Abstract
Toll-like receptor (TLR)-7 agonists show prominent Th1-biased immunostimulatory activities. A TLR7-active N1-(4-aminomethyl)benzyl substituted imidazoquinoline 1 served as a convenient precursor for the syntheses of isothiocyanate and maleimide derivatives for covalent attachment to free amine and thiol groups of peptides and proteins. 1 was also amenable to direct reductive amination with maltoheptaose without significant loss of activity. Covalent conjugation of the isothiocyanate derivative 2 to α-lactalbumin could be achieved under mild, non-denaturing conditions, in a controlled manner and with full preservation of antigenicity. The self-adjuvanting α-lactalbumin construct induced robust, high-affinity immunoglobulin titers in murine models. The premise of covalently decorating protein antigens with adjuvants offers the possibility of drastically reducing systemic exposure of the adjuvant, and yet eliciting strong, Th1-biased immune responses.
Keywords: Imidazoquinoline, Toll-like receptor, TLR7, Self-adjuvanting vaccine, ELISA, Adjuvant
Toll-like receptors (TLRs) are pattern recognition receptors that recognize specific molecular patterns present in molecules that are broadly shared by pathogens, but are structurally distinct from host molecules.1 There are 10 TLRs in the human genome.2 The ligands for these receptors are highly conserved microbial molecules such as lipopolysaccharides (LPS) (recognized by TLR4), lipopeptides (TLR2 in combination with TLR1 or TLR6), flagellin (TLR5), single stranded RNA (TLR7 and TLR8), double stranded RNA (TLR3), CpG motif-containing DNA (recognized by TLR9), and profilin present on uropathogenic bacteria (TLR 11).3 The activation of TLRs by their cognate ligands leads to activation of innate immune effector mechanisms, including the production of pro-inflammatory cytokines, up-regulation of MHC molecules and co-stimulatory signals in antigen-presenting cells, resulting in amplification of specific adaptive immune responses involving both T- and B-cell effector functions.4–6 Thus, TLR stimuli serve to link innate and adaptive immunity4 and can therefore be exploited as powerful adjuvants in eliciting both primary and anamnestic immune responses.
One aspect of our work in the area of evaluating TLR agonists as vaccine adjuvants7–9 focuses on developing self-adjuvanting vaccine constructs, i.e., antigen covalently coupled to a suitable adjuvant. The premise of covalently decorating protein antigens with potential adjuvants offers the possibility of drastically reducing systemic exposure of the adjuvant, and yet maintaining relatively high local concentrations at the site of vaccination.10 Most self-adjuvanting vaccine constructs to date have utilized TLR-2 agonistic 2,3-bis-(palmitoyloxy)propyl-cysteinyl peptides as the adjuvant.11–16 The conjugation of the poorly soluble lipopeptide adjuvant to antigen has limited this approach to peptide12–15;17 or glycopeptide16 antigens, since native proteins are often irrevocably denatured under the coupling conditions employed, with potential loss of key epitopes. These limitations have recently been addressed by appending to the lipopeptide a long, water-solubilizing poly-lysine or polyethylene glycol moiety, and terminating in a free thiol.18 However, in addition to the potential problem of oxidation of lipopeptide thiol to the disulfide, free exposed thiols in proteins are rare.19 Furthermore, TLR2 ligation has been associated with Th2 and Th17 responses20;21 which may, in many instances, be undesirable.
