Abstract
Studies were undertaken to characterize and determine the pathogenic mechanisms involved in a newly described systemic disease in Homarus americanus (American lobster) caused by a Vibrio fluvialis-like microorganism. Nineteen isolates were obtained from eight of nine lobsters sampled. Biochemically, the isolates resembled V. fluvialis, and the isolates grew optimally at 20°C; none could grow at temperatures above 23°C. The type strain (1AMA) displayed a thermal reduction time (D value) of 5.77 min at 37°C. All of the isolates required at least 1% NaCl for growth. Collectively, the data suggest that these isolates may embody a new biotype. Pulsed-field gel electrophoresis (PFGE) analysis of the isolates revealed five closely related subgroups. Some isolates produced a sheep hemagglutinin that was neither an outer membrane protein nor a metalloprotease. Several isolates possessed capsules. The isolates were highly susceptible to a variety of antibiotics tested. However, six isolates were resistant to erythromycin. Seventeen isolates harbored plasmids. Lobster challenge studies revealed that the 50% lethal dose of a plasmid-positive strain was 100-fold lower than that of a plasmid-negative strain, suggesting that the plasmid may enhance the pathogenicity of these microorganisms in lobsters. Microorganisms that were recovered from experimentally infected lobsters exhibited biochemical and PFGE profiles that were indistinguishable from those of the challenge strain. Tissue affinity studies demonstrated that the challenge microorganisms accumulated in heart and midgut tissues as well as in the hemolymph. Culture supernatants and polymyxin B lysates of the strains caused elongation of CHO cells in tissue culture, suggesting the presence of a hitherto unknown enterotoxin. Both plasmid-positive and plasmid-negative strains caused significant dose-related intestinal fluid accumulations in suckling mice. Absence of viable organisms in the intestinal contents of mice suggests that these microorganisms cause diarrhea in mice by intoxication rather than by an infectious process. Further, these results support the thermal reduction data at 37°C and suggest that the mechanism(s) that led to fluid accumulation in mice differs from the disease process observed in lobsters by requiring neither the persistence of viable microorganisms nor the presence of plasmids. In summary, results of lobster studies satisfy Koch's postulates at the organismal and molecular levels; the findings support the hypothesis that these V. fluvialis-like organisms were responsible for the originally described systemic disease, which is now called limp lobster disease.
Catastrophic losses of Homarus americanus (American lobster) have been most consistently associated with gaffkemia, a disease caused by Aerococcus viridans subsp. homari (52, 59). Recognized more than half a century ago as a cause of heavy mortalities in natural populations and impounded lobsters on the east coast of North America, infection usually results when A. viridans breaches the integument through wounds. Noted symptoms include variably pink coloration of the ventral abdomen, pink-colored hemolymph, prolonged hemolymph clotting times, and a drastic reduction in the number of hemocytes. In the fall of 1997 and 1998, an unexplained, highly invasive, non-gaffkemic-like disease, now called limp lobster disease, emerged in H. americanus lobsters that were harvested from the Gulf of Maine coastal waters (B. D. Tall, M. Crosby, D. Prince, G. Clerge, D. Lightner, L. Mohney, M. Dey, F. Khambaty, K. Lampel, J. W. Bier, B. Eribo, and R. C. Bayer., Abstr. Natl. Shellfisheries Assoc. Annu. Meet., p. 43, 1999). Economic losses exceeding $2.5 million threatened the $136 million-a-year industry. Lobsters with this syndrome display weakness, lethargy, and slow or ineffectual responses to sensory stimuli. Vibrio fluvialis-like organisms were isolated from ill lobsters. Originally, the syndrome was thought to be associated only with impounded lobsters, but ill animals harvested from traps in the later stages of the disease outbreak were also found. Vibrio infections have been reported in lobsters that were held for extended periods of time. The first account of vibriosis in impounded lobsters was that of Brinkley et al. (16), who reported the isolation of both Vibrio parahaemolyticus and Vibrio alginolyticus from moribund aquarium-held lobsters. Recently, “luminous vibriosis” attributable to Vibrio harveyi appeared in phyllosoma larvae of the packhorse rock lobster (Jasus verreauxi) raised in an experimental culture facility (22).
