Abstract
The Nuclear factor I (NFI) transcription factor family consists of four genes (Nfia, Nfib, Nfic and Nfix) that regulate the development of multiple organ systems in mice and humans. Nfib is expressed in both lung mesenchyme and epithelium and mice lacking Nfib have severe lung maturation defects and die at birth. Here we continue our analysis of the phenotype of Nfib−/− lungs and show that Nfib specifically in lung mesenchyme controls late epithelial and mesenchymal cell proliferation and differentiation. There are more PCNA, BrdU, PHH3 and Ki67 positive cells in Nfib−/− lungs than in wild type lungs at E18.5 and this increase in proliferation marker expression is seen in both epithelial and mesenchymal cells. The loss of Nfib in all lung cells decreases the expression of markers for alveolar epithelial cells (Aqp5 & Sftpc), Clara cells (Scgb1a1) and ciliated cells (Foxj1) in E18.5 lungs. To test for a specific role of Nfib in lung mesenchyme we generated and analyzed Nfibflox/flox, Dermo1-Cre mice. Loss of Nfib only in mesenchyme results in decreased Aqp5, Sftpc and Foxj1 expression, increased cell proliferation, and a defect in sacculation similar to that seen in Nfib−/− mice. In contrast, mesenchyme specific loss of Nfib had no effect on the expression of Scgb1a1 in the airway. Microarray and QPCR analyses indicate that the loss of Nfib in lung mesenchyme affects the expression of genes associated with extracellular matrix, cell adhesion and FGF signaling which could affect distal lung maturation. Our data indicate that mesenchymal Nfib regulates both mesenchymal and epithelial cell proliferation through multiple pathways and that mesenchymal NFI-B-mediated signals are essential for the maturation of distal lung epithelium.
Keywords: NFI-B, lung development, mesenchymal-epithelial induction, FGF, Elastin
Introduction
The Nuclear factor I (NFI) family of transcription factors functions both in mammalian development and in adenoviral DNA replication (de Jong and van der Vliet, 1999; Nagata et al., 1982; Nagata et al., 1983). In vertebrates, the NFI family consists of four genes, Nfia, Nfib, Nfic and Nfix, that encode proteins which bind as homo- and heterodimers to the consensus sequence TTGGC(N5)GCCAA on duplex DNA (Gronostajski, 1986; Meisterernst et al., 1988). NFI-binding sites have been identified in the promoter, enhancer and silencer regions of more than 100 genes expressed in multiple organs, including brain, lung, liver and intestine (Gronostajski, 2000). Here we address the role of Nfib in lung development.
Lung immaturity is a major problem in premature infants. It is associated with respiratory distress syndrome, an acute lung problem that presents shortly after birth, and bronchopulmonary dysplasia, a chronic lung disease of premature infants (Coalson et al., 1999; Jobe, 2005). Previous studies of mice lacking Nfib revealed defects in lung development and Nfib−/− mice die at birth with immature lungs (Grunder et al., 2002; Steele-Perkins et al., 2005). At E18.5 Nfib−/− lungs lack saccules, have an increased DNA content, and decreased levels of surfactant protein transcripts. This phenotype indicates that Nfib plays an important role in lung maturation. Previous studies also showed that NFI-B can directly regulate the expression of genes in lung epithelium (Bachurski et al., 2003). However, the specific cell types in which Nfib is required for normal lung development, and the critical target genes regulated by Nfib during lung maturation remain unknown.
Lung development is regulated by mesenchymal-epithelial interactions (Deimling et al., 2007; Demayo et al., 2002; Morrisey and Hogan, 2010). Some signals from epithelial and/or mesenchymal cells that control smooth muscle cell differentiation and vasculogenesis have been well characterized, including the VEGF-A (Akeson et al., 2003; White et al., 2007; Zeng et al., 1998), PDGF (Hellstrom et al., 1999; Li and Hoyle, 2001), and WNT (Cohen et al., 2009; Li et al., 2002; Shu et al., 2002) signaling pathways. In contrast, the signaling pathways from mesenchymal cells that influence epithelial cell proliferation and differentiation are less well understood. One mesenchymal-expressed factor that is known to affect lung epithelial cell proliferation and differentiation is FGF-10. Fgf10 is expressed in mesenchyme and Fgf10−/− mice were characterized by the absence of lungs (Min et al., 1998). Overexpression of FGF-10 in late lung development results in the impairment of lung branching and attenuation of distal epithelial cell differentiation (Clark et al., 2001; Nyeng et al., 2008), suggesting that FGF-10 must be tightly regulated for normal lung development. However, much more information is needed regarding how mesenchyme regulates lung maturation.
Here we demonstrate an essential role of Nfib in lung maturation using Nfib−/− mice and mice in which Nfib is deleted specifically in mesenchyme using Dermo1-Cre. We show that while Nfib is expressed in both mesenchyme and epithelium during lung development, loss of Nfib specifically in mesenchyme affects both mesenchymal and epithelial cell proliferation, and distal epithelial cell differentiation. These data demonstrate a heretofore unrecognized pathway of mesenchymal regulation of late epithelial maturation. In addition, microarray and QPCR analyses were used to identify biochemical pathways regulated by NFI-B that appear important for lung maturation.
Materials and Methods
Histology and Immunohistochemistry
Fetal lungs were dissected and fixed in 4% paraformaldehyde overnight at 4°C. After paraffin embedding, 4 µm sections were cut and stained with hematoxylin and eosin. For immunohistochemistry, antibodies against the following proteins were used: CC10 (Santa Cruz T-18, 1:200), pro-SPC (Chemicon AB3786, 1:2000), PCNA (Santa Cruz FL-261, 1:200), phospho-histone H3 (Sigma HTA28, 1:200), cleaved Caspase-3 (Cell Signaling #9661, 1:200), Ki67 (Abcam ab15580, 1:400), TTF-1 (Dako 8G7G3/1, 1:200), Vimentin (Sigma LN-6, 1:400), Caveolin-1 (Santa Cruz N-20, 1:200), smooth muscle actin (SMA) (Sigma 1A4, 1:5000), AQP5 (Alomone labs AQP-005, 1:200) and NFI-B (Active Motif 1:1000). In general, primary antibodies were incubated with slides overnight at 4°C. After a PBS wash, slides were incubated with secondary antibodies (Alexa 488 or Alexa 568, Invitrogen) for 1 hour at room temperature and auto-fluorescence was eliminated with 0.5% Sudan black B. TOPRO3 (Invitrogen, 1:5000) was used as a nuclear counterstain on some sections. For BrdU staining, pregnant mice were injected with 0.1 mg/g BrdU 2 hours before sacrifice, lungs were fixed and processed, and sections were stained using a BrdU staining kit (Zymed).
Statistics
Comparisons were made between at least three KO and littermate control mice. Statistical significance was assessed using the two-tailed student t-test in Microsoft Excel. P values of less than 0.05 were considered statistically significant. Values for all experiments are expressed as the mean +/− SD. In all QPCR analyses triplicate PCR reactions from each of at least 3 biological replicates were analyzed. For statistical quantification of cell number using immunohistochemistry, 15 random fields were imaged from 3 mice of each genotype. A minimum of 100 positive cells were counted for each condition. Intra-litter comparisons of each genotype were used whenever possible and multiple litters were assessed for consistency of phenotype. The numbers of fluorescent and total cells were quantified using Image J software.
RNA extraction and RT-QPCR
RNA was extracted with TRIzol reagent (Invitrogen) and 2–5 µg was used for random hexamer primed cDNA synthesis with Superscript II (Invitrogen). Transcript levels were quantified by quantitative PCR (QPCR) with a Bio-Rad iCycler real-time PCR machine using SYBR Green as described previously (Steele-Perkins et al., 2005). All results were normalized to β2-microglobulin levels. Sequences of the primers are available upon request.
Chromatin immunoprecipitation (ChIP) assay
E16.5 lungs were isolated from wild type embryos, minced and fixed with 1% formaldehyde. Chromatin was sheared to ∼ 200–500 bp using a Branson Sonifier 250 sonicator. Immunoprecipitation was performed with a ChIP assay kit (Upstate Biotechnology) and αNFI-B antibody (Geneka Biotechnology). After immunoprecipitation and reversal of cross-links the chromatin was subjected to PCR and QPCR. ChIP analysis was performed on E16.5 lung in an effort to identify potential early mesenchymal targets of Nfib that could contribute to the later morphological phenotype seen at E18.5. The large number of biochemical and morphological differences between E18.5 wild-type and mutant lungs and the increase in epithelial NFIB expression could complicate the identification of direct mesenchymal NFIB targets at this stage.
Transient Transfection assays
Mouse embryo fibroblasts (MEFs) were isolated from WT E13 embryos as described previously (Plasari et al., 2009). Cells were transfected using Lipofectamine as recommended by the manufacturer with vectors expressing GFP or NFIB2 from the CMV promoter (Chaudhry et al., 1999). After 48 hours cells were harvested using Trizol, RNA was isolated, cDNA was prepared using random primers and QPCR was performed with primers specific for Elastin as described above.
Gene targeting and mouse strains
A targeting vector was constructed with a 4.4 kb 5' homology arm containing all of exon 2 of Nfib and 300 bp of intron 2, with a loxP site inserted 363 bp 5' to the start of exon 2. The 5' arm was followed by an FRT-flanked PGK-neo expression cassette in the opposite transcriptional orientation, a 3' loxP site, the contiguous 3.6 kb 3' homology arm and a PGK-diphtheria toxin A chain cassette in the opposite transcriptional orientation (Fig. S1A) (Campbell et al., 2008). Cre-mediated recombination deletes all of exon 2, 363 bp of intron 1 and 300 bp of intron 2. This vector was linearized with AscI, electroporated into J1 ES cells and G418-resistant colonies were picked, expanded and banked until PCR screening was completed. Five of six correctly targeted clones (6 targeted/256 total) were thawed, targeting was confirmed, and two were expanded and injected into C57Bl/6 blastocysts to generate chimeric animals. Chimeras were bred with C57Bl/6 females and agouti progeny were screened for the presence of the targeted conditional KO allele by PCR. Nfibflox/flox mice survive birth with no overt lung phenotype, grow to adulthood and at E18.5 express ∼70–80% of wild type levels of Nfib.
Dermo1-Cre mice were kindly provided by David Ornitz (Yin et al., 2008). Nfibflox/flox mice were crossed with Dermo1-Cre mice to generate Nfibflox/+, Dermo1-Cre offspring. Nfibflox/+, Dermo1-Cre mice were then crossed to Nfibflox/flox mice to generate mesenchymal-specific Nfib null mice. All mice were genotyped using PCR and sequences of the primers are available upon request. All protocols were approved by the IACUC at Roswell Park Cancer Institute.