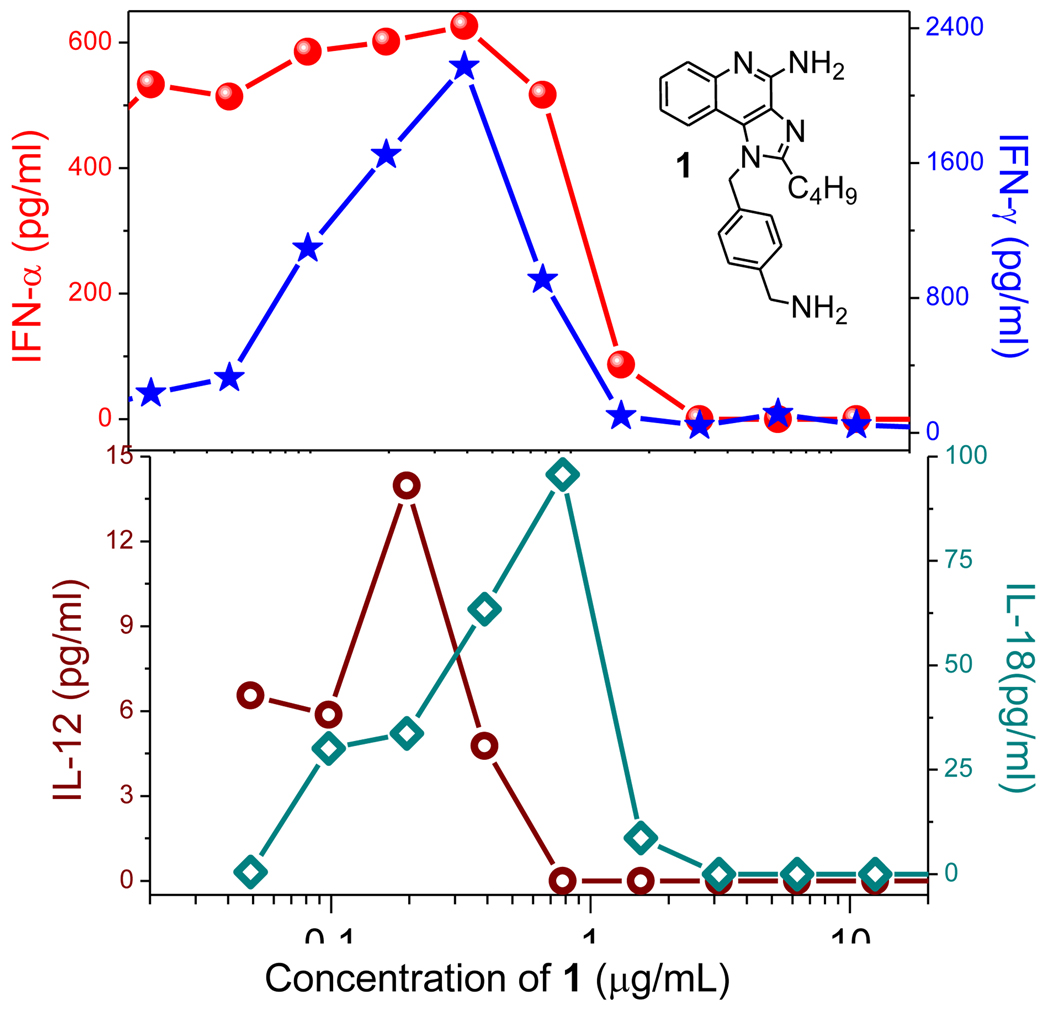
We have surveyed TLR agonists in a series of hierarchical assays including primary TLR-reporter gene assays, secondary indices of immune activation such as cytokine induction and activation of lymphocytic subsets in whole human blood, and tertiary screens characterizing transcriptomal activation patterns with a view to identifying optimal immunostimulatory chemotypes.7 Of all the innate immune stimuli examined, we found that TLR7 agonists were extraordinarily potent, stimulating virtually all subsets of lymphocytes, and yet without inducing dominant proinflammatory cytokine responses.7 Desirous of specifically identifying chemotypes with strong Th1-biased immunostimulatory signatures, we implemented additional screens examining the induction of Type I22–24 and Type II25;26 interferons (IFN-α/β and IFN-γ, respectively), Interleukin-12 (IL-12),27;28 and Interleukin-18 (IL-18) using human PBMCs,29–31 all of which are strongly associated with dominant Th1 outcomes. These experiments were most instructive in that they enabled us to determine that of all of the diverse chemotypes of our rapidly expanding libraries of TLR agonists,8;9;32;33 an N1-(4-aminomethyl)benzyl substituted imidazoquinoline 1 displayed a prominent Th1 bias (Fig. 1).
Figure 1.
Induction of IFN-α, IFN-γ (top) and IL-12 and IL-18 (bottom) by 1 in human PBMCs. IFN and cytokine levels were quantified by ELISA. Results of a representative experiment are shown.
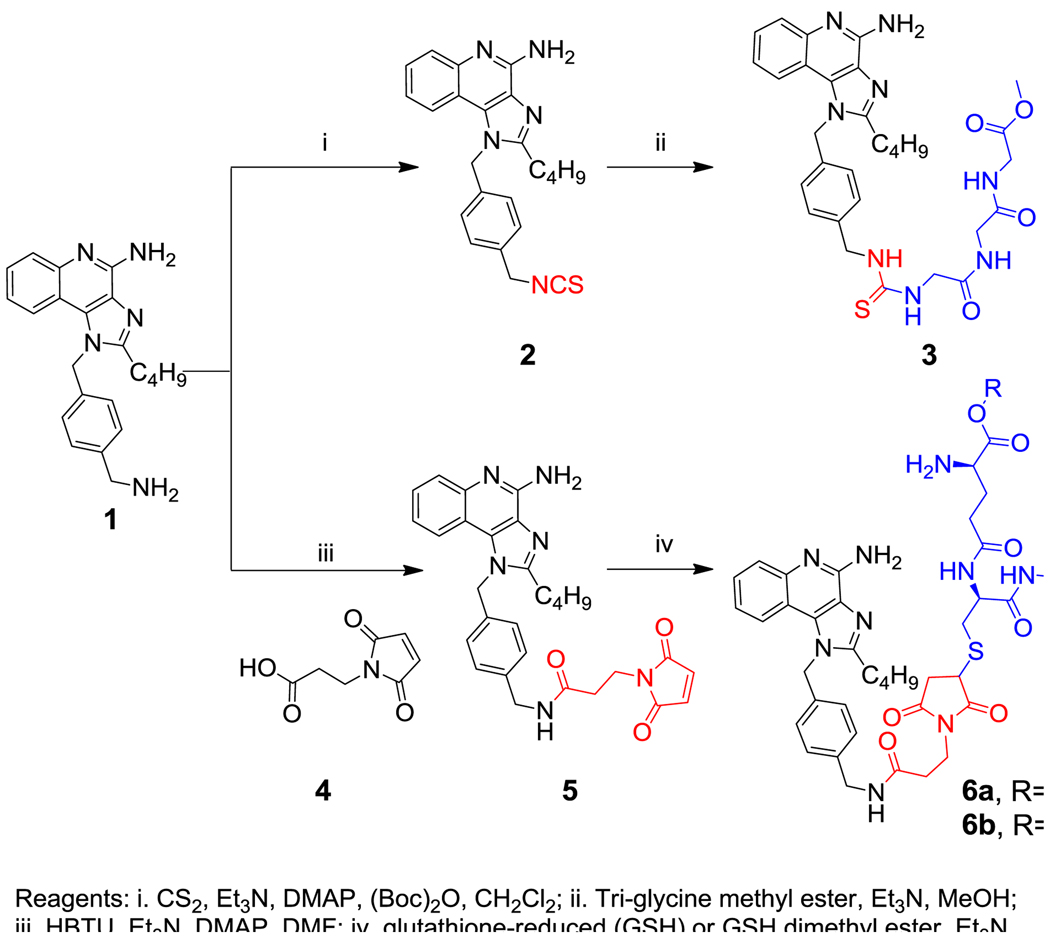
Because the aqueous solubility of 1 and several of its congeners were excellent, we elected to evaluate direct covalent coupling to free amines and thiols on protein antigens via the introduction of conventional isothiocyanate and maleimide electrophilic handles on the imidazoquinoline scaffold. Our earlier attempts at incorporating the electrophilic isothiocyanate functionality on the C2-sidechain for covalent adduction to peptides and proteins had met with failure in that such adducts were bereft of TLR7-agonistic activity,32 and it occurred to us that not only could the free primary amine on the N1 substituent of 1 be converted directly to the corresponding isothiocyanate (2, Scheme 1), but also an additional, thiol-specific maleimide electrophile could be attached via a spacer group (5, Scheme 1). The isothiocyanate derivative 2 reacted well with a tri-glycine methyl ester model peptide, yielding the adduct 3 (Scheme 1), which was purified to homogeneity, and found to be active (Fig. 2). Facile adduction of the maleimide derivative 5 with glutathione (reduced) also afforded the 6a in near-quantitative yields.