We present in this report the first description of the isolation of V. fluvialis-like bacteria from lobsters with limp lobster disease. Because of possible human disease implications, studies were undertaken to determine the clinical and microbial characteristics of this newly recognized marine Vibrio pathogen. Although the emergence of this pathogen poses a significant economic threat that merits additional studies, the causative V. fluvialis-like strains are probably not infectious for humans. Understanding how this organism is able to overcome species barriers and adapt to new hosts is crucial to the production of disease-free seafoods.
MATERIALS AND METHODS
Preliminary bacterial isolation and identification.
In October 1998, hemolymph aspirates from six weakened (i.e., showing signs of limp lobster disease) lobsters were aseptically collected by a process involving disinfection of the surface of the ill lobster's exoskeleton in the vicinity of the junction between the second and third abdominal somites. This was followed by insertion of a 26-gauge needle attached to a 1-ml tuberculin syringe through the somite junction into the dorsal sinus and withdrawal of hemolymph. The hemolymph samples were shipped in syringes to the Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration (FDA), in an insulated mailing container accommodating refrigerated cold packs. The samples arrived at the FDA within 18 h, and the container had an internal temperature of approximately 12°C. The samples were cultured onto two plates each of thiosulfate citrate bile sucrose (TCBS) medium (Difco) and Marine agar (MA) (20 ppt; Difco). The plates were incubated for 18 h, one at 20°C and the other at 30°C. An aliquot of each sample was also inoculated into two duplicate enrichment tubes containing 5 ml of marine broth (MB), pH 8.4 (20 ppt; Difco). One of the MB enrichment cultures was incubated at 20°C and the other at 30°C, both for 18 h. Each enrichment broth tube was used to inoculate duplicate sets of three types of isolation media: TCBS, tryptic soy agar supplemented with 1% NaCl (TSA-S; Difco) and 10 μg of ampicillin per ml, and MA. Each set of plates was incubated for 18 h, one at 20°C and the other at 30°C. Sixteen isolates were obtained from five culture-positive hemolymph samples. Three other isolates were provisionally identified and sent to the FDA for identity confirmation (D. Bouchard; MicroTechnologies, Hancock, Maine).
All of the isolates were identified as V. fluvialis-like organisms by using a standard set of biochemical tests with the conventional API 20E microtube test panel (1% NaCl used as diluent; BioMérieux, Inc., Hazelwood, Mo.) as a focal point of analysis (Table 1). Analytab Products profiles were referred to the analytical profile index or were identified by consultation with the technical services group of BioMérieux Vitek, Inc., who analyzed the profiles by using the APILAB software version 3.2.2 (APILAB Plus; BioMérieux). Cultures were maintained on TSA-S; frozen cultures were stored at −70°C in tryptic soy broth (Difco) supplemented with 1% NaCl (TSB-S) and 25% glycerol. For biotyping, the isolates were identified phenotypically by using a standard set of biochemical tests as described by Baumann and Schubert (12), which included gas production from d-glucose and the utilization of salicin, d-glucuronate, and glutarate. Unless otherwise noted, the growth media just described, TSA-S or TSB-S, were used as the routine culture media.
TABLE 1.
