (Rhodothermus), Escherichia coli (E. coli) and Neisseria meningitidis (Neisseria) colored by the Consurf scores. Structural elements are labeled and highlighted in boxes colored as in Figure 1. (B) Conservation scores mapped on a surface representation of the rmBamD structure showing a groove between the extended loop in TPR3 and the C-terminal domain lined with slowly evolving residues.
To evaluate conserved features in BamD we subjected the aligned sequences to analysis with the Consurf server, which uses Bayesian methods to score the rate of evolution at each aligned position 35. Figure 3A shows the alignment of rmBamD with E. coli and Neisseria sequences for reference. The results mapped on the structure of rmBamD (Figure 3B), show that the N-terminal domain is generally more conserved than the C-terminal domain. The extended loop connecting the A and B helices of TPR3 together with TPR4 form a deep groov. e lined with conserved residues suggestive of a protein:protein interaction surface. Consistent with this hypothesis, C-terminal truncations of E. coli BamD resulted in impaired binding to BamA as well as the accessory lipoproteins BamC and BamE 9; 17. These truncations map to the beginning of TPR5 in rmBamD and would not only eliminate the entire TPR5 but likely result in destabilization of TPR4 as well. Therefore, we suggest that the extended loop of TPR3 together with the C-terminal domain of BamD harbor the determinants for interaction with other BAM components.
Potential roles for BamD in OMP biogenesis
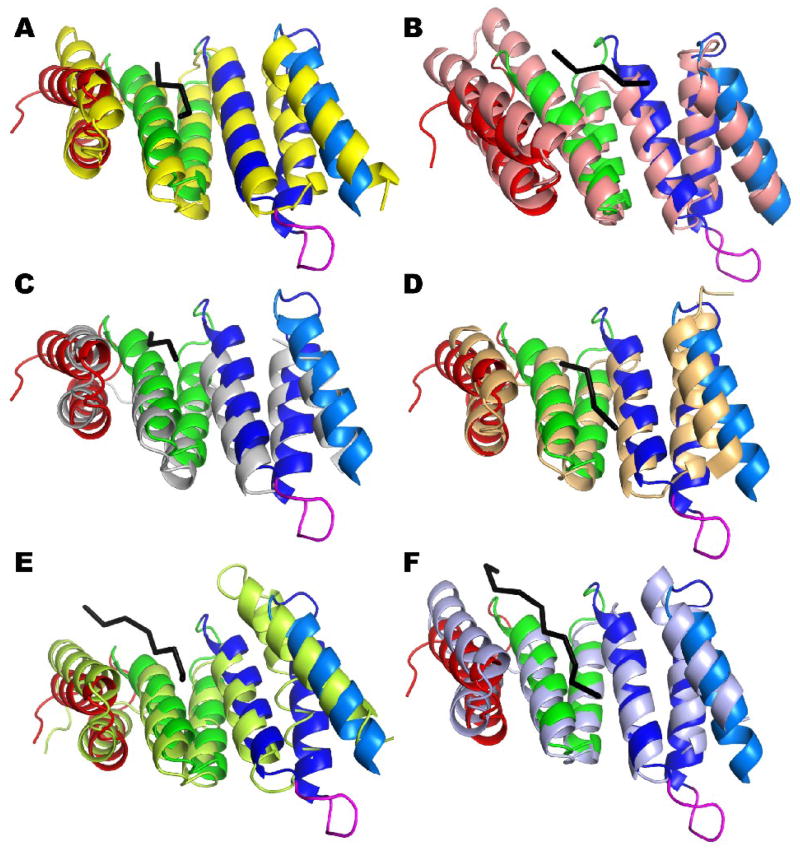
The structure of rmBamD N-terminal domain is strikingly similar to a number of proteins involved in recognition of C-terminal targeting signal sequences (Figure 4). The right-handed twist in the TPR tandem creates a concave face lined by the TPR’s the A helices. TPR domains in proteins such as the Hsp-Organizing-Protein (Hop), Tom70 (a component of the mitochondrial Translocase of the Outer Membrane) and FKBP52 use this concave surface to recruit molecular chaperones Hsp70 or Hsp90 by binding their C-terminal tails in an extended conformation 36; 37; 38 (Figure 4A, B and C). Similarly, the C-terminal Peroxisomal Targeting Signal-1, present at the C-terminus of proteins destined to the peroxisome, is bound by PEX5 in the concave face of its TPR domain (Figure 4D), although a second cluster of TPRs also contributes to the binding 39. Notably, the targeting signal peptides are all bound in a similar extended conformation in these proteins. Since the structure of the N-terminus of rmBamD is very similar to TPR domains in these proteins (Figure 4), BamD may have an analogous role, recognizing the C-terminal consensus sequence observed in the vast majority of OMPs: starting at the C-terminus, positions 1, 3, 5, 7 and 9 are hydrophobic amino acids with 1 being almost always F or W and 3 preferentially Y 13. Given the conservation of this C-terminal signature in OMPs across species, it is likely that they are important for one or more mechanistic steps of OMP folding and insertion. BamD is ideally suited to recognize the OMP signal.