Microarray analysis
Total RNA was isolated from three different Nfibflox/flox and Nfibflox/flox, Dermo1-Cre lungs at E18.5 using TRIzol reagent (Invitrogen) and then purified using an RNeasy Mini Kit (Qiagen). The purified RNA was examined using an Agilent Bioanalyzer 2100 (Agilent Technologies) and then labeled and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays using the manufacturer's protocol. Three independent sets of biological replicates (Nfibflox/flox and Nfibflox/flox, Dermo1-Cre lungs) were used. Scanned microarray images were imported into GeneChip Operating Software (GCOS, Affymetrix) to generate raw signal values for each probe. The MAS5.0 algorithm in the ‘Affy’ package of Bioconductor in the R statistical computing environment was used to generate expression summary values, followed by trimmed mean global normalization to bring the mean expression values of all six GeneChips to the same scale (Gentleman et al., 2004). For data quality control, MAS5.0 present/absent calls were used to filter out probe sets whose expression intensities were close to background noise across the majority of samples. Specifically, filtering of three ‘present calls’ was applied to either the Nfibflox/flox or Nfibflox/flox, Dermo1-Cre group, with 25,086 unique transcripts passing the quality control. We used the linear model implemented in Limma program to calculate the level of gene differential expression (Smyth, 2004).Transcripts that were altered at a P-value less than 0.05 and with at least 1.5-fold expression change between Nfibflox/flox, Dermo1-Cre and Nfibflox/flox mice were considered significant and used for further analysis. These differentially expressed genes were analyzed for statistically enriched gene ontology terms using the NCBI DAVID package with the default setting (Huang da et al., 2007). The mRNA expression profiling datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) data repository (http://www.ncbi.nlm.nih.gov/geo/) under Accession number GSE24465.
Results
NFI-B is expressed predominantly in mesenchyme at E14.5 and E16.5, and in both epithelial cells and mesenchyme at E18.5
We showed previously by lacZ staining in heterozygous NfiblacZ/+ mice that Nfib is expressed almost exclusively in mesenchyme at E14.5-E16.5 but is expressed in both mesenchyme and epithelium by E18.5 (Steele-Perkins et al., 2005). To assess the expression pattern of NFI-B in WT mice, aNFI-B antibodies were used for immunohistochemistry on sections of E14.5, E16.5 and E18.5 lungs. As expected, from E14.5 to E16.5, NFI-B was expressed almost exclusively in the nuclei of mesenchymal cells surrounding bronchial tubules (dashed lines, Fig. 1 A , B). At E18.5, dual immunostaining showed that NFI-B was colocalized with both Thyroid Transcription Factor 1 (TTF-1) (arrows in Fig. 1C, magnified 5X in inset), a bronchiolar and alveolar type II epithelial cell marker, and Vimentin (arrows in Fig. 1D, magnified 5X in inset), a mesenchymal cell marker. These data suggest that NFI-B likely function primarily in mesenchyme until E14.5 and in both mesenchyme and epithelium after E16.5.
Fig. 1.
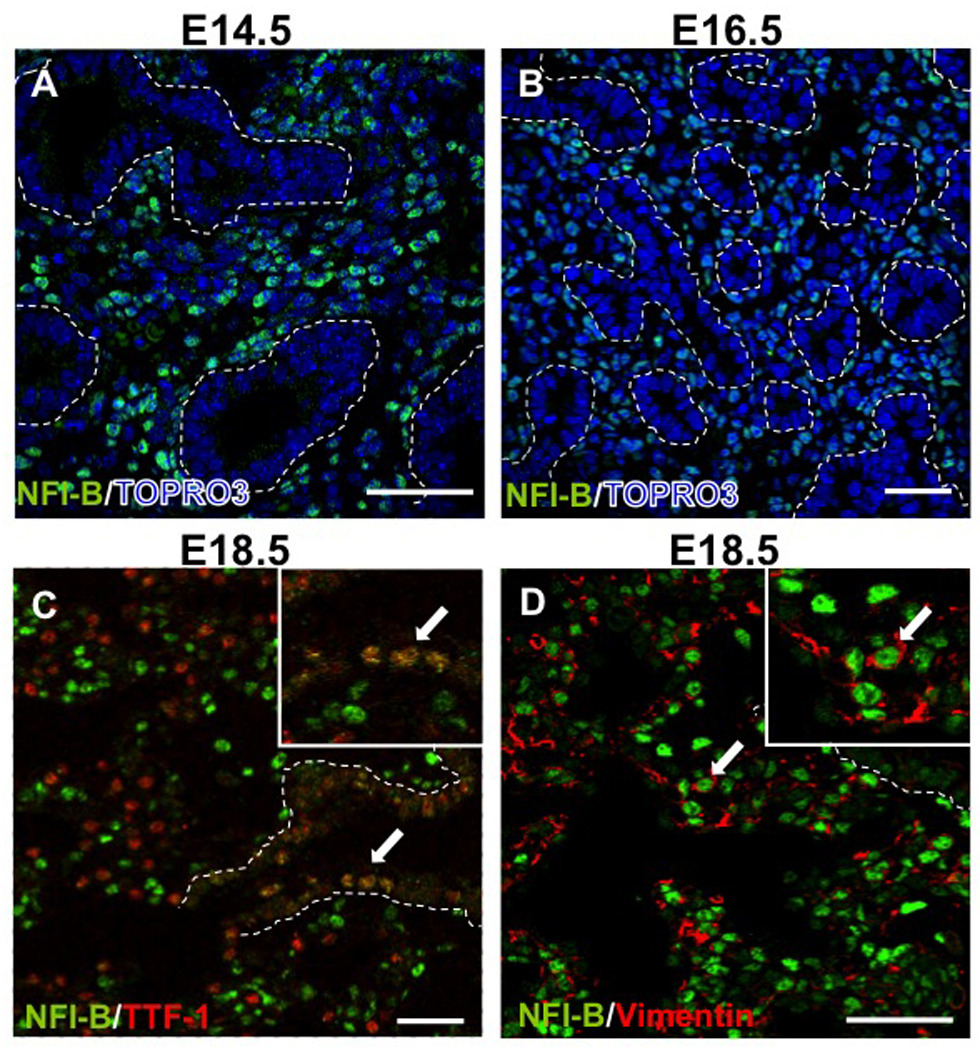
Expression of NFI-B in mesenchyme and epithelium during lung development. Paraffin sections of E14.5 (A), E16.5 (B), and E18.5 (C, D) lungs were immunostained for NFI-B expression. At E14.5 and E16.5, NFI-B was expressed predominantly in mesenchyme. At E18.5, NFI-B was expressed in both epithelial cells (TTF-1 positive cells) (C) and mesenchymal (Vimentin positive cells) (D). The dashed lines denote the border between epithelial and mesenchymal cells in bronchioles. Arrows denote the colocalization of NFI-B and TTF-1 (C) or NFI-B and Vimentin (D). Insets in C and D show a 5X magnification. TOPRO-3 shows nuclear staining. Scale bars, 50 µm.
Loss of Nfib increases cell proliferation but doesn’t affect apoptosis
We showed previously that the DNA content/total body weight of E18.5 Nfib−/− lungs was about double that of WT lungs, suggesting changes in either cell proliferation or apoptosis late in lung development (Steele-Perkins et al., 2005). To assess cell proliferation, antibodies against proliferating cell nuclear antigen (PCNA), Ki67 or phospho-histone H3 (PHH3) were used to stain E18.5 lung sections from Nfib+/+ and Nfib−/− embryos. Multiple markers were used to ensure that any change in proliferation detected was not marker-dependent. The number of PCNA positive cells was increased in Nfib−/− lungs compared with Nfib+/+ lungs (Fig. S2A, B) and quantification of these data (PCNA positive nuclei/ total nuclei) supported this conclusion (Fig. S2C). In addition, there were more Ki67 positive cells and PHH3 positive cells in Nfib−/− lungs than in Nfib+/+ lungs (Fig S2G, H, J, K) and quantification showed an increase in the number of cells expressing multiple proliferation markers in Nfib−/− lungs (Fig. S2I, L). To further assess cell proliferation, bromodeoxyuridine (BrdU) was injected into mice 2 hours before sacrifice and then detected with αBrdU antibodies. BrdU staining was increased significantly in Nfib−/− lungs compared to Nfib+/+ lungs (Fig. S2D, E, F). These data indicate that loss of Nfib results in dramatically increased cell proliferation at E18.5.
Apoptosis in E18.5 lungs was examined by immunostaining of cleaved Caspase 3. The number of cleaved Caspase 3 positive cells is similar in Nfib+/+ and Nfib−/− lungs (Fig. S3 A, B, C). Together, these data indicate that loss of Nfib increases lung DNA content through increasing cell proliferation with no affect on apoptosis.
Loss of Nfib increases both TTF-1 positive and TTF-1 negative cell proliferation
To identify the hyperproliferative cells in Nfib−/− lungs at E18.5, αTTF-1 antibody was used to label epithelial cells. Dual immunostaining for PCNA and TTF-1 revealed more double positive cells (PCNA+TTF-1+ cells) (arrows in Fig. 2A, B, magnified 5X in insets) in Nfib−/− lungs than in Nfib+/+ lungs. Quantification of these data (PCNA+TTF-1+ cells/ total TTF-1+ cells) (Fig. 2C) supported this conclusion. We also examined TTF-1− cell proliferation. There was a substantial increase in TTF1−PCNA+ cells, indicating an increase in TTF-1− cell proliferation (Fig. 2C). Dual immunostaining for Ki67 and TTF-1 was used to further assess TTF-1+ and TTF-1− cell proliferation. These data indicate that more of both Ki67+TTF-1+ and Ki67+TTF-1− cells are present in Nfib−/− lungs than in Nfib+/+ lungs (Fig. 2D–F, magnified 5X in insets). These data are consistent with our previous PCNA analysis and show that loss of Nfib leads to an increase in proliferation of both epithelial and mesenchymal cells during lung development.
Fig. 2.
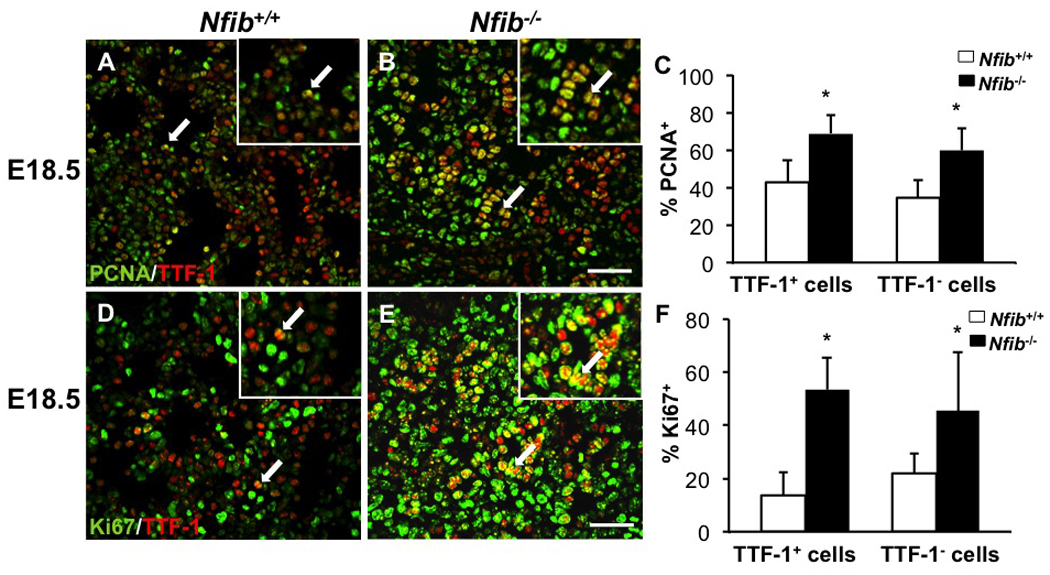
Loss of Nfib increases both epithelial (TTF-1+) and mesenchymal (TTF-1−) cell proliferation. Paraffin sections of E18.5 lungs from Nfib+/+ and Nfib−/− embryos were stained for both PCNA and TTF-1 (A, B) or for both Ki67 and TTF-1 (D, E). Immunostaining reveals there are more PCNA+TTF-1+ cells (arrows) and Ki67+TTF-1+ cells (arrows) in Nfib−/− lungs compared with Nfib+/+ lungs. Insets in panels A, B, D and E show a 5X magnification. Quantification (C, F) indicates there are more PCNA+TTF-1− cells and Ki67+TTF-1− cells in Nfib−/− lungs compared with Nfib+/+ lungs. Arrows denote the colocalization of PCNA and TTF-1 or Ki67 and TTF-1 and the region of magnification; *P<0.05; Scale bars, 50 µm.