Characteristics of V. fluvialis-like isolates obtained from ill lobstersa
| Isolate | Plasmid (no. of bands) | PFGE type | Sheep HA | Hemolysin | API profile no. | TCBS growth 48 h | Salt tolerance at concn:
|
Biotype
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2% | 4% | 8% | Salicin | Gas glucose | |||||||
| 1AMA | + (3) | A | + | − | 3044104 | G | − | + | + | − | + | + |
| 2E1MA | + (2) | B | + | − | 3044126 | Y | − | + | + | − | + | + |
| 3DMA | + (2) | B | + | − | 3044126 | Y | − | + | + | − | + | + |
| 4D119MA | + (2) | B | + | − | 3044126 | Y | − | + | + | − | + | + |
| 5E2MA | + (2) | B | + | − | 3044126 | G | − | + | + | − | + | + |
| 6A1MA | + (3) | A | + | − | 3044104 | G | − | + | + | − | + | + |
| 7E1AMA | + (2) | B | + | − | 3044126 | Y | − | + | + | − | + | + |
| 8D122MA | + | B | + | − | 3044126 | NG | − | + | + | − | + | + |
| 9DMA | + (3) | B | + | − | 3044126 | G | − | + | + | − | + | + |
| 10C119MA | + (2) | B | + | − | 3044126 | Y | − | + | + | − | + | + |
| 11EMA | + (2) | B | + | − | 3044126 | G | − | + | + | − | + | + |
| 15A4TSA | + (3) | A | + | − | 3044104 | G | − | + | + | − | + | + |
| 18C110MA | + (2) | B | + | − | 3044126 | Y | − | + | + | − | + | + |
| 27F3MA | + (2) | C | − | + | 3046105 | G | − | + | + | + | + | − |
| 28F4MA | + (2) | C | − | + | 3046105 | G | − | + | + | − | + | − |
| 31F7G | − | UT | − | + | 3046105 | G | − | + | + | + | + | − |
| DB6 | − | A1 | + | − | 3046104 | G | − | + | + | − | + | + |
| DB7 | + | B | − | − | 3246124 | Y | − | + | + | − | + | + |
| DB8 | + | A2 | + | − | 3246104 | G | − | + | − | − | − | − |
Abbreviations: G, sucrose-negative colonies; Y, sucrose-positive colonies; NG, no growth; UT, untypeable; HA, hemagglutinin. +, positive for indicated attribute; −, negative for indicated attribute.
Antibiotic susceptibility to ampicillin, apramycin, chloramphenicol, ciprofloxacin, carbenicillin, cephalothin, erythromycin, gentamicin, imipenem, kanamycin, naladixic acid, rifampin, sulfisoxazole, streptomycin, tetracycline, trimethroprin, trimethroprim-sulfamethoxazole, and norfloxacin was determined by the Bauer-Kirby antibiotic disk diffusion method, which was performed according to the recommendations of the National Committee for Clinical Laboratory Standards using Mueller-Hinton agar (Difco) containing 1% NaCl (47, 65).
Crude hemagglutinin preparation.
A crude potassium thiocyanate (KSCN) hemagglutinin preparation was isolated from V. fluvialis strain 1AMA by using the procedure described previously by Tall et al. (61).
Hemagglutination assay.
Hemagglutination assays on cells grown on TSA-S plates at 20°C as described above were performed by the procedure described by Tall et al. (61). For use in the assay, sheep, chicken, bovine, rabbit, guinea pig, and human A, O, and B erythrocytes (RBCs) were suspended in 0.9% NaCl to a final concentration of 0.3%. Bacterial cells were suspended in saline to an A660 of 0.5. One hundred-microliter aliquots of each bacterial cell suspension were diluted twofold in 0.9% saline in wells of a 96-well round-bottomed microtiter plate (Nunc, Thomas Scientific, Inc., Swedesboro, N.J.). One hundred-microliter aliquots of the diluted cell samples were mixed with 100 μl of washed RBCs, followed by incubation for 4 h at 30°C. Negative controls consisted of 100 μl of saline mixed with equal volumes of RBCs. The microtiter plates were observed macroscopically for agglutination of the RBCs. KSCNextracts were also analyzed in a similar fashion.
Preparation of intact DNA for PFGE analysis.