Scheme 1.
Syntheses of isothiocyanate and maleimide derivatives of 1, and their corresponding model peptide conjugates.
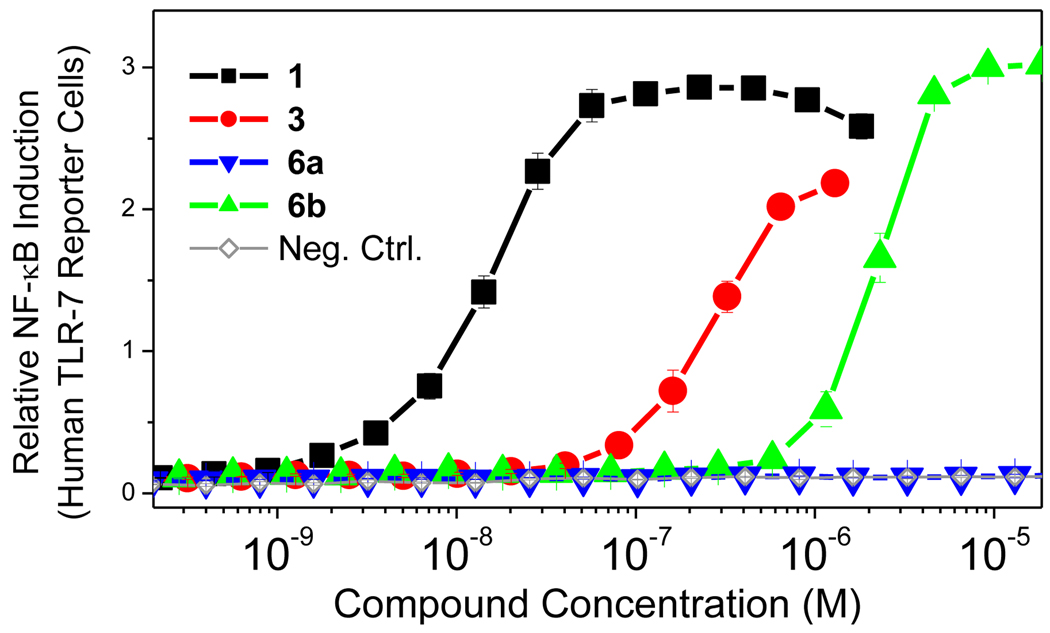
Figure 2.
TLR7-agonistic activities of imidazoquinoline analogues in a human TLR7-specific reporter gene assay.
The imidazoquinolines themselves are small, non-polar, and basic, and therefore gain access to the endolysosomal compartment in which TLR7 is predominantly sequestered. The human embryonic kidney reporter cell lines stably transfected with TLR7 (and reporter secreted alkaline phosphatase genes) that we employ in our primary screen are not professional phagocytic cells, and we were concerned if the trans-membrane permeability of the bulky, dianionic adduct 6a would be sufficient to trigger activation, and we therefore also obtained the conjugate of 5 with the dimethyl ester of reduced glutathione (6b). As could be predicted, the adducts 3 and 6b retained activity (EC50: 269 nM and 2.2 µM), while 6a was completely inactive (Fig. 2), indicating that trans-cellular transport of the polar adduct with two net negative charges was insufficient.
We elected to work with bovine α-lactalbumin as a model antigen for self-adjuvanting vaccine constructs not only because it lent itself eminently well to rigorous characterization by electrospray ionization mass spectrometry (ESI-MS) methods (Fig. 3), but also because it is being evaluated as a potential antigen for breast cancer vaccines.34
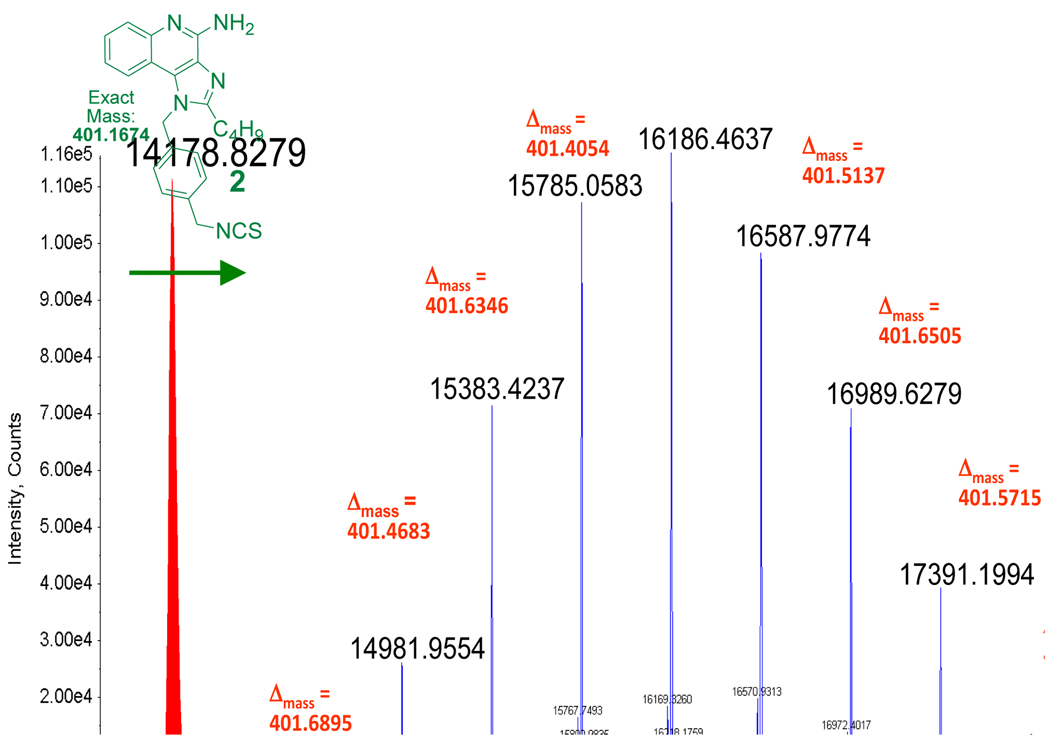
Figure 3.