Loss of Nfib results in decreased epithelial Type I, type II, ciliated cell and Clara cell differentiation
To confirm and extend our previous findings that Nfib is essential for epithelial cell differentiation, we performed both RT-QPCR and immunohistochemistry for several cell-type-specific markers. Aquaporin 5 (Aqp5), an alveolar type I cell marker, was essentially absent from Nfib−/− lungs compared with Nfib+/+ lungs (Fig. 3A, B), consistent with the result of RT-QPCR (Fig. 3G). Expression of pro-surfactant protein C (Sftpc), an alveolar type II cell marker, was also decreased in Nfib−/− lungs (Fig. 3C, D) and confirmed by RT-QPCR (Fig. 3G). Since alveolar type II cells potentially differentiate into type I alveolar cells during lung injury and lung development (Sugahara et al., 2006), loss of Nfib could affect alveolar type II cell differentiation directly and indirectly reduce alveolar type I cell number. Both Immunostaining and RT-QPCR for CC10 (Scgb1a1), a Clara cell marker, showed a dramatic decrease in CC10 expression in Nfib−/− lungs versus Nfib+/+ lungs (Fig. 3E, F, G). In addition, the levels of Foxj1 transcripts (a ciliated cell marker) were dramatically reduced in the Nfib−/− lungs (Fig. 3G). These data indicate that loss of Nfib results in defects in both distal and proximal epithelial cell differentiation.
Fig. 3.
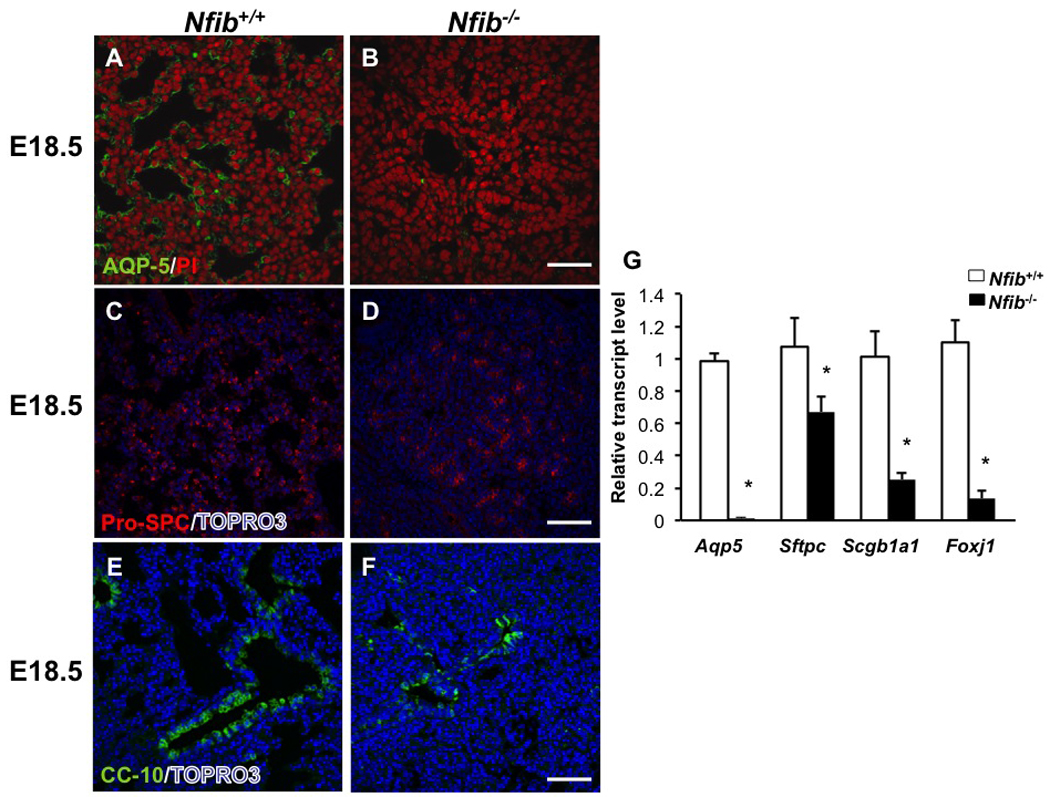
Nfib−/− lungs exhibit decreased epithelial cell differentiation at E18.5.
E18.5 lung sections from Nfib+/+ and Nfib−/− embryos were stained for AQP5 (type I epithelial cell marker) (A, B), pro-SPC (type II epithelial cell marker) (C, D) and CC10 (Clara cell marker) (E, F). Loss of Nfib at E18.5 lungs led to decreased expression of all three epithelial cell markers. RT-QPCR analysis (G) reveals Aqp5, Sftpc and Scgb1a1 and Foxj1 expression is significantly decreased in Nfib−/− lungs at E18.5. PI and TOPRO3 staining show nuclei; *P<0.05; Scale bars, 50 µm.
Dermo1-Cre deletes a floxed Nfib allele specifically in lung mesenchyme
Because Nfib is required for lung maturation and is expressed almost exclusively in mesenchyme at E14.5, we proposed that Nfib in lung mesenchyme may regulate the phenotype seen in Nfib−/− lungs. To test this hypothesis, we created a conditional Nfib allele (Nfibflox) such that exposure to Cre deletes exon 2 and ∼660 bp of adjacent intronic sequence (Fig. S1A). Nfibflox/flox mice have no overt lung phenotype, survive to adulthood, are fertile, and at E18.5 express at least ∼70–80% of wild type levels of Nfib (data not shown). To determine the effect of deleting Nfib exclusively in the mesenchyme, Dermo1-Cre mice (here termed D1-Cre) were crossed with Nfibflox/flox mice to generate Nfibflox/+, D1-Cre offspring. Nfibflox/+, D1-Cre mice were then crossed to Nfibflox/flox mice to generate mesenchymal-specific Nfib null mice (here termed Nfibflox/flox, D1-Cre). By E16.5, D1-Cre had efficiently eliminated NFI-B expression in lung mesenchyme (Fig. 4A, B arrows, magnified 5X in insets). At E18.5, D1-Cre had no obvious affect on NFI-B expression in epithelium (arrows in Fig. 4C, D, magnified 5X in insets), indicating that Nfib was deleted specifically in the mesenchyme of Nfibflox/flox, D1-Cre mice. These data are consistent with previous studies showing that D1-Cre specifically deletes floxed alleles only in lung mesenchyme, but not lung epithelium (De Langhe et al., 2008; White et al., 2006; Yin et al., 2008).
Fig. 4.
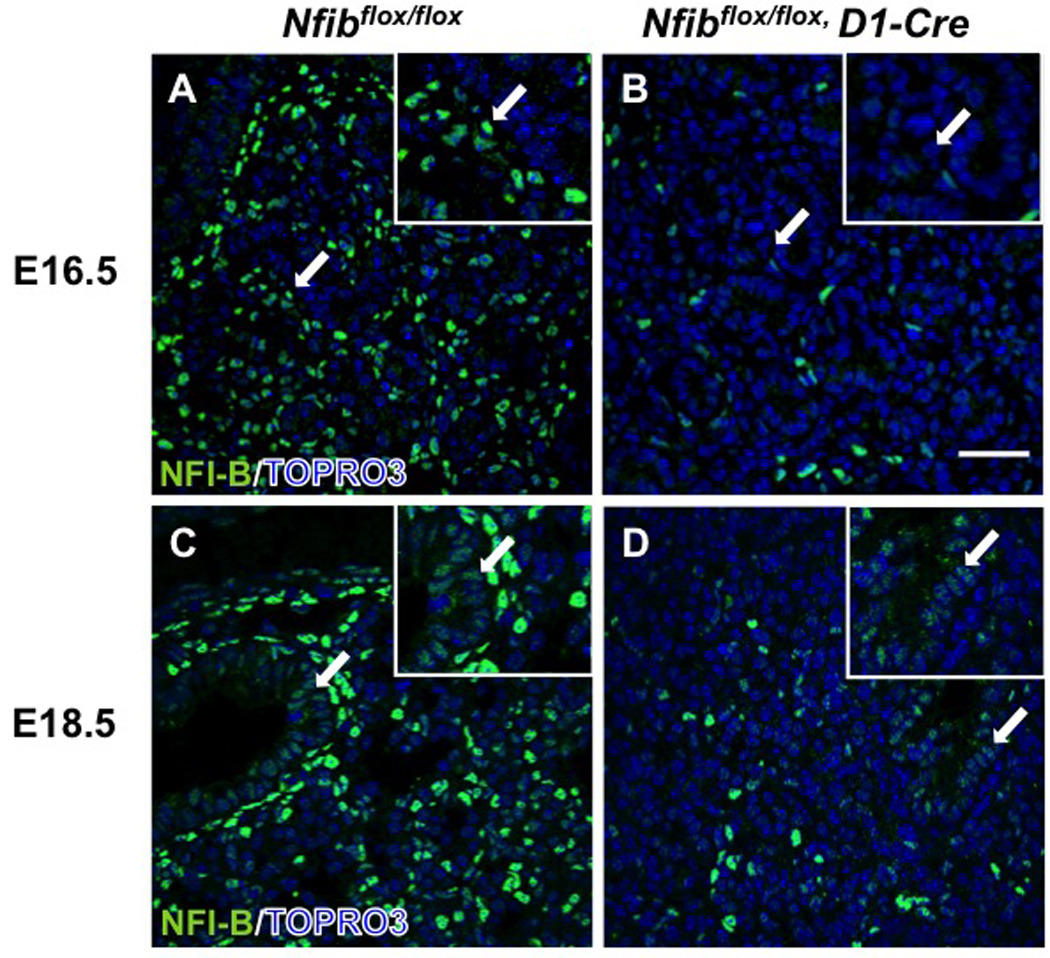
Dermo1-Cre reduces NFI-B expression in Nfibflox/flox lungs in mesenchyme, but not in epithelium. Lung sections from E16.5 Nfibflox/flox (A) and Nfibflox/flox, D1-Cre (B) embryos stained for NFI-B showed Dermo1-Cre eliminated almost all NFI-B in mesenchyme at E16.5. Immunostaining for NFI-B (C, D) in E18.5 Nfibflox/flox and Nfibflox/flox, D1-Cre lungs reveals Dermo1-Cre eliminated NFI-B expression throughout the lung mesenchyme at E18.5 but didn’t affect NFI-B expression in the epithelial cells (arrows). Insets show a 5X magnification. Arrows denote NFI-B in bronchiolar epithelial cells and the region of magnification; Scale bar, 50 µm.