Agarose plugs containing DNA were prepared as described previously by McCarthy and Khambaty (44). Strains were grown at 20°C in TSB-S on a shaker at 100 rpm; 1.0 ml of each culture was harvested at a cell density of ≈0.6 at A610. Washed cells in 1.0 ml of ice-cold Pett V plus E buffer (10 mM Tris, 1.0 M NaCl, 20 mM EDTA [pH 8.0]) were embedded into 1% PFGE-grade agarose plugs, which after solidification were lysed in EC lysis buffer (1 M NaCl, 6 mM Tris, 0.1 M EDTA, 0.5% Sarkosyl, 0.2% sodium deoxycholate, 1 mg of lysozyme per ml [pH 8.0]) at 37°C for 1 h, followed by a wash step in ESP buffer (0.5 M EDTA, 1% Sarkosyl, 1 mg of proteinase K per ml [pH 9.0] incubated at 55°C for 8 h), followed by another two washing steps with phenylmethylsulfonyl fluoride solution (10 mM Tris HCl, 1 mM EDTA, 1.5 mM phenylmethylsulfonyl fluoride [pH 8.0]) for 1 h each. The plugs were rinsed three more times with T10E50 buffer (10 mM Tris HCl, 50 mM EDTA [pH 8.0]) for 1 h each and stored at 4°C in the final wash solution.
PFGE analysis.
Agarose slices (2 mm) for use in the analysis were rinsed in sterile distilled water and equilibrated in the appropriate restriction enzyme digestion buffer, followed by digestion for 6 h in fresh restriction enzyme digestion buffer (according to the technical reference guide for each enzyme) containing 20 to 30 U of either NotI, ApaI, or SmaI restriction enzymes (New England Biolabs, Beverly, Mass.). Electrophoresis was carried out by transferring washed slices (0.5× Tris-borate-EDTA [TBE]) to a 1% PFGE-grade agarose gel and then sealing the wells. The DNA in the gel was resolved using a Bio-Rad CHEF-DRII PFGE apparatus (Bio-Rad Laboratories, Hercules, Calif.). Restriction fragments were separated over a size range of approximately 25 to 800 kb using 0.5× TBE. The CHEF Mapper was programmed with a voltage gradient of 6 V/cm and a switch time ramped linearly from 2 to 10 s over the course of 23 h. The gel was stained with ethidium bromide (1 μg of ethidium bromide per ml) and photographed by using UV transillumination. The DNA fingerprints were digitized by using an Imagestore 7500 gel documentation system (Ultra Violet Products, Inc., Upland, Calif.) and saved as TIFF files. The fingerprints generated were evaluated with Molecular Analyst DST version 1.6 software (Bio-Rad). To allow comparisons between different gels, DNA fragments on each gel were normalized by using a XbaI-digested DNA from a Salmonella enterica Newport sample as the molecular weight standard. A 1.5% band tolerance was selected for use during comparisons of DNA profiles. Cluster analysis was performed by the unweighted pair-group method using arithmetic averages, and DNA relatedness was calculated based on the Dice coefficient.
Plasmid isolation and analysis.
Plasmids were isolated from TSB-S overnight 20°C cultures by using a Wizard Miniprep kit (Promega, Madison, Wis.); the final volume was 45 μl. Purified plasmids were subjected to electrophoresis through a 1% agarose gel in either 1× Tris-acetate-EDTA buffer (pH 8) or 1× TBE buffer (pH 8).
CHO cell elongation assay.
The ability of the enterotoxin to elongate CHO cells was estimated by a modification (36) of a procedure described previously by Guerrant et al. (26). One CHO cell unit was defined as the reciprocal of the dilution that caused elongation of 50% of the cells contained in a well of a 96-well plate. Controls included similarly obtained supernatants from a culture grown at 37°C of a known CHO cell-elongating Vibrio cholerae strain, CVD103-HgR, as well as uninoculated culture medium with and without polymyxin B (2 mg/ml).
Lobster challenge studies.