Figure 4. The N-terminal domain of rmBamD is similar to proteins binding extended polypeptides.
The N-terminal domain of rmBamD (colored as in Figure 1) is superimposed to TPR domains of Hop (A, yellow), FKBP52 (B, salmon), Tom70(C, gray) and PEX5 (D, light orange); as well as the chaperones PcrH (E, light green) and IpgC (light blue). All these proteins bind extended polypeptides (shown as black C-alpha traces) in the concave face of the TPR tandem.
The N-terminal domain of BamD is also structurally similar to chaperones of the Type Three Secretion System (TTSS) such as IpgC, SycD and PcrH 40; 41; 42 (Figure 4E and F). Crystal structures of IpgC and PcrH in complex with their targets show the concave face of the chaperone 3-TPR tandem binding 8–13 amino acids long peptides in extended conformations, spanning the entire face of the TPR tandem (Figure 4E and F) 41; 42. By analogy to these proteins it is possible that BamD binds unfolded OMPs in an extended conformation prior to folding and insertion into the Outer Membrane. Interestingly, the receptor for β-barrel protein import in mitochondria (Tom70) also has TPR domains that interact with precursor β-barrels prior to translocation through the TOM pore, in addition to the TPRs binding Hsp70 and Hsp90 mentioned above 43; 44.
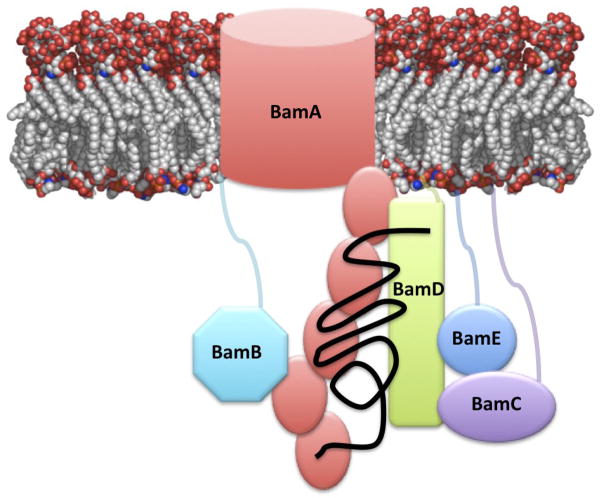
BamD is associated with the membrane by its lipidated amino terminal cysteine 34. All but 7 amino acids at the N-terminus are defined in the structure of rmBamD. Therefore the N-terminal domain must be proximal to the membrane. Conversely, BamC and BamE, the other two BAM lipoproteins known to interact directly with BamD, have disordered N-terminal extensions 25; 27; 45; 46. Both BamE and BamC are thought to bind the C-terminus of BamD 9; 17. Therefore, currently available data on the BAM complex components can be consolidated into a working model described in Figure 5. BamA, with its periplasmic five-POTRA domain recognizes nascent OMPs 14 and may nucleate formation or β-strands or β-hairpins in its substrates 20; 21; 22; 23. BamA interacts directly with BamD, whose C-terminal domain provides a scaffold for the binding of BamC and BamE, which in turn stabilize the complex with BamA. C-terminal truncations of BamD are thus tolerated in E. coli, but result in loss of BamC and weakened binding of BamE to the complex 9; 17. The N-terminal domain of BamD plays an essential role in BAM function. We suggest that it may be required to recognize the C-terminal consensus signal in OMPs or perhaps interact with unfolded OMP precursors. BamB interacts directly with BamA independently of BamCDE 9; 11; 17. As a non-essential component of BAM, BamB may increase the efficiency of the complex or assist in the folding and insertion of a specific subset of substrates either recognizing their C-terminal OMP signal or nucleating β-strand formation 24.
Figure 5. Working model of the BAM complex.
BamA and BamD constitute the core of the complex. BamA is proposed to recognize and help fold nascent OMPs with its five POTRA repeats (show as orange ovals). The N-terminal domain of BamD may assist in this folding role, or may recognize the consensus sequence at the C-terminus of OMPs. BamC and E interact with the C-terminal domain of BamD and help stabilize the complex. BamB interacts with BamA independently of BamC, D and E.