Perinatal lethality and sacculation defects in Nfibflox/flox, D1-Cre mice
No live Nfibflox/flox, D1-Cre mice were recovered at P0 from 4 litters (Table 1), while all other genotypes were found in appropriate numbers. This lethality occurs perinatally since the number of animals of each genotype recovered at E16.5 and E18.5 was near the predicted Mendelian frequency of 1:1:1:1 (12 Nfibflox/+ mice, 11 Nfibflox/+, D1-Cre mice, 8 Nfibflox/flox mice and 12 Nfibflox/flox, D1-Cre mice, 5 litters). Perinatal lethality is consistent with our previous results with Nfib−/− mice (Steele-Perkins et al., 2005).
TABLE 1.
Loss of Nfib in mesenchyme results in perinatal lethality
| No. of live animals of each genotype from Nfibflox/+, D1-CreXNfibflox/flox matings | ||||
|---|---|---|---|---|
| Genotype | Nfibflox/+ | Nfibflox/flox | Nfibflox/+, D1-Cre | Nfibflox/flox, D1-Cre |
| Observed | 10 | 14 | 9 | 0 |
| Expected | 10 | 10 | 10 | 10 |
4 litters were examined in this experiment. The expected numbers are based on the number of Nfibflox/+ progeny obtained.
Our and other studies previously found a defect in sacculation in Nfib−/− mice (Grunder et al., 2002; Steele-Perkins et al., 2005). Therefore, we examined the morphology and histology of Nfibflox/flox, D1-Cre lungs at E18.5. Like Nfib−/− lungs, Nfibflox/flox, D1-Cre lungs were larger than Nfibflox/flox lungs (Fig. 5A). Histological analysis of E18.5 lungs showed that lungs from Nfibflox/flox, D1-Cre mice had a severe defect in sacculation, while those from control Nfibflox/flox mice were well sacculated (Fig. 5D, E). In addition to the defect in sacculation, we found aberrant clefts in Nfibflox/flox, D1-Cre lungs at E16.5 and E18.5 (arrows in Fig. 5C, F). Similar clefts were seen previously in Nfib−/− lungs (Steele-Perkins et al., 2005). These data show that the Nfibflox/flox, D1-Cre lungs share multiple aberrant morphological changes with Nfib−/− lungs.
Fig. 5.
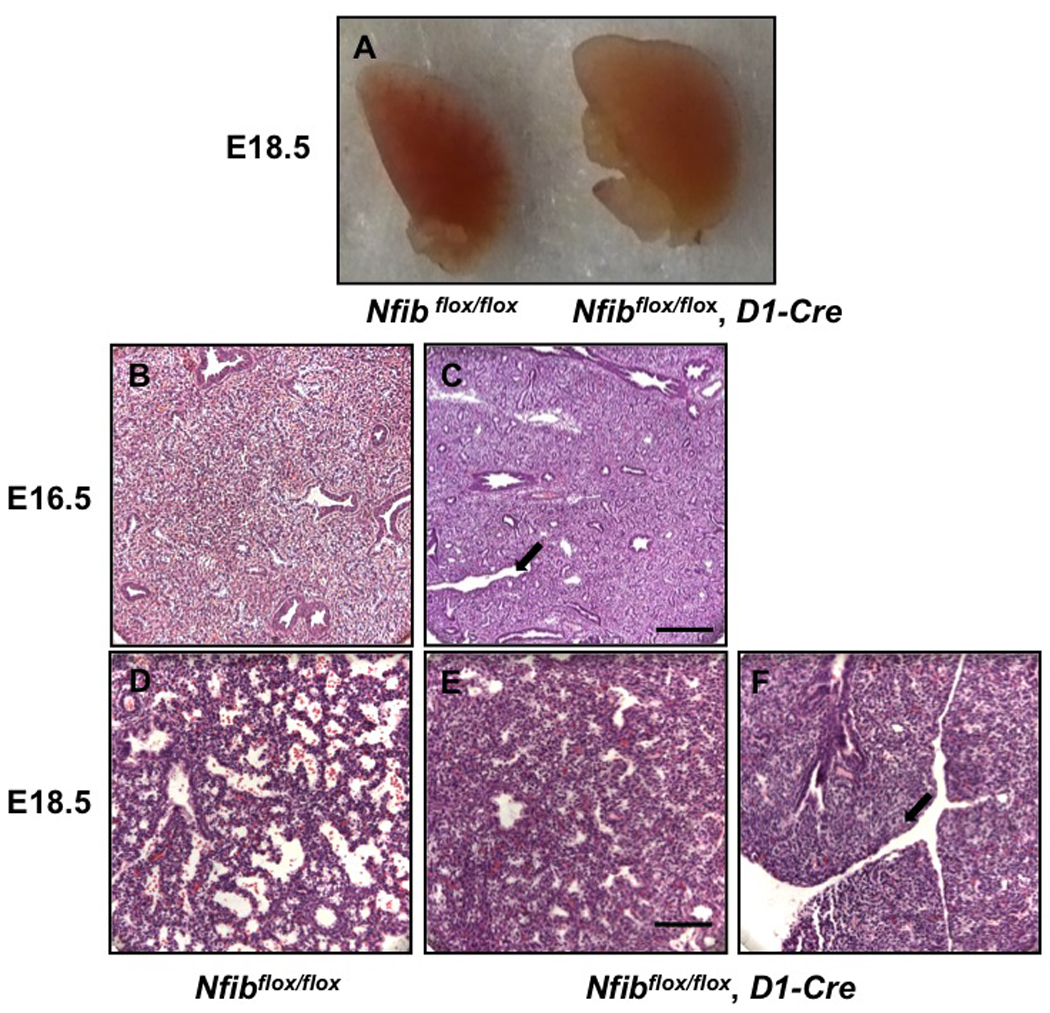
Sacculation and lung morphology is affected by loss of Nfib in mesenchyme. Dissected left lobes of Nfibflox/flox and Nfibflox/flox, D1-Cre lungs are shown unstained at E18.5 (A), and H&E stained at E16.5 (B, C) and E18.5 (D, E, F). Nfibflox/flox, D1-Cre lungs were larger than Nfibflox/flox lungs (A). H&E staining shows abnormal clefts (arrows in C, F) in Nfibflox/flox, D1-Cre lungs at E16.5 (C) and E18.5 (F). At E18.5, there is increased mesenchyme thickness and reduced sacculation in Nfibflox/flox, D1-Cre lungs. Scale bar, 50 µm.
Loss of Nfib in mesenchyme increases cell proliferation
To determine whether Nfib in lung mesenchyme is required for the control of cell proliferation during lung maturation, we performed PHH3 immunostaining on E18.5 Nfibflox/flox and Nfibflox/flox, D1-Cre lung sections. There were more PHH3 positive cells in Nfibflox/flox, D1-Cre lungs than in Nfibflox/flox lungs (Fig. S4A, B), indicating a higher rate of proliferation at E18.5 in the former. Ki67 staining was also performed on E16 and E18.5 lung samples. These data show more Ki67 positive cells in Nfibflox/flox, D1-Cre lung at both E16.5 and E18.5 (Fig. S4D–G). Quantification of the immunostaining (Fig. S4C, H) was consistent with the data from Nfib−/− lungs (Fig. S2C–L) in showing an increase in cell proliferation.
Loss of Nfib in mesenchyme increases both TTF-1 positive and TTF-1 negative cell proliferation
To determine whether the increased proliferation in Nfibflox/flox, D1-Cre lungs was predominantly in the epithelial or mesenchymal compartments, we performed dual staining for TTF-1 and Ki67 on sections of E16 and E18.5 lungs. Notably, more Ki67+TTF-1+ cells were present in Nfibflox/flox, D1-Cre lungs relative to Nfibflox/flox lungs indicating increased proliferation of epithelial cells (arrows in Fig. 6A–D, magnified 5X in insets). Quantification confirmed an increase in the percentage of Ki67+TTF-1+ cells (Fig. 6E). Furthermore, we determined the percentage of Ki67+TTF-1− cells at E16.5 and E18.5. The percentage of Ki67+TTF-1− mesenchymal cells in Nfibflox/flox, D1-Cre lungs was higher at E16.5 and then decreased to a level similar to that seen in Nfibflox/flox lungs by E18.5 (Fig. 6F). These data indicate that loss of Nfib in mesenchyme results in an increase in both TTF-1+and TTF-1− cell proliferation during lung development. However, at E18.5 the apparent rate of TTF-1− cell proliferation appears similar in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs, which is different from that seen in Nfib+/+ vs. Nfib−/− lungs (Fig. 2D–F). We discuss below possible reasons for these relatively minor differences in apparent TTF-1− cell proliferation.
Fig. 6.
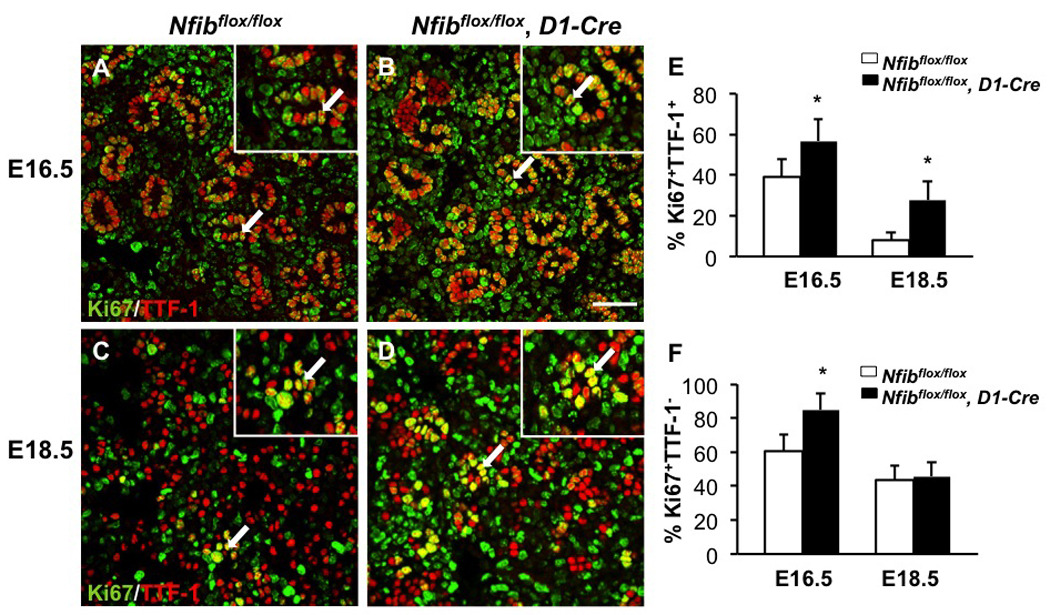
Loss of Nfib in mesenchyme increases TTF-1 positive and TTF-1 negative cell proliferation. Paraffin sections of E18.5 and E16.5 lungs from Nfibflox/flox and Nfibflox/flox, D1-Cre embryos were stained for both Ki67 and TTF-1 (A, B, C, D). Insets show a 5X magnification. Immunofluorescence staining reveals there were more Ki67+TTF-1+ cells (arrows) in Nfibflox/flox, D1-Cre lungs compared with Nfibflox/flox lungs at E16.5 and E18.5 (E). The percentage of Ki67+TTF-1− cells was also higher in Nfibflox/flox, D1-Cre lungs at E16.5 (F). In contrast, the percentage of Ki67+TTF-1− cells was similar between Nfibflox/flox and Nfibflox/flox, D1-Cre lungs at E18.5 (F). Arrows denote the colocalization of Ki67 and TTF-1 and the region of magnification; *P<0.05; Scale bar, 50 µm.