Deconvoluted positive-mode ESI-MS spectra of native bovine α-lactalbumin (left) showing a mass of 14178.83 Da, and α-lactalbumin reacted with 5 eq. of 2, resulting in a stochastic coupling of the adjuvant with the centroid of the mass distribution corresponding to exactly 5 units of imidazoquinoline per protein molecule.
We reacted 5 equivalents of 2 with bovine α-lactalbumin in isotonic aqueous buffer at pH 8.5. Direct LC-ESI-MS evidence was obtained for covalent adduction of 2 with the protein, indicating a remarkably beautiful and precise Gaussian distribution of adducted species, with the preponderant conjugate corresponding to a 1:5 molar ratio of protein:2 (Fig. 3).
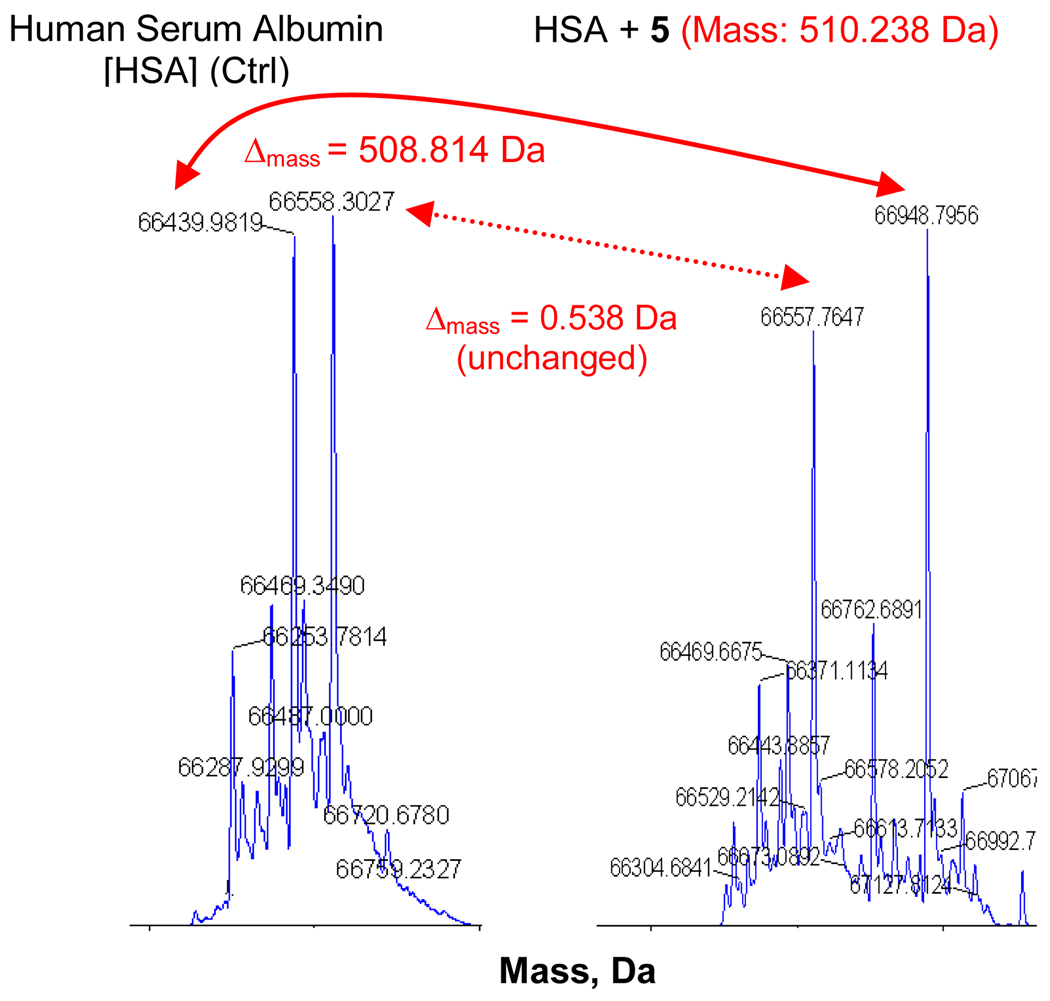
Encouraged by these results, we also attempted to conjugate 5 with human serum albumin (HSA), a 66 kDa protein with a single free thiol. We elected to use clinical grade (meant for human parenteral use, formulated with amino acids) HSA rather than the purer, ‘essentially fatty acid free’ protein available commercially. Furthermore, HSA is a carrier protein which binds promiscuously to a vast range of ligands including heavy metals, bilirubin, fatty acids, etc. We anticipated for these reasons, that deconvoluted direct electrospray-time-of-flight mass (ESI-TOF) spectra would be polydisperse and microheterogeneous. In the HSA-alone control sample, we found two major peaks corresponding to 66439.9819 and 66558.3027 Da (Fig. 4). Reaction of HSA with 5 produced a shift in one of the peaks with a Δmass of 508.814 Da which corresponds to the maleimide derivative 5 within instrument error (1.424 Da at 66KDa = 21.2 parts per million). The other peak at 66558 Da had remained unaltered upon addition of excess thiol-specific maleimide analogue 5, suggesting that the thiol was unreactive (Fig. 4). An examination of the difference between the species with the free, reactive thiol (66439 Da) and the unreactive thiol (66558 Da) in the control sample suggests that the ‘blocking’ group is cysteine (expected exact mass of cysteine − 1 proton [disulfide] = 118.02; observed Δmass = 118.321 Da).
Figure 4.
Covalent coupling of the thiol-specific maleimide derivative 5 with human serum albumin showing addition of a single equivalent of 5 to albumin, as examined by LC-ESI-TOF. An excess (5 equiv.) of 5 was used.