Materials and Methods
Protein Cloning, Expression and Purification
The gene fragment coding for mature BamD (amino acids 23–280) was PCR amplified from Rhodothermus marinus genomic DNA. The primers incorporated NdeI and XhoI restriction sites at the 5’ and 3’ ends of the fragment respectively and also mutated the N-terminal cysteine (C23) to alanine. The amplified fragment was ligated into a modified pET28 vector that incorporates an N-terminal His-tag followed by the recognitions site for the TEV protease. The resulting plasmid (pMS716) thus drives the expression of a His-tagged mature rmBamD C23A. Upon cleavage with TEV protease, amino acid GHM from the tag remain fused to the N-terminus of the protein.
E.coli Rosetta (DE3) cells (Novagen) were transformed with plasmid pMS716 and selected in LB agar plates containing 50 μg/ml kanamycin. A single colony was used to inoculate 100 ml of LB supplemented with 50 μg/ml kanamycin and the culture grown overnight. A 10 ml aliquot was then diluted into 1L of LB with kanamycin and allowed to grow at 37º C to an OD600 of 0.6. Expression was induced by addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG, Gold Bio Inc.) and incubation for 3 hours at 37º C. Cells were harvested by centrifugation, resuspended in a buffer A (25 mM TrisHCl pH 8.0, 150 mM NaCl) supplemented with a protease inhibitor cocktail (Complete EDTA-free, Roche) and frozen until used. After thawing, cells were lysed on ice by sonication. Cell debris were removed by centrifugation at 16,000 rpm for 30 min at 4º C and the supernatant applied to a 10 ml Ni-NTA column (Qiagen) pre-equilibrated with buffer A. The protein was allowed to bind the Ni-NTA beads in batch for 2 hours at 4º C. The resin was collected in a column and washed with 5 column volumes of buffer A, followed by a wash with 5 column volumes of buffer A containing 25 mM imidazole. The protein was then eluted with buffer A supplemented with 200 mM imidazole. TEV protease was added to the elution pool and dialyzed overnight at 4º C in Buffer A supplemented with 10 mM DL-Dithiothreitol (Gold Bio Inc.) to clave the His tag. The cleaved protein was concentrated and loaded on a size exclusion column (HiLoad 26/60 Superdex 200, Amersham Pharmacia Biotech) pre-equilibrated with buffer A and eluted in the same buffer. Purified rmBamD protein was concentrated to 15 mg/ml and stored at -80º C until needed.
Seleno-methionine substituted rmBamD was prepared as described above with the following modifications to the bacterial cultures. Overnight cultures of E. coli Rosetta (DE3) transformed with pMS716 were spun down and resuspended in 3 L M9 minimal medium supplemented with 50 μg/ml Kan, and 4.0 g/l glucose. Cultures were grown at 37º C to OD600 of 0.4. Methionine synthesis was then inhibited by addition of 100 mg/l D-lysine, D-phenylalanine, and D-threonine; 50 mg/l D-isoleucine and D-valine. The culture was also supplemented with 120 mg/l of seleno-methionine (Acros Organics) and incubation continued at 37º C until OD600 was 0.6. Cultures were cooled on an ice bath for 10 minutes, expression induced by addition of 1 mM IPTG followed by incubation overnight at 20º C. Protein purification was performed as describe above. Final protein concentration was 7.5 mg/ml.
Protein Crystallization
Crystallization screening with purified rmBamD was carried out using numerous commercial and homemade screens at various protein concentrations. Several conditions yielded crystals, but best conditions appeared in Wizard screens (Emerald Biosystems). Further optimization was performed using the hanging drop vapor diffusion method (2.0 μl protein: 2.0 μl precipitant) at 16º C. The best native crystals were obtained form a precipitant solution containing 3.25 M NaCl and 0.1 M Tris pH 8.0 and a protein solution at 7.5 mg/ml concentration. Crystals were harvested, transferred directly into 4.0 M malonate pH 7.0 and flash frozen in a stream of N2 at 100 K. Crystals of seleno-methionine substituted rmBamD were obtained in 2.7 M NaCl, 0.1 M Tris pH 7.5, and 0.2 M MgCl2. Cryo-protection was achieved in 4 M malonate as well.
Structure Determination
A dataset to 2.7Å resolution collected at the peak wavelength for selenium from seleno-methionine substituted rmBamD crystals was used to solve the structure by Single Anomalous Dispersion using the AutoSol module of Phenix 47. Three selenium sites were readily identified (Rhodothermus marinus BamD has three non-terminal methionines) and used to calculate SAD phases followed by density modification. The resulting electron density map was readily interpretable and a partial model was automatically built into this map with the AutoBuild module of Phenix. Manual rebuilding using Coot 48 interspersed with model refinement in Phenix were carried out until no further improvement of the R-factor could be achieved. Amino acids 30 to 280 could be unambiguously modeled and refined. The final model statistics are shown in Table 1.