Loss of Nfib in mesenchyme affects epithelial cell differentiation and surfactant gene expression
As done with Nfib−/− lungs, we assessed multiple epithelial cell differentiation markers in Nfibflox/flox, D1-Cre lungs by immunostaining. AQP5 and pro-SPC staining was significantly reduced in Nfibflox/flox, D1-Cre lungs relative to Nfibflox/flox lungs (Fig. 7A–D). RT-QPCR analysis showed that Aqp5 and Sftpc transcript levels were also significantly decreased with loss of Nfib in the mesenchyme (Fig. 7G). In addition, as was seen in germline Nfib KO lungs (Fig. 3G) the expression of the ciliated cell marker Foxj1 was substantially decreased in the Nfibflox/flox, D1-Cre lungs (Fig. 7G). In contrast, Scgb1a1 (CC10) expression was similar in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs as assessed by both immunostaining (Fig. 7E–F) and RT-QPCR (Fig. 7G). These data show that mesenchymal Nfib is clearly required for Type I, Type II and ciliated cell differentiation, but fail to support a role of mesenchymal expression of Nfib in the regulation of Clara cell differentiation.
Fig. 7.
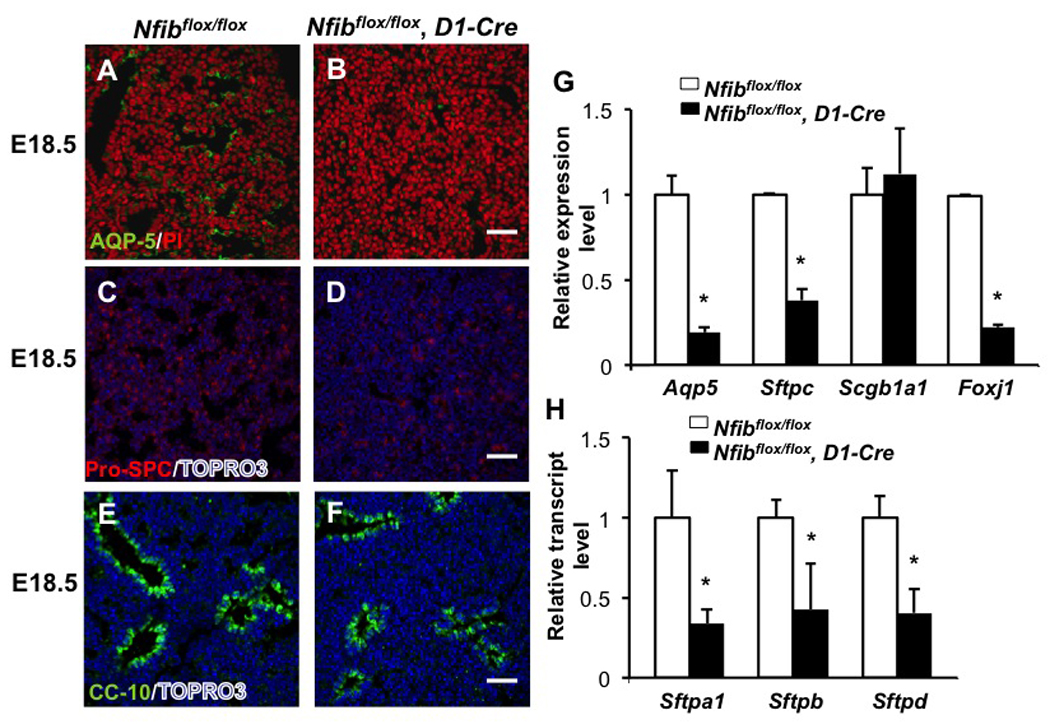
Loss of Nfib in mesenchyme affects type I and type II epithelial cell differentiation. E18.5 lung sections from Nfibflox/flox and Nfibflox/flox, D1-Cre embryos were stained for AQP5 (A, B), pro-SPC (C, D) and CC10 (E, F). AQP5, pro-SPC and Foxj1 transcript expression was also reduced in Nfibflox/flox, D1-Cre lungs, as shown by RT-QPCR (I). In contrast, immunostaining and RT-QPCR revealed CC 10 (Scgb1a1) protein and transcript levels were similar in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs. Along with reduced Sftpc expression, Sftpa1, Sftpb and Sftpd expression was significantly reduced in Nfibflox/flox, D1-Cre lungs (H). *P<0.05; Scale bars, 50 µm.
RT-QPCR analysis showed that mRNA levels of the other surfactant proteins, including surfactant protein A (Sftpa1), surfactant protein B (Sftpb) and surfactant protein D (Sftpd), were dramatically decreased in Nfibflox/flox, D1-Cre lungs compared with Nfibflox/flox lungs at E18.5 (Fig. 7H). Thus, loss of Nfib in mesenchyme results in defects in distal epithelial cell differentiation as assessed by surfactant protein gene expression.
Loss of Nfib in mesenchyme affects the expression of genes related to lipid production, extracellular matrix, cell adhesion and the FGF signaling pathway
To assess potential target genes of NFI-B that could affect cell proliferation and distal epithelial cell differentiation during lung maturation, we performed microarray analysis on RNA from E18.5 Nfibflox/flox and Nfibflox/flox, D1-Cre lungs. Genes with expression changes of 1.5 fold or greater and p-values of <0.05 were selected for further analysis (see Table S1).
The expression of genes involved in a number of biological processes, including extracellular matrix and cell adhesion appear affected by loss of Nfib in lung mesenchyme (Fig. 8A). To confirm these expression data, the transcript levels of several genes, including Elastin (Eln), Fibronectin (Fn1), collagen, type IV, alpha 3 (Col4a3) and Ephrin A3 (Efna3), were examined using RT-QPCR. The RT-QPCR data are consistent with our microarray data (Fig. 8B–E), indicating an effect on genes involved in extracellular matrix production and cell adhesion in Nfibflox/flox, D1-Cre mice. The transcripts levels of Tenascin C (Tnc), an extracellular matrix protein involved in lung vasculogenesis and smooth muscle cell differentiation (Cohen et al., 2009) were also examined. There was no significant change of Tnc transcript levels between Nfibflox/flox and Nfibflox/flox, D1-Cre lungs (fold change=1, P=0.5) (Fig. S5).
Fig. 8.
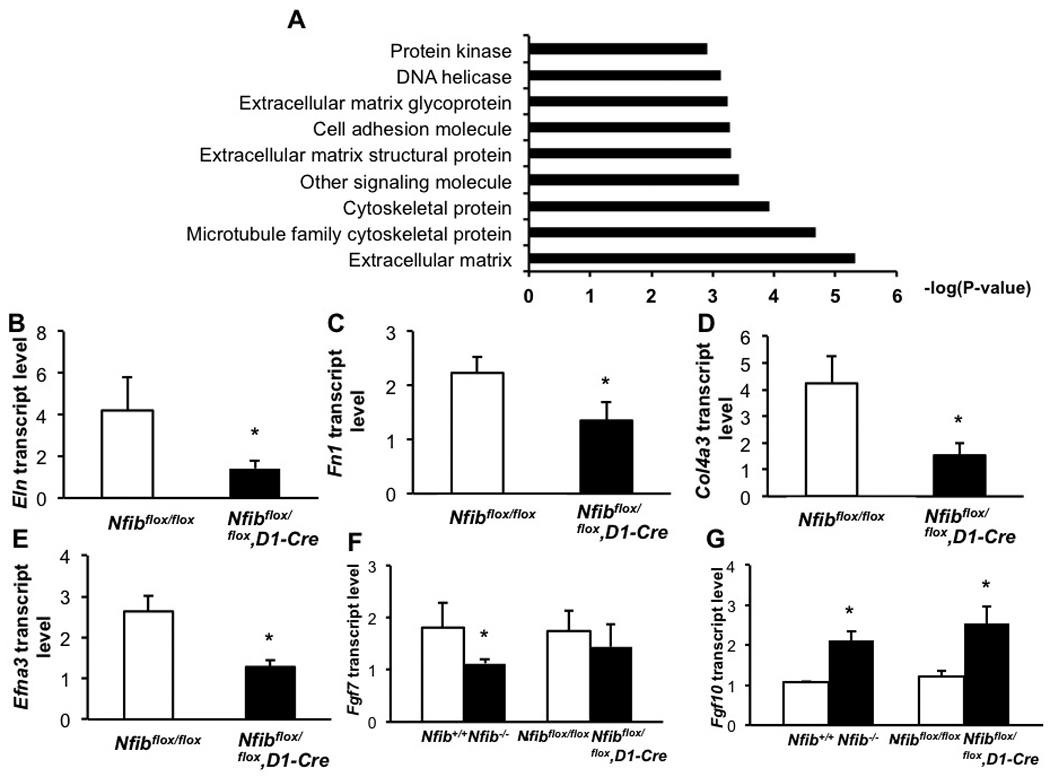
Microarray and RT-QPCR analysis show changes in the expression of genes involved in extracellular matrix, cell adhesion and FGF signaling pathway in Nfib−/− E18.5 lungs.
(A) Microarray studies show that loss of Nfib in mesenchyme influences the expression of genes associated with extracellular matrix and cell adhesion. Decreased expression of several genes from the microarray data were confirmed by RT-QPCR from lung RNA isolated from E18.5 Nfib mesenchyme-specific knockout and WT mice (B–E). In addition, the expression of Fgf7 and Fgf10 were measured using RT-QPCR from lung RNA isolated from E18.5 Nfib knockout, mesenchyme-specific knockout and WT mice (F–G). *P<0.05.
In addition, we examined the expression of Fgf7 and Fgf10 transcripts because both of these FGFs are expressed in lung mesenchyme and have been implicated in epithelial cell differentiation as part of a paracrine pathway (Min et al., 1998; Nyeng et al., 2008; Shannon et al., 1999). Interestingly, Fgf7 expression is significantly decreased in Nfib−/− lungs (fold change=−1.6, P=0.03) but essentially unchanged in Nfibflox/flox, D1-Cre lungs (fold change=−1.2, p=0.2) (Fig. 8F). Increased levels of Fgf10 transcripts were detected in both Nfib−/− and Nfibflox/flox, D1-Cre lungs (Fig. 8G), suggesting an excess of FGF-10 might attenuate epithelial cell differentiation and trigger cell proliferation, as was seen previously in Fgf10 overexpressing mice (Nyeng et al., 2008).
Finally, lipid and surfactant production are critical for lung function. Our microarray analysis also found decreased expression of several genes related to lipid and surfactant protein production (Table S2), suggesting that multiple pathways influencing epithelial cell function and differentiation are altered in Nfibflox/flox, D1-Cre lungs.