Aside from engineering otherwise feebly immunogenic peptide and subunit protein vaccines for the induction of strong CTL responses, we are also interested in polysaccharide vaccines which have proved enormously useful in the prevention of infections by bacterial pathogens such as N. meningitidis and H. influenzae.35–37
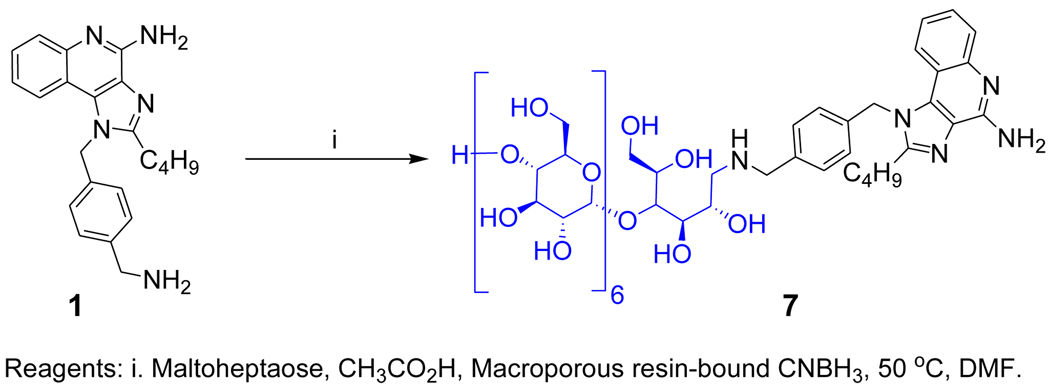
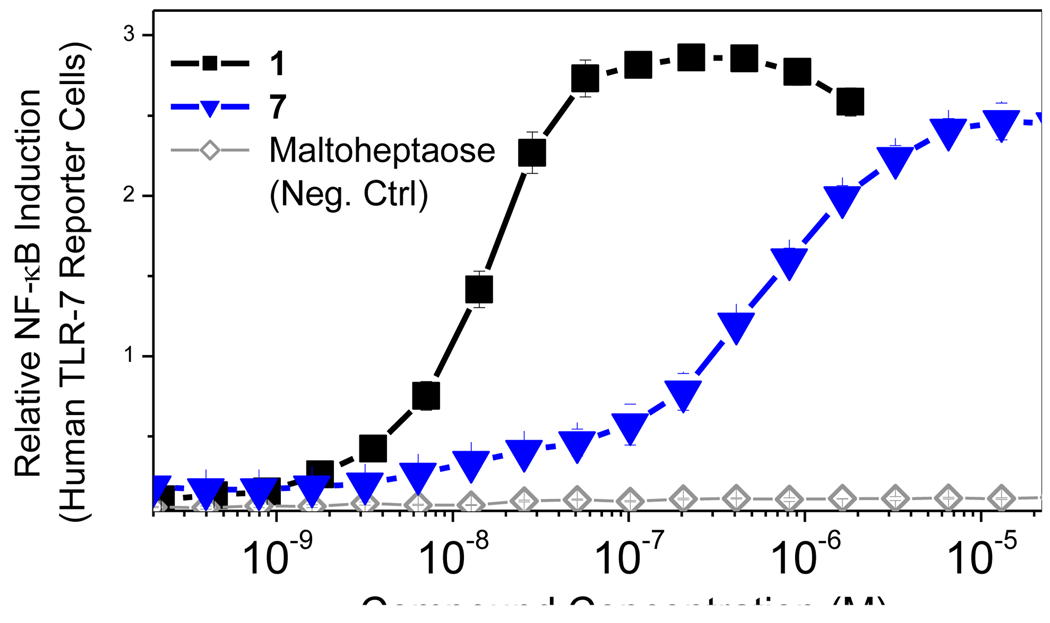
Bacterial polysaccharides, unlike conventional protein antigens, have been considered classic T cell-independent antigens that do not elicit cell-mediated immune responses but rather elicit non-anamnestic responses characterized by low-affinity IgM and restricted classes of IgG immunoglobulins without the recruitment of T cell help. Conversion to canonical, T lymphocyte-dependent responses require their covalent coupling to immunogenic ‘carrier proteins’ such as diphtheria toxoid.38;39 This appears not to be the case for zwitterionic polysaccharides, however, which elicit potent CD4+ T cell responses.40;41 We are keen to re-examine the structural determinants of T-dependent and -independent humoral responses, especially in light of recent findings of intrinsic TLR2 activation by zwitterionic polysaccharides,42;43 and we are currently testing the hypothesis that polysaccharides with covalently linked TLR stimuli may be superior to carrier proteins. The free primary amine on the N1 substituent of 1 lent itself eminently well to direct reductive amination with maltoheptaose, a model oligosaccharide with a reducing terminal maltose unit (7, Scheme 2) which, gratifyingly, was found to be active in TLR7 assays (EC50: 528 nM; Fig. 5). We are currently exploring conjugating N. meningitidis Group C polysaccharide44 with 1.
Scheme 2.
Syntheses of the maltoheptaose conjugate.
Figure 5.
TLR7-agonistic activities of imidazoquinoline-maltoheptaose conjugate in a human TLR7-specific reporter gene assay.