To satisfy Koch's postulates, healthy lobsters (weight, 450 to 500 g each), free of pathogens, were separated into groups of six and were allowed to acclimate at 20°C in eight separate self-contained aquaria containing artificial seawater (20 ppt) for 24 h. The aquaria were housed in the aquaculture facility at the University of Maine at Orono. V. fluvialis-like strains 1AMA (plasmid positive) and 31 (plasmid negative) were inoculated onto TSA-S plates and incubated for 18 h at 20°C. A cell suspension containing approximately 1010 CFU/ml (i.e., having an A660 of 0.3 when diluted 1:100) was made in 2% saline. Dilutions of this cell suspension were prepared and used in an experiment designed to determine the 50% lethal dose (LD50). Lobsters were challenged by injection into the dorsal sinus. Finally, animals in a control group were given sterile 2% saline only. The lobsters were then placed back into the aquaria, observed, and monitored for signs of illness. The remaining portion of inoculum was diluted and plated onto TSA-S for viability and inoculum size determination. Hemolymph was aseptically removed from ill and control lobsters, diluted 10,000-fold in 2% saline, and plated onto both a MA and a TCBS agar plate. In one experiment, pooled samples of heart and midgut tissues (weight of each sample, 5 g total, from five animals) were placed in 50 ml of 2% saline, blended in a sterile Warring blender for 1 min at a high speed, and diluted 10,000-fold. One hundred-microliter aliquots were spread plated onto MA and TCBS agar plates. Suspected V. fluvialis-like colonies were isolated and identified by API 20E, and V. fluvialis-positive colonies were further analyzed by PFGE.
Suckling mouse assay.
V. fluvialis cells were tested for their ability to cause fluid accumulation in a suckling mouse as previously described by Kothary et al. (35, 36) in accordance with Institutional Animal Care and Use Committee-approved protocol number 301. Comparatively, V. cholerae O1 strain N16961 and V. parahaemolyticus strain TX 2103 were also analyzed. The optical absorbances (A650) of the suspensions were adjusted to 50, 10, 1, and 0.1 U prior to oral feeding (50 μl). Estimates of the numbers of CFU in these suspensions were obtained by quantitative spread plating onto TSA-S. Pregnant Institute of Cancer Research mice were obtained from Harlan Sprague Dawley (Indianapolis, Ind.). After 6 h, the mice were sacrificed by cervical dislocation and the fluid accumulation ratio was determined. The fluid accumulation ratio was expressed as 1,000 times the ratio of the weight of the stomach plus intestine to the remaining body weight. Six mice were used for each inoculum tested in the assay. The stomach plus intestine from some of the mice were examined for the presence of live organisms.
Electron microscopy.
For negative staining, a cell suspension containing approximately 109 cells/ml was prepared by suspending cells from TSA-S plates, cultured as described above, in 1 ml of fixative (3% glutaraldehyde in 0.1 M sodium cacodylate [pH 7.2]) contained in an Eppendorf tube. After a 1-h incubation at room temperature, the cells were washed twice in 0.01 M phosphate-buffered saline (pH 7.4) (Sigma Chemical Co.), and 15 μl was applied to the coated surface of a 300-mesh carbon-coated copper grid. After allowing the cells to settle on the grid for 1 min, excess liquid was removed and the specimens were stained for 1 min with 15 μl of 1% phosphotungstic acid adjusted to pH 6.8 using 1 M NaOH. Excess stain was removed, and the specimens were allowed to air dry.
In order to visualize the acidic mucosubstances by transmission electron microscopy, samples were treated by using an Alcian Blue staining procedure described by Fassel et al. (24a). Ultrathin Eponate sections made using a Leica Ultracut S ultramicrotome were doubly stained with aqueous 3% uranyl acetate and Reynolds' lead citrate (53). The specimens were examined using a Philips 400 transmission electron microscope operating at an accelerating voltage of 80 kV.
Heat kill (decimal reduction) studies.