Elastin is an NFI-B target gene during lung development
We next asked whether any of the genes whose expression was affected by loss of Nfib might be direct NFI-B target genes. A NFI binding site between −401 and −415 in the human Elastin gene was characterized previously using in vitro DNA-binding assays and transient transfection analysis (Degterev and Foster, 1999). We examined the corresponding sequence of the mouse Eln promoter and identified 3 putative NFI binding sites located between −467bp to −344bp in a region highly homologous to the human ELN promoter (Fig. 9A). To test whether these sites are bound by NFI-B in vivo in E16.5 mouse lungs we performed ChIP analysis. Both standard PCR and QPCR indicate that NFI-B binding occurs in the region containing these three predicted NFI-B binding sites (Fig. 9B–C). In contrast, there is no enrichment in the region of the first intron of β2-microglobulin, the negative control region used in our ChIP assay (Fig. 9B–C). These data indicate that NFI-B binds to this region of the mouse Eln promoter at E16.5. To determine whether NFI-B could activate endogenous Eln expression we transfected MEFs with either a control GFP vector or a vector expressing NFI-B2 from the CMV promoter (Fig. 9D). The NFI-B2 isoform was used because it is the major spliced isoform of NFIB in lung (data not shown). Transient expression of NFI-B2 increased Eln expression by ∼4 fold in MEFs when compared to the control GFP vector. Taken together these data indicate that Nfib can regulate Eln expression, most likely through the conserved NFI binding sites in the promoter region. Since Eln is expressed exclusively in mesenchyme (Mariani et al., 1997; Pierce et al., 2006), we propose Eln as a mesenchyme-specific target gene of Nfib. We are currently investigating which other genes whose expression is affected by loss of Nfib may be direct targets of NFI-B.
Fig. 9.
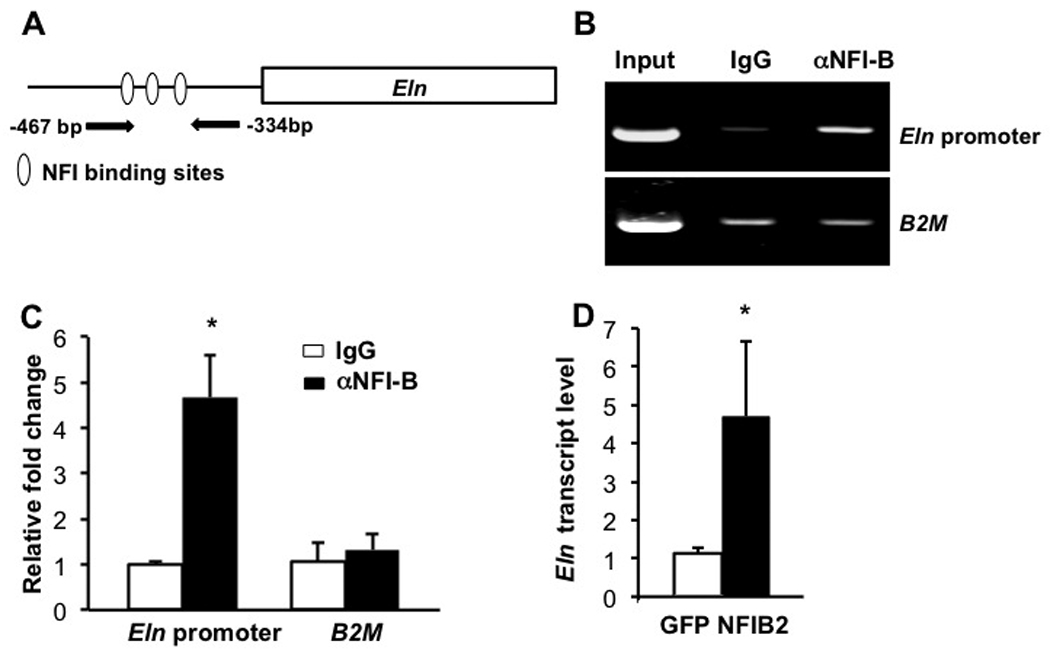
The promoter region of Eln is bound by NFI-B during lung development and NFI-B can activate endogenous Eln expression.
Three putative NFI binding sites were found in the mouse Eln promoter (A). ChIP-PCR (B) and ChIP-QPCR (C) were performed as described in methods and indicate that NFI-B in binds to the mouse Eln promoter in E16.5 mouse lungs. Transient expression of NFI-B in MEFs increases the expression of endogenous Eln expression (D). *P<0.05.
Discussion
Here we show a critical role for mesenchymal expression of NFI-B in regulating fetal lung maturation. We confirmed that NFI-B is expressed predominantly in lung mesenchyme at E14.5 and in both epithelial cells and mesenchyme at E18.5 (Fig. 1). Loss of Nfib results in lung hyperplasia and inhibition of Clara cell, ciliated cell, and distal epithelial cell differentiation (Figs. 2&3, Fig. S2). Further, the data from our conditional KO mice show that the loss of Nfib in only mesenchyme appears responsible for the majority of defects seen in Nfib−/− lungs, with the exception of the inhibition of Clara cell differentiation (Figs. 4–7, Fig. S4). Finally, the microarray analysis reveals that Nfib in mesenchyme may directly regulate the expression of genes related to the synthesis of extracellular matrix, cell adhesion and the FGF signaling pathway (Figs. 8&9). Taken together, these data provide new insights into the role of mesenchymal Nfib in the regulation of cell proliferation and maturation in the lung.
NFI-B regulates lung maturation and cell proliferation
Previous studies showed that loss of Nfib in all cells in the lung prevents normal lung maturation (Grunder et al., 2002; Steele-Perkins et al., 2005). Here, similar though not identical defects in lung maturation were found in Nfibflox/flox, D1-Cre lungs, indicating that Nfib expression specifically in mesenchyme is required for normal lung maturation. In contrast, CC10 expression was decreased in Nfib−/− lungs but unchanged in Nfibflox/flox, D1-Cre lungs, suggesting a possible direct effect of NFI-B on Clara cell differentiation. Further loss-of function and gain-of-function studies on Nfib in lung epithelium will be needed to address this possibility.
Cell proliferation and relatively low levels of apoptosis occur throughout embryonic lung maturation (Stiles et al., 2001). A fraction of lung mesenchymal cells undergo apoptosis during the pseudoglandular and saccular stages of lung development, and this process is more obvious around birth (Kresch et al., 1998; Scavo et al., 1998). In our Nfib-deficient lungs, there is no change in the number of cleaved Caspase 3 positive cells at E18.5 (Fig. S2A–C). In contrast, an increase in cell proliferation marker expression was observed in both germline Nfib-deficient and mesenchyme-specific Nfib deficient lungs (Figs. 2&6, Fig. S2&Fig. S4). These data strongly indicate that an increase in cell proliferation rather than a decrease of apoptosis is the cause of the increase in DNA content and lung size in Nfib-deficient mice. We observed increases in proliferation marker expression in both TTF+ and TTF− cells in germline Nfib-deficient lungs at E18.5 (Fig. 2). Interestingly, in the mesenchyme-specific Nfib-deficient lungs there was an increase in proliferation marker expression in both cell populations at E16.5, but no apparent increase in expression in mesenchymal cells at E18.5 (Fig. 6). The lack of increased proliferation marker expression in the TTF− cells at E18.5, despite the clear loss of Nfib in the TTF− cells, may indicate that Nfib in TTF+ lung epithelium can influence the proliferation of TTF− cells during lung maturation. Further studies where Nfib is deleted specifically in epithelial cells are needed to address this issue.
Prenatal glucocorticoid treatment has been a standard therapy to stimulate lung maturation in premature infants (Crowley, 1995; Liggins, 1968, 1969; Liggins and Howie, 1972). Glucocorticoid receptor (GR)−/− (Cole et al., 1995) and corticotrophin-releasing hormone (Crh)−/− (Muglia et al., 1999) mice display defective lungs with few or no saccules and an increase in cell proliferation, similar in many respects to the phenotype of Nfib-deficient lungs. While the expression levels of these two genes is not reduced in Nfib-deficient lungs (data not shown), it is possible that NFI-B may function downstream of GR signaling and/or may cooperate with GR to promote lung maturation. Indeed the transcription of several GR-regulated genes has been shown to be co-regulated both positively and negatively by NFI proteins (Chaudhry et al., 1999; Hebbar and Archer, 2007). Since microarray data from E18.5, GR−/− lungs has been published (Bird et al., 2007), it will be of interest to identify common lung maturation-related genes in the Nfib−/− and GR−/− microarray data which could relate to the common morphological changes seen in these mutant lungs
Mesenchymal NFI-B regulates epithelial cell differentiation
Defects of distal epithelial cell differentiation were found in both Nfib−/− and Nfibflox/flox, D1-Cre lungs, indicating NFI-B may regulate this process through paracrine signaling. Among the genes whose transcript levels are reduced in Nfibflox/flox, D1-Cre lungs is Fibronectin 1 (Fn1) Fn1 is necessary for epithelial cell differentiation in primary cultures of lung epithelial cells (Isakson et al., 2001; Olsen et al., 2005; Roman, 1997). It is possible that decreased mesenchymal expression of Fn1 could affect distal epithelial cell differentiation in our Nfib-deficient mice. Eln transcript expression was decreased ∼3 fold in Nfib-deficient lungs (fold change=−3.0, P=0.007). Elastin synthesis is required for breath and lung extension (Shifren and Mecham, 2006; Starcher, 2000) and Eln−/− mice die by P3.5 with lung alveolar defects (Wendel et al., 2000). In addition, mechanical stretch has been demonstrated to promote type II epithelial cell differentiation (Sanchez-Esteban et al., 2001; Wang et al., 2009). While Eln−/− mice show no lung maturation defects at E18.5 (Wendel et al., 2000), it is possible that the reduction in Eln expression, in conjunction with the decreased expression of Fn1 and other genes, could inhibit lung extension in Nfib-deficient mice and thus inhibit epithelial cell differentiation. Previous studies showed that the human ELN promoter contained an NFI binding site that was required for NFI-induced expression of the promoter in transient reporter assays (Degterev and Foster, 1999). Here we have demonstrated that NFI-B binds to the Eln promoter in mouse lungs in vivo at E16.5 and can activate endogenous Eln expression (Fig. 9). It will be important in future studies to determine how direct binding of NFI-B to the Eln promoter in vivo regulates Eln expression and whether decreased Eln expression in Nfib−/− lungs directly or indirectly influences lung maturation. In addition, it will be important to assess gene expression profiles at multiple stages of lung development to determine how many of the changes in gene expression seen in the E18.5 Nfibflox/flox, D1-Cre lungs reflect direct targets of Nfib that could mediate the lung-maturation phenotype, versus secondary changes due to the altered differentiation state of the mutant lungs.
While the initial microarray data showed no significant difference in the levels of Fgf10 transcripts (fold change=1.2, P=0.13), subsequent QPCR analysis showed that Fgf10 expression was consistently increased in both Nfib−/− (fold change=2.0, P=0.0007) and Nfibflox/flox, D1-Cre lungs (fold change=2.1, P=0.003). Such discrepancies indicate the importance of careful testing of individual candidate genes from microarray analysis by QPCR. The overexpression of Fgf10 during lung development results in some changes similar to those seen in Nfib-deficient lungs, including lung hyperplasia and inhibition of distal epithelial cell differentiation (Clark et al., 2001; Nyeng et al., 2008). However, the defects in branching morphogenesis, increase in pro-SPC and TTF-1 expression, and the formation of a layer of smooth muscle cells around the lung perimeter seen in lungs in which Fgf10 is highly overexpressed were not observed in Nfib-deficient lungs. This may be due to differences in the timing, degree and spatial distribution of Fgf10 overexpression in the two systems. In future studies, it will be useful to determine whether reduction of Fgf10 expression can partially rescue the defects of Nfib-deficient lungs by crossing Fgf10−/+ mice with Nfib−/+ mice.