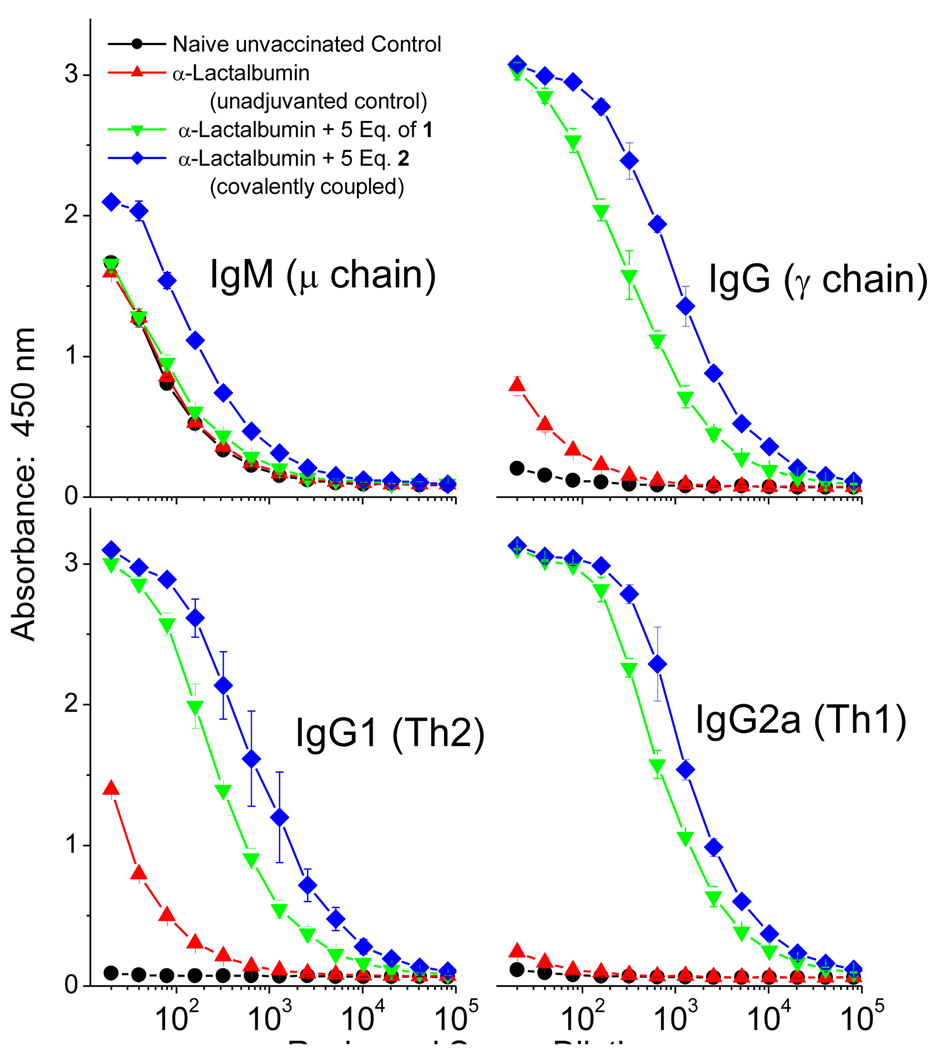
It was of particular interest to us to evaluate the well-characterized 2: α-lactalbumin conjugate (Fig. 3) as a test-case for self-adjuvanting subunit vaccine construct. We specifically asked whether the conjugation procedure would preserve antigenicity of the protein, and whether the covalently-adducted construct would be superior to a physical mixture of α-lactalbumin and 1. Cohorts (5 per group) of outbred CF-1 mice were immunized with 50 µg per animal of α-lactalbumin, or 50 µg of α-lactalbumin covalently conjugated with 5 equivalents of 2, or a mixture of 50 µg of α-lactalbumin and 5 equivalents of 1. The animals were boosted once after two weeks following the priming dose, and bled after an additional week. α-lactalbumin-specific IgM, IgG, as well as IgG1 and IgG2a (isotypes characteristic of Th2 and Th1 responses,45 respectively) were quantified by ELISA. No apparent systemic or local adverse effects were noted in animals. Weight gain and general health of the vaccinated cohorts were comparable to unvaccinated controls. As shown in Fig. 6, dramatic enhancements in antibody titers were observed with both covalently- and non-covalently adjuvanted protein (relative to α-lactalbumin alone).
Figure 6.
Immunoglobulin profiles in outbred CF-1 mice immunized on Day 0 with 50 µg/animal of α-lactalbumin, or α-lactalbumin covalently coupled with 5 equivalents of 2, or α-lactalbumin mixed with 5 equivalents of 1. Animals (5 per cohort) were boosted once on Day 14 exactly as mentioned above, and bled on Day 21. α-lactalbumin-specific immunoglobulin levels were quantified by standard antibody-capture ELISA, performed in liquid handler-assisted 384-well format.
We also observed modest, but consistent, and statistically significant differences in titers between the covalently coupled self-adjuvanting construct, and mixture of antigen and adjuvant, indicating that self-adjuvanting subunit protein vaccines may indeed be generated with full preservation of antigenicity.
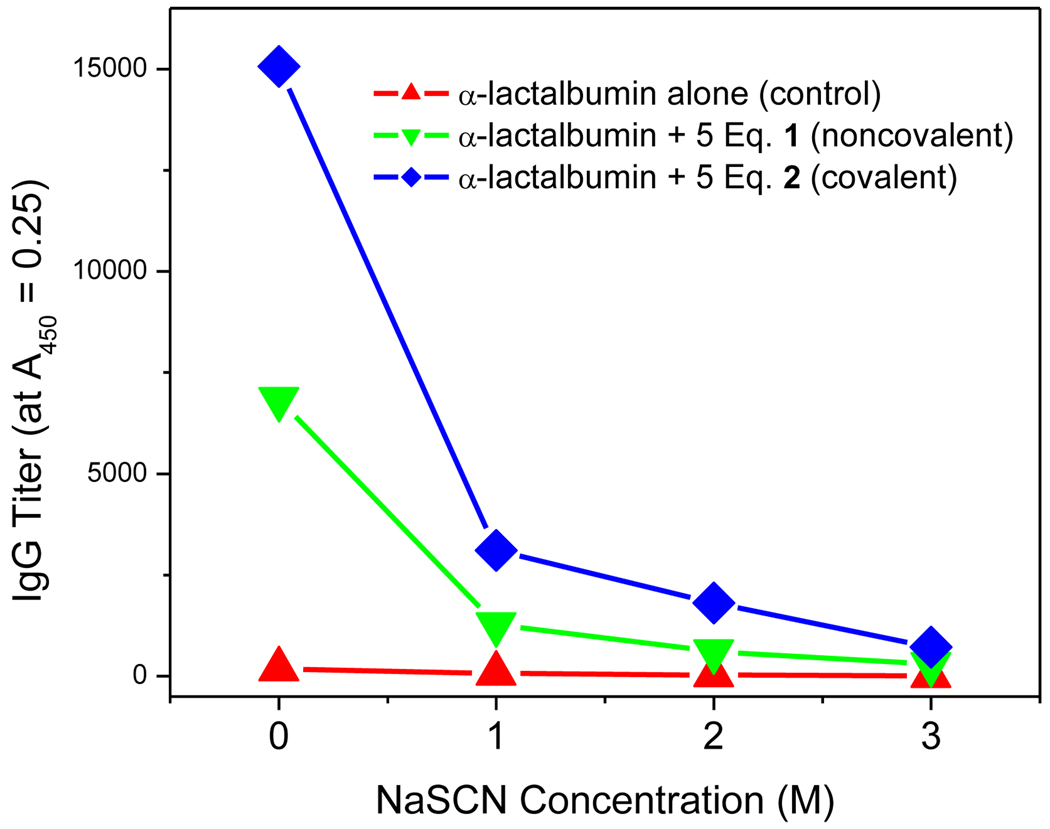
Examination of the affinity of antigen-specific IgG using conventional chaotropic ELISA46;47 also indicates higher quality IgG (Fig. 7) elicited by the self-adjuvanting construct.