Acknowledgments
This research was supported by NIH grant AI080709 to MCS. Part of this work was performed at the Advanced Light Source, funded by the Department of Energy. Structural biology research at the University of Colorado at Boulder is supported in part by the William M Keck Foundation.
Footnotes
Accession Numbers
Atomic coordinates and structures factors have been deposited in the Protein DataBank with accession number 3QKY.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–63. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem. 2004;73:467–89. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 5.Dong CJ, Beis K, Nesper J, Brunkan-LaMontagne AL, Clarke BR, Whitfield C, Naismith JH. Wza the translocon for E-coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins RF, Derrick JP. Wza: a new structural paradigm for outer membrane secretory proteins? Trends Microbiol. 2007;15:96–100. doi: 10.1016/j.tim.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–45. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–5. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–84. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–2. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–8. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 14.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–5. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 15.Voulhoux R, Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol. 2004;155:129–35. doi: 10.1016/j.resmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–25. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 17.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–64. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 18.Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol. 2005;187:4552–61. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatsos X, Perry AJ, Anwari K, Dolezal P, Wolynec PP, Likic VA, Purcell AW, Buchanan SK, Lithgow T. Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiol Rev. 2008;32:995–1009. doi: 10.1111/j.1574-6976.2008.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–4. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 21.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–81. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and Flexibility of the Complete Periplasmic Domain of BamA. The Protein Insertion Machine of the Outer Membrane. Structure. 2010 doi: 10.1016/j.str.2010.08.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–27. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 24.Heuck A, Schleiffer A, Clausen T. Augmenting beta-Augmentation: Structural Basis of How BamB Binds BamA and May Support Folding of Outer Membrane Proteins. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Kim KH, Kang HS, Okon M, Escobar-Cabrera E, McIntosh LP, Paetzel M. Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex. Biochemistry. 2011 doi: 10.1021/bi101659u. [DOI] [PubMed] [Google Scholar]
- 26.Kim KH, Paetzel M. Crystal Structure of Escherichia coli BamB, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Knowles TJ, Browning DF, Jeeves M, Maderbocus R, Rajesh S, Sridhar P, Manoli E, Emery D, Sommer U, Spencer A, Leyton DL, Squire D, Chaudhuri RR, Viant MR, Cunningham AF, Henderson IR, Overduin M. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 2011 doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–62. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Main ER, Xiong Y, Cocco MJ, D'Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 30.Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2010 doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volokhina EB, Beckers F, Tommassen J, Bos MP. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J Bacteriol. 2009;191:7074–85. doi: 10.1128/JB.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–11. [PubMed] [Google Scholar]
- 33.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fussenegger M, Facius D, Meier J, Meyer TF. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol. 1996;19:1095–105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- 35.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Suppl):W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 37.Wu B, Li P, Liu Y, Lou Z, Ding Y, Shu C, Ye S, Bartlam M, Shen B, Rao Z. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc Natl Acad Sci U S A. 2004;101:8348–53. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 2006;13:589–93. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- 39.Gatto GJ, Jr, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–5. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 40.Buttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J Mol Biol. 2008;375:997–1012. doi: 10.1016/j.jmb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Job V, Mattei PJ, Lemaire D, Attree I, Dessen A. Structural basis of chaperone recognition of type III secretion system minor translocator proteins. J Biol Chem. 2010;285:23224–32. doi: 10.1074/jbc.M110.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci U S A. 2009;106:9661–6. doi: 10.1073/pnas.0812900106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan NC, Likic VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol. 2006;358:1010–22. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- 44.Mills RD, Trewhella J, Qiu TW, Welte T, Ryan TM, Hanley T, Knott RB, Lithgow T, Mulhern TD. Domain organization of the monomeric form of the Tom70 mitochondrial import receptor. J Mol Biol. 2009;388:1043–58. doi: 10.1016/j.jmb.2009.03.070. [DOI] [PubMed] [Google Scholar]
- 45.Albrecht R, Zeth K. Crystallization and preliminary X-ray data collection of the Escherichia coli lipoproteins BamC, BamD and BamE. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1586–90. doi: 10.1107/S1744309110034160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knowles TJ, McClelland DM, Rajesh S, Henderson IR, Overduin M. Secondary structure and (1)H, (13)C and (15)N backbone resonance assignments of BamC, a component of the outer membrane protein assembly machinery in Escherichia coli. Biomol NMR Assign. 2009;3:203–6. doi: 10.1007/s12104-009-9175-3. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]