Previous studies on lung maturation focused primarily on genes expressed in lung epithelial cells, such as Klf5 (Wan et al., 2008), Tgfb1 (Zhou et al., 1996), Cebpa (Berg et al., 2006; Martis et al., 2006), Vegf (Zeng et al., 1998), T1a (Millien et al., 2006), GR (Manwani et al., 2009), Carm1 (O’Brien et al., 2010), Pparγ(Simon et al., 2006), Alk5 (Xing et al., 2010) and Pdgfa (Li and Hoyle, 2001). There is far less data on how gene expression in mesenchyme regulates lung maturation. Here we found that Nfib in lung mesenchyme is necessary for lung maturation and epithelial cell differentiation. These studies suggest that Nfib in lung mesenchyme regulates cell proliferation and distal epithelial cell differentiation through regulation of the expression of genes related to extracellular matrix deposition, cell adhesion and/or FGF signaling. Future studies will focus on defining in more detail the molecular mechanisms by which NFI-B in mesenchyme affects epithelial cell differentiation and both epithelial and mesenchymal cell proliferation during lung maturation.
Supplementary Material
Fig. S1. Generating a conditional knockout allele of Nfib. (A) The wild-type Nfib allele (Wild Type Allele, WT) was modified using a targeting vector with a 4.4 kb 5' homology arm that contained all of exon 2 of Nfib and 300 bp of intron 2, with a loxP site inserted 363 bp 5' to the start of exon 2. The 5' arm was followed by an FRT-flanked PGK-neo expression cassette in the opposite transcriptional orientation (NEO), a 3' loxP site, the contiguous 3.6 kb 3' homology arm and a PGK-diphtheria toxin A chain cassette (DT) in the opposite transcriptional orientation (Targeting Vector). Cre-mediated recombination deletes all of exon 2, 363 bp of intron 1 and 300 bp of intron 2 (After Cre, KO). The correctly targeted floxed allele (Floxed Allele, FA) was detected by PCR using primers overlapping the LoxP sites and within flanking regions outside of the targeting vector (5' PCR FA and 3' PCR FA). WT, FA, and KO alleles were followed using PCR primers flanking the 5'-most loxP site (WT-PCR, FA-PCR and KO-PCR, respectively). (B) PCR analysis of WT, FA and KO alleles in tail DNA from mice. Agarose gel of PCR analysis of tail DNAs from mice containing WT (+/+) lane 7, heterozygous KO (del/+) lanes 1 and 2, and heterozygous conditional floxed alleles (flox/+) lanes 2, 4, 5 and 6. The WT, FA and KO alleles yield PCR products of 219, 400, and 498bp, respectively.
Fig. S2. Loss of Nfib promotes increased cell proliferation during lung development. Paraffin sections of E18.5 lungs from Nfib+/+ and Nfib−/− embryos were stained for several cell proliferation markers: PCNA (A and B), BrdU (D, E), Ki67 (G, H) or phospho-histone H3 (PHH3) (J, K), and counterstained with hematoxylin or TOPRO-3. Arrows denote positive cells in main panels and insets (5X magnification). Immunostaining showed increased expression of 4 different cell proliferation markers in Nfib−/− lungs compared with Nfib+/+ lungs. Quantification of cell proliferation marker positive cells/total cell numbers of Nfib+/+ and Nfib−/− lungs (C, F, I, L) are shown.*P<0.05; Scale bars, 50 µm.
Fig. S3. Loss of Nfib does not affect cell apoptosis during lung development. Lung sections from Nfib+/+ (A) and Nfib−/− mice (B) were stained with anti-cleaved Caspase 3 antibodies, an apoptotic cell marker, to determine whether loss of Nfib affect cell apoptosis during lung development. Arrows denote positive cells in main panels and insets (5X magnification). (C) Quantification of cleaved Caspase 3 staining.
Fig. S4. Loss of Nfib in mesenchyme increases cell proliferation. Paraffin sections of E18.5 lungs were stained for cell proliferation marker: phospho-histone H3 (PHH3) (A, B) and counterstained with TOPRO-3. There are more PHH3 positive cells in Nfibflox/flox, D1-Cre lungs than in Nfibflox/flox lungs at E18.5. E18.5 and E16.5 lung sections were stained with αKi67 antibody. Ki67 positive cells are increased in Nfibflox/flox, D1-Cre lungs at E16.5 and E18.5 (D, E, F, G). Arrows denote region of magnification in insets (5X magnification). Data were quantified in C and H. *P<0.05; Scale bars, 50 µm.
Fig. S5. Tnc expression was similar in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs. The expression of Tnc in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs at E18.5 was measured by RT-QPCR.
Acknowledgements
This work was supported in part by NIH/NHLBI grant HL080624 to RMG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akeson AL, Greenberg JM, Cameron JE, Thompson FY, Brooks SK, Wiginton D, Whitsett JA. Temporal and spatial regulation of VEGF-A controls vascular patterning in the embryonic lung. Dev Biol. 2003;264:443–455. doi: 10.1016/j.ydbio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Bachurski CJ, Yang GH, Currier TA, Gronostajski RM, Hong D. Nuclear factor I/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. Mol Cell Biol. 2003;23:9014–9024. doi: 10.1128/MCB.23.24.9014-9024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Didon L, Nord M. Ectopic expression of C/EBPalpha in the lung epithelium disrupts late lung development. Am J Physiol Lung Cell Mol Physiol. 2006;291:L683–L693. doi: 10.1152/ajplung.00497.2005. [DOI] [PubMed] [Google Scholar]
- Bird AD, Tan KH, Olsson PF, Zieba M, Flecknoe SJ, Liddicoat DR, Mollard R, Hooper SB, Cole TJ. Identification of glucocorticoid-regulated genes that control cell proliferation during murine respiratory development. J Physiol. 2007;585:187–201. doi: 10.1113/jphysiol.2007.136796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CE, Piper M, Plachez C, Yeh YT, Baizer JS, Osinski JM, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor Nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry AZ, Vitullo AD, Gronostajski RM. Nuclear factor I-mediated repression of the mouse mammary tumor virus promoter is abrogated by the coactivators p300/CBP and SRC-1. J Biol Chem. 1999;274:7072–7081. doi: 10.1074/jbc.274.11.7072. [DOI] [PubMed] [Google Scholar]
- Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT, Whitsett JA. FGF-10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol Lung Cell Mol Physiol. 2001;280:L705–L715. doi: 10.1152/ajplung.2001.280.4.L705. [DOI] [PubMed] [Google Scholar]
- Coalson JJ, Winter VT, Siler-Khodr T, Yoder BA. Neonatal chronic lung disease in extremely immature baboons. Am J Respir Crit Care Med. 1999;160:1333–1346. doi: 10.1164/ajrccm.160.4.9810071. [DOI] [PubMed] [Google Scholar]
- Cohen ED, Ihida-Stansbury K, Lu MM, Panettieri RA, Jones PL, Morrisey EE. Wnt signaling regulates smooth muscle precursor development in the mouse lung via a tenascin C/PDGFR pathway. J Clin Invest. 2009;119:2538–2549. doi: 10.1172/JCI38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- Crowley PA. Antenatal corticosteroid therapy: a meta-analysis of the randomized trials, 1972 to 1994. Am J Obstet Gynecol. 1995;173:322–335. doi: 10.1016/0002-9378(95)90222-8. [DOI] [PubMed] [Google Scholar]
- de Jong RN, van der Vliet PC. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene. 1999;236:1–12. doi: 10.1016/s0378-1119(99)00249-8. [DOI] [PubMed] [Google Scholar]
- De Langhe SP, Carraro G, Tefft D, Li C, Xu X, Chai Y, Minoo P, Hajihosseini MK, Drouin J, Kaartinen V, Bellusci S. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PLoS One. 2008;3:e1516. doi: 10.1371/journal.pone.0001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Foster JA. The role of NF-1 factors in regulation of elastin gene transcription. Matrix Biol. 1999;18:295–307. doi: 10.1016/s0945-053x(99)00020-7. [DOI] [PubMed] [Google Scholar]
- Deimling J, Thompson K, Tseu I, Wang J, Keijzer R, Tanswell AK, Post M. Mesenchymal maintenance of distal epithelial cell phenotype during late fetal lung development. Am J Physiol Lung Cell Mol Physiol. 2007;292:L725–L741. doi: 10.1152/ajplung.00221.2006. [DOI] [PubMed] [Google Scholar]
- Demayo F, Minoo P, Plopper CG, Schuger L, Shannon J, Torday JS. Mesenchymal-epithelial interactions in lung development and repair: are modeling and remodeling the same process? Am J Physiol Lung Cell Mol Physiol. 2002;283:L510–L517. doi: 10.1152/ajplung.00144.2002. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM. Analysis of nuclear factor I binding to DNA using degenerate oligonucleotides. Nucleic Acids Res. 1986;14:9117–9132. doi: 10.1093/nar/14.22.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- Grunder A, Ebel TT, Mallo M, Schwarzkopf G, Shimizu T, Sippel AE, Schrewe H. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech Dev. 2002;112:69–77. doi: 10.1016/s0925-4773(01)00640-2. [DOI] [PubMed] [Google Scholar]
- Hebbar PB, Archer TK. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem. 2007;282:8284–8291. doi: 10.1074/jbc.M610554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom M, Kalen M, Lindahl P, Abramsson A, Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakson BE, Lubman RL, Seedorf GJ, Boitano S. Modulation of pulmonary alveolar type II cell phenotype and communication by extracellular matrix and KGF. American Journal of Physiology: Cell Physiology. 2001;281:C1291–C1299. doi: 10.1152/ajpcell.2001.281.4.C1291. [DOI] [PubMed] [Google Scholar]
- Jobe AH. Antenatal associations with lung maturation and infection. J Perinatol. 2005;25 Suppl 2:S31–S35. doi: 10.1038/sj.jp.7211317. [DOI] [PubMed] [Google Scholar]
- Kresch MJ, Christian C, Wu F, Hussain N. Ontogeny of apoptosis during lung development. Pediatr Res. 1998;43:426–431. doi: 10.1203/00006450-199803000-00020. [DOI] [PubMed] [Google Scholar]
- Li C, Xiao J, Hormi K, Borok Z, Minoo P. Wnt5a participates in distal lung morphogenesis. Dev Biol. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- Li J, Hoyle GW. Overexpression of PDGF-A in the lung epithelium of transgenic mice produces a lethal phenotype associated with hyperplasia of mesenchymal cells. Dev Biol. 2001;239:338–349. doi: 10.1006/dbio.2001.0441. [DOI] [PubMed] [Google Scholar]
- Liggins GC. Premature parturition after infusion of corticotrophin or cortisol into foetal lambs. J Endocrinol. 1968;42:323–329. doi: 10.1677/joe.0.0420323. [DOI] [PubMed] [Google Scholar]
- Liggins GC. Premature delivery of foetal lambs infused with glucocorticoids. J Endocrinol. 1969;45:515–523. doi: 10.1677/joe.0.0450515. [DOI] [PubMed] [Google Scholar]