Figure 7.
Affinity IgG ELISA showing antibody titer as a function of chaotrope (NaSCN) concentration. IgG titers on the ordinate axis were calculated from absorbance values at 0.25 (which corresponds to 3σ above that of naïve controls).
In conclusion, our continuing exploration of the TLR7-agonistic imidazoquinoline chemotype in tandem with expanded secondary and tertiary screens designed to specifically evaluate Th1-orienting immunostimulatory profiles have enabled the identification of 1, whose free amine group can be conveniently exploited in constructing covalent conjugates with peptides, proteins, as well as polysaccharides with preservation of immunostimulatory activity.
The feasibility of covalently coupling a small-molecule TLR7 agonist to proteins under mild, non-denaturing conditions to yield self-adjuvanting subunit vaccines is evident, and may have considerable practical value in significantly reducing systemic exposure of the adjuvant, and yet inducing high innate and adaptive immune responses. We are currently examining CD8+ CTL responses elicited by such constructs.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIAID contract HHSN272200900033C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Experimental methods and characterization of compounds (1H, 13C, and mass spectra).
References
- 1.Kawai T, Akira S. Semin.Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai Y, Takeuchi O, Akira S. J.Infect.Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- 3.Akira S. Immunol.Rev. 2009;227:5–8. doi: 10.1111/j.1600-065X.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K, Kaisho T. Nature Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 5.Cottalorda A, Verschelde C, Marcais A, Tomkowiak M, Musette P, Uematsu S, Akira S, Marvel J, Bonnefoy-Berard N. Eur.J.Immunol. 2006;36:1684–1693. doi: 10.1002/eji.200636181. [DOI] [PubMed] [Google Scholar]
- 6.Kaisho T, Akira S. Biochim.Biophys.Acta. 2002;1589:1–13. doi: 10.1016/s0167-4889(01)00182-3. [DOI] [PubMed] [Google Scholar]
- 7.Hood JD, Warshakoon HJ, Kimbrell MR, Shukla NM, Malladi S, Wang X, David SA. Hum.Vaccin. 2010;6:1–14. doi: 10.4161/hv.6.4.10866. [DOI] [PubMed] [Google Scholar]
- 8.Shukla NM, Kimbrell MR, Malladi SS, David SA. Bioorg.Med.Chem.Lett. 2009;19:2211–2214. doi: 10.1016/j.bmcl.2009.02.100. [DOI] [PubMed] [Google Scholar]
- 9.Warshakoon HJ, Hood JD, Kimbrell MR, Malladi S, Wu WY, Shukla NM, Agnihotri G, Sil D, David SA. Hum.Vaccin. 2009;5:381–394. doi: 10.4161/hv.5.6.8175. [DOI] [PubMed] [Google Scholar]
- 10.Bachmann MF, Jennings GT. Nat.Rev.Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 11.Zeng W, Ghosh S, Lau YF, Brown LE, Jackson DC. J.Immunol. 2002;169:4905–4912. doi: 10.4049/jimmunol.169.9.4905. [DOI] [PubMed] [Google Scholar]
- 12.Bettahi I, Zhang X, Afifi RE, BenMohamed L. Viral Immunol. 2006;19:220–236. doi: 10.1089/vim.2006.19.220. [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Aal AB, Batzloff MR, Fujita Y, Barozzi N, Faria A, Simerska P, Moyle PM, Good MF, Toth I. J.Med.Chem. 2008;51:167–172. doi: 10.1021/jm701091d. [DOI] [PubMed] [Google Scholar]
- 14.Moyle PM, Toth I. Curr.Med.Chem. 2008;15:506–516. doi: 10.2174/092986708783503249. [DOI] [PubMed] [Google Scholar]
- 15.Zeng W, Horrocks KJ, Robevska G, Wong CY, Azzopardi K, Tauschek M, Robins-Browne RM, Jackson DC. J.Biol.Chem. 2011 doi: 10.1074/jbc.M111.227744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson BL, Day S, Malins LR, Apostolopoulos V, Payne RJ. Angew.Chem.Int.Ed Engl. 2011;50:1635–1639. doi: 10.1002/anie.201006115. [DOI] [PubMed] [Google Scholar]
- 17.Zeng W, Ghosh S, Lau YF, Brown LE, Jackson DC. J.Immunol. 2002;169:4905–4912. doi: 10.4049/jimmunol.169.9.4905. [DOI] [PubMed] [Google Scholar]
- 18.Zeng W, Eriksson EM, Lew A, Jackson DC. Mol.Immunol. 2011;48:490–496. doi: 10.1016/j.molimm.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Hill BG, Reily C, Oh JY, Johnson MS, Landar A. Free Radic.Biol.Med. 2009;47:675–683. doi: 10.1016/j.freeradbiomed.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wenink MH, Santegoets KC, Broen JC, van Bon L, Abdollahi-Roodsaz S, Popa C, Huijbens R, Remijn T, Lubberts E, van Riel PL, van den Berg WB, Radstake TR. J.Immunol. 2009;183:6960–6970. doi: 10.4049/jimmunol.0900713. [DOI] [PubMed] [Google Scholar]
- 21.Aliahmadi E, Gramlich R, Grutzkau A, Hitzler M, Kruger M, Baumgrass R, Schreiner M, Wittig B, Wanner R, Peiser M. Eur.J.Immunol. 2009;39:1221–1230. doi: 10.1002/eji.200838742. [DOI] [PubMed] [Google Scholar]
- 22.Bracci L, La SV, Belardelli F, Proietti E. Expert.Rev.Vaccines. 2008;7:373–381. doi: 10.1586/14760584.7.3.373. [DOI] [PubMed] [Google Scholar]
- 23.Tovey MG, Lallemand C, Thyphronitis G. Biol.Chem. 2008;389:541–545. doi: 10.1515/bc.2008.051. [DOI] [PubMed] [Google Scholar]
- 24.Rajagopal D, Paturel C, Morel Y, Uematsu S, Akira S, Diebold SS. Blood. 2010;115:1949–1957. doi: 10.1182/blood-2009-08-238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berenson LS, Ota N, Murphy KM. Immunol.Rev. 2004;202:157–174. doi: 10.1111/j.0105-2896.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- 26.Pulendran B. Immunol.Res. 2004;29:187–196. doi: 10.1385/IR:29:1-3:187. [DOI] [PubMed] [Google Scholar]
- 27.Gately MK, Brunda MJ. Cancer Treat.Res. 1995;80:341–366. doi: 10.1007/978-1-4613-1241-3_14. [DOI] [PubMed] [Google Scholar]
- 28.Scott P, Trinchieri G. Semin.Immunol. 1997;9:285–291. doi: 10.1006/smim.1997.0084. [DOI] [PubMed] [Google Scholar]
- 29.Eberl M, Beck E, Coulson PS, Okamura H, Wilson RA, Mountford AP. Vaccine. 2000;18:2002–2008. doi: 10.1016/s0264-410x(99)00532-0. [DOI] [PubMed] [Google Scholar]
- 30.Tough DF, Zhang X, Sprent J. J.Immunol. 2001;166:6007–6011. doi: 10.4049/jimmunol.166.10.6007. [DOI] [PubMed] [Google Scholar]
- 31.Marshall DJ, Rudnick KA, McCarthy SG, Mateo LR, Harris MC, McCauley C, Snyder LA. Vaccine. 2006;24:244–253. doi: 10.1016/j.vaccine.2005.07.087. [DOI] [PubMed] [Google Scholar]
- 32.Shukla NM, Malladi SS, Mutz CA, Balakrishna R, David SA. J.Med.Chem. 2010;53:4450–4465. doi: 10.1021/jm100358c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Li R, Malladi SS, Warshakoon HJ, Kimbrell MR, Amolins MW, Ukani R, Datta A, David SA. J.Med.Chem. 2010;53:3198–3213. doi: 10.1021/jm901839g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaini R, Kesaraju P, Johnson JM, Altuntas CZ, Jane-Wit D, Tuohy VK. Nat.Med. 2010;16:799–803. doi: 10.1038/nm.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappuoli R. Vaccine. 2001;19:2319–2322. doi: 10.1016/s0264-410x(00)00552-1. [DOI] [PubMed] [Google Scholar]
- 36.Mawas F, Peyre M, Beignon AS, Frost L, Del Giudice G, Rappuoli R, Muller S, Sesardic D, Partidos CD. J.Infect.Dis. 2004;190:1177–1182. doi: 10.1086/423327. [DOI] [PubMed] [Google Scholar]
- 37.Broker M, Dull PM, Rappuoli R, Costantino P. Vaccine. 2009;27:5574–5580. doi: 10.1016/j.vaccine.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 38.Falugi F, Petracca R, Mariani M, Luzzi E, Mancianti S, Carinci V, Melli ML, Finco O, Wack A, Di Tommaso A, De Magistris MT, Costantino P, Del Giudice G, Abrignani S, Rappuoli R, Grandi G. Eur.J.Immunol. 2001;31:3816–3824. doi: 10.1002/1521-4141(200112)31:12<3816::AID-IMMU3816>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 39.Costantino P, Viti S, Podda A, Velmonte MA, Nencioni L, Rappuoli R. Vaccine. 1992;10:691–698. doi: 10.1016/0264-410x(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 40.Avci FY, Kasper DL. Annu.Rev.Immunol. 2010;28:107–130. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- 41.Kalka-Moll WM, Tzianabos AO, Bryant PW, Niemeyer M, Ploegh HL, Kasper DL. J.Immunol. 2002;169:6149–6153. doi: 10.4049/jimmunol.169.11.6149. [DOI] [PubMed] [Google Scholar]
- 42.Gallorini S, Berti F, Mancuso G, Cozzi R, Tortoli M, Volpini G, Telford JL, Beninati C, Maione D, Wack A. Proc.Natl.Acad.Sci.U.S.A. 2009;106:17481–17486. doi: 10.1073/pnas.0903313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallorini S, Berti F, Parente P, Baronio R, Aprea S, D'Oro U, Pizza M, Telford JL, Wack A. J.Immunol. 2007;179:8208–8215. doi: 10.4049/jimmunol.179.12.8208. [DOI] [PubMed] [Google Scholar]
- 44.Costantino P, Viti S, Podda A, Velmonte MA, Nencioni L, Rappuoli R. Vaccine. 1992;10:691–698. doi: 10.1016/0264-410x(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan C, Valdez JC, Chandrasekaran R, Eibel H, Mikecz K, Glant TT, Finnegan A. Arthritis Res. 2002;4:54–58. doi: 10.1186/ar383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pullen GR, Fitzgerald MG, Hosking CS. J.Immunol.Methods. 1986;86:83–87. doi: 10.1016/0022-1759(86)90268-1. [DOI] [PubMed] [Google Scholar]
- 47.Macdonald RA, Hosking CS, Jones CL. J.Immunol.Methods. 1988;106:191–194. doi: 10.1016/0022-1759(88)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.