- Liggins GC, Howie RN. A controlled trial of antepartum glucocorticoid treatment for prevention of the respiratory distress syndrome in premature infants. Pediatrics. 1972;50:515–525. [PubMed] [Google Scholar]
- Manwani N, Gagnon S, Post M, Joza S, Muglia L, Cornejo S, Kaplan F, Sweezey NB. Reduced Viability of Mice with Lung Epithelial-Specific Knockout of Glucocorticoid Receptor. Am J Respir Cell Mol Biol. 2009 doi: 10.1165/rcmb.2009-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani TJ, Sandefur S, Pierce RA. Elastin in lung development. Exp Lung Res. 1997;23:131–145. doi: 10.3109/01902149709074026. [DOI] [PubMed] [Google Scholar]
- Martis PC, Whitsett JA, Xu Y, Perl AK, Wan H, Ikegami M. C/EBPalpha is required for lung maturation at birth. Development. 2006;133:1155–1164. doi: 10.1242/dev.02273. [DOI] [PubMed] [Google Scholar]
- Meisterernst M, Gander I, Rogge L, Winnacker EL. A quantitative analysis of nuclear factor I/DNA interactions. Nucleic Acids Res. 1988;16:4419–4435. doi: 10.1093/nar/16.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millien G, Spira A, Hinds A, Wang J, Williams MC, Ramirez MI. Alterations in gene expression in T1 alpha null lung: a model of deficient alveolar sac development. BMC Dev Biol. 2006;6:35. doi: 10.1186/1471-213X-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia LJ, Bae DS, Brown TT, Vogt SK, Alvarez JG, Sunday ME, Majzoub JA. Proliferation and differentiation defects during lung development in corticotropin-releasing hormone-deficient mice. Am J Respir Cell Mol Biol. 1999;20:181–188. doi: 10.1165/ajrcmb.20.2.3381. [DOI] [PubMed] [Google Scholar]
- Nagata K, Guggenheimer RA, Enomoto T, Lichy JH, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Guggenheimer RA, Hurwitz J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1983;80:6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyeng P, Norgaard GA, Kobberup S, Jensen J. FGF10 maintains distal lung bud epithelium and excessive signaling leads to progenitor state arrest, distalization, and goblet cell metaplasia. BMC Dev Biol. 2008;8:2. doi: 10.1186/1471-213X-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KB, Alberich-Jorda M, Yadav N, Kocher O, Diruscio A, Ebralidze A, Levantini E, Sng NJ, Bhasin M, Caron T, Kim D, Steidl U, Huang G, Halmos B, Rodig SJ, Bedford MT, Tenen DG, Kobayashi S. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010;137:2147–2156. doi: 10.1242/dev.037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CO, Isakson BE, Seedorf GJ, Lubman RL, Boitano S. Extracellular matrix-driven alveolar epithelial cell differentiation in vitro. Exp Lung Res. 2005;31:461–482. doi: 10.1080/01902140590918830. [DOI] [PubMed] [Google Scholar]
- Pierce RA, Moore CH, Arikan MC. Positive transcriptional regulatory element located within exon 1 of elastin gene. Am J Physiol Lung Cell Mol Physiol. 2006;291:L391–L399. doi: 10.1152/ajplung.00441.2004. [DOI] [PubMed] [Google Scholar]
- Plasari G, Calabrese A, Dusserre Y, Gronostajski RM, McNair A, Michalik L, Mermod N. Nuclear factor I-C links platelet-derived growth factor and transforming growth factor beta1 signaling to skin wound healing progression. Mol Cell Biol. 2009;29:6006–6017. doi: 10.1128/MCB.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman J. Fibronectin and fibronectin receptors in lung development. Exp Lung Res. 1997;23:147–159. doi: 10.3109/01902149709074027. [DOI] [PubMed] [Google Scholar]
- Sanchez-Esteban J, Cicchiello LA, Wang Y, Tsai SW, Williams LK, Torday JS, Rubin LP. Mechanical stretch promotes alveolar epithelial type II cell differentiation. J Appl Physiol. 2001;91:589–595. doi: 10.1152/jappl.2001.91.2.589. [DOI] [PubMed] [Google Scholar]
- Scavo LM, Ertsey R, Chapin CJ, Allen L, Kitterman JA. Apoptosis in the development of rat and human fetal lungs. Am J Respir Cell Mol Biol. 1998;18:21–31. doi: 10.1165/ajrcmb.18.1.2744. [DOI] [PubMed] [Google Scholar]
- Shannon JM, Gebb SA, Nielsen LD. Induction of alveolar type II cell differentiation in embryonic tracheal epithelium in mesenchyme-free culture. Development. 1999;126:1675–1688. doi: 10.1242/dev.126.8.1675. [DOI] [PubMed] [Google Scholar]
- Shifren A, Mecham RP. The stumbling block in lung repair of emphysema: elastic fiber assembly. Proc Am Thorac Soc. 2006;3:428–433. doi: 10.1513/pats.200601-009AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Jiang YQ, Lu MM, Morrisey EE. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129:4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- Simon DM, Arikan MC, Srisuma S, Bhattacharya S, Tsai LW, Ingenito EP, Gonzalez F, Shapiro SD, Mariani TJ. Epithelial cell PPAR[gamma] contributes to normal lung maturation. FASEB J. 2006;20:1507–1509. doi: 10.1096/fj.05-5410fje. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Starcher BC. Lung elastin and matrix. Chest. 2000;117:229S–234S. doi: 10.1378/chest.117.5_suppl_1.229s-a. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles AD, Chrysis D, Jarvis HW, Brighton B, Moats-Staats BM. Programmed cell death in normal fetal rat lung development. Exp Lung Res. 2001;27:569–587. doi: 10.1080/019021401753181836. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Tokumine J, Teruya K, Oshiro T. Alveolar epithelial cells: differentiation and lung injury. Respirology. 2006;11 Suppl:S28–S31. doi: 10.1111/j.1440-1843.2006.00804.x. [DOI] [PubMed] [Google Scholar]
- Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Maciejewski BS, Soto-Reyes D, Lee HS, Warburton D, Sanchez-Esteban J. Mechanical stretch promotes fetal type II epithelial cell differentiation via shedding of HB-EGF and TGF-alpha. J Physiol. 2009;587:1739–1753. doi: 10.1113/jphysiol.2008.163899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel DP, Taylor DG, Albertine KH, Keating MT, Li DY. Impaired distal airway development in mice lacking elastin. Am J Respir Cell Mol Biol. 2000;23:320–326. doi: 10.1165/ajrcmb.23.3.3906. [DOI] [PubMed] [Google Scholar]
- White AC, Lavine KJ, Ornitz DM. FGF9 and SHH regulate mesenchymal Vegfa expression and development of the pulmonary capillary network. Development. 2007;134:3743–3752. doi: 10.1242/dev.004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- Xing Y, Li C, Li A, Sridurongrit S, Tiozzo C, Bellusci S, Borok Z, Kaartinen V, Minoo P. Signaling via Alk5 controls the ontogeny of lung Clara cells. Development. 2010;137:825–833. doi: 10.1242/dev.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM. An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol. 2008;319:426–436. doi: 10.1016/j.ydbio.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Wert SE, Federici R, Peters KG, Whitsett JA. VEGF enhances pulmonary vasculogenesis and disrupts lung morphogenesis in vivo. Dev Dyn. 1998;211:215–227. doi: 10.1002/(SICI)1097-0177(199803)211:3<215::AID-AJA3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Zhou L, Dey CR, Wert SE, Whitsett JA. Arrested lung morphogenesis in transgenic mice bearing an SP-C-TGF-beta 1 chimeric gene. Dev Biol. 1996;175:227–238. doi: 10.1006/dbio.1996.0110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Generating a conditional knockout allele of Nfib. (A) The wild-type Nfib allele (Wild Type Allele, WT) was modified using a targeting vector with a 4.4 kb 5' homology arm that contained all of exon 2 of Nfib and 300 bp of intron 2, with a loxP site inserted 363 bp 5' to the start of exon 2. The 5' arm was followed by an FRT-flanked PGK-neo expression cassette in the opposite transcriptional orientation (NEO), a 3' loxP site, the contiguous 3.6 kb 3' homology arm and a PGK-diphtheria toxin A chain cassette (DT) in the opposite transcriptional orientation (Targeting Vector). Cre-mediated recombination deletes all of exon 2, 363 bp of intron 1 and 300 bp of intron 2 (After Cre, KO). The correctly targeted floxed allele (Floxed Allele, FA) was detected by PCR using primers overlapping the LoxP sites and within flanking regions outside of the targeting vector (5' PCR FA and 3' PCR FA). WT, FA, and KO alleles were followed using PCR primers flanking the 5'-most loxP site (WT-PCR, FA-PCR and KO-PCR, respectively). (B) PCR analysis of WT, FA and KO alleles in tail DNA from mice. Agarose gel of PCR analysis of tail DNAs from mice containing WT (+/+) lane 7, heterozygous KO (del/+) lanes 1 and 2, and heterozygous conditional floxed alleles (flox/+) lanes 2, 4, 5 and 6. The WT, FA and KO alleles yield PCR products of 219, 400, and 498bp, respectively.
Fig. S2. Loss of Nfib promotes increased cell proliferation during lung development. Paraffin sections of E18.5 lungs from Nfib+/+ and Nfib−/− embryos were stained for several cell proliferation markers: PCNA (A and B), BrdU (D, E), Ki67 (G, H) or phospho-histone H3 (PHH3) (J, K), and counterstained with hematoxylin or TOPRO-3. Arrows denote positive cells in main panels and insets (5X magnification). Immunostaining showed increased expression of 4 different cell proliferation markers in Nfib−/− lungs compared with Nfib+/+ lungs. Quantification of cell proliferation marker positive cells/total cell numbers of Nfib+/+ and Nfib−/− lungs (C, F, I, L) are shown.*P<0.05; Scale bars, 50 µm.
Fig. S3. Loss of Nfib does not affect cell apoptosis during lung development. Lung sections from Nfib+/+ (A) and Nfib−/− mice (B) were stained with anti-cleaved Caspase 3 antibodies, an apoptotic cell marker, to determine whether loss of Nfib affect cell apoptosis during lung development. Arrows denote positive cells in main panels and insets (5X magnification). (C) Quantification of cleaved Caspase 3 staining.
Fig. S4. Loss of Nfib in mesenchyme increases cell proliferation. Paraffin sections of E18.5 lungs were stained for cell proliferation marker: phospho-histone H3 (PHH3) (A, B) and counterstained with TOPRO-3. There are more PHH3 positive cells in Nfibflox/flox, D1-Cre lungs than in Nfibflox/flox lungs at E18.5. E18.5 and E16.5 lung sections were stained with αKi67 antibody. Ki67 positive cells are increased in Nfibflox/flox, D1-Cre lungs at E16.5 and E18.5 (D, E, F, G). Arrows denote region of magnification in insets (5X magnification). Data were quantified in C and H. *P<0.05; Scale bars, 50 µm.
Fig. S5. Tnc expression was similar in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs. The expression of Tnc in Nfibflox/flox and Nfibflox/flox, D1-Cre lungs at E18.5 was measured by RT-QPCR.