Decimal reduction times (D37) were determined for V. fluvialis-like strain 1AMA by preparing a cell suspension in 1% saline (10 ml) containing approximately 108 CFU/ml (0.3 U at A660). The cell suspension was then exposed to a temperature of 37°C by using an immersed coil apparatus (Protol Instrument, Stirlingshire, United Kingdom). At 5-min intervals up to and including 60 min, survivors were plated onto TSA-S at each sampling interval using an Autoplate 4000 spiral plater (Spiral Biotech, Bethesda, Md.). Following incubation at 20°C for 24 h, residual cell populations were determined using a Laser Colony scanner, Model 500A (Spiral Biotech). D37 values from each of three replicates were combined to yield the average D37 value.
Outer membrane preparation.
Outer membrane preparations for V. tubiashii, V. vulnificus, and V. fluvialis were prepared using the LiC2H3O2-LiCl procedure described previously by Johnston et al. (30) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE analysis.
To determine the outer membrane protein profiles and the protein profile of the hemagglutinin, SDS-PAGE was performed as described by Laemmli (38), using 8 to 25% gradient gels in a PhastSystem (Amersham Pharmacia Biotech, Piscataway, N.J.).
Molecular weight estimations.
The molecular weights of the denatured and reduced outer membrane preparations and hemagglutinin were estimated by the relative mobility method of Weber et al. (68).
RESULTS
Characterization of the V. fluvialis-like isolates isolated from lobsters with limp lobster disease.
Five of six hemolymph samples originally obtained from ill lobsters showing signs of limp lobster disease yielded primarily V. fluvialis-like organisms. Sixteen isolates were obtained from this set of samples. One hemolymph sample did not yield bacteria upon being cultured. Three other isolates, each from a different ill lobster, were sent to the Center for Food Safety and Applied Nutrition for identity confirmation. Identification of the combined collection of 19 isolates was achieved by using a number of biochemical tests, including the API 20E identification system. Table 1 presents a summary of the diagnostic biochemical properties of the isolates.
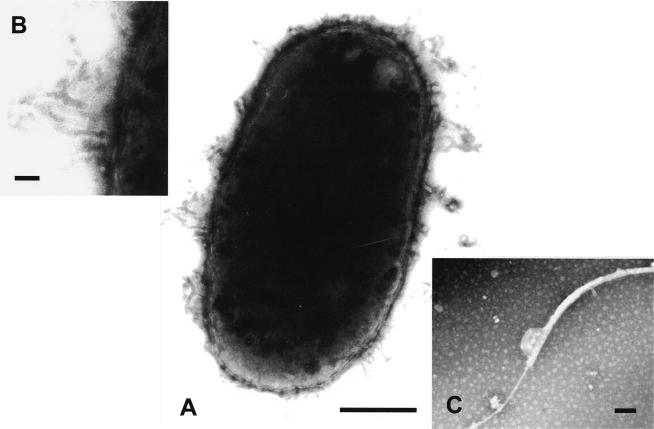
Morphologically, the V. fluvialis-like isolates were gram-negative, straight- to curved rod-shaped bacteria possessing the typical ultrastructure (0.5 to 0.8 μm in width and 1.2 to 3 μm in length) of members of the family Vibrionaceae (Fig. 1). They were motile by means of polar flagella. Additionally, the lobster isolates possessed numerous tubular appendages (Fig. 1) similar to those expressed by Vibrio campbellii (12). However, the appendages observed for the lobster isolates were neither as long nor as numerous as those described for V. campbellii.
FIG. 1.
(A) Transmission electron photomicrograph of a 1% phosphotungstic acid negatively stained preparation of Vibrio fluvialis-like strain 1AMA showing typical Vibrio-like ultrastructure. Bar, 0.5 μm. (B) A higher magnification of a tubular structure typically seen in these organisms. Bar, 0.5 μm. (C) The single polar ensheathed flagellum. Bar, 0.1 μm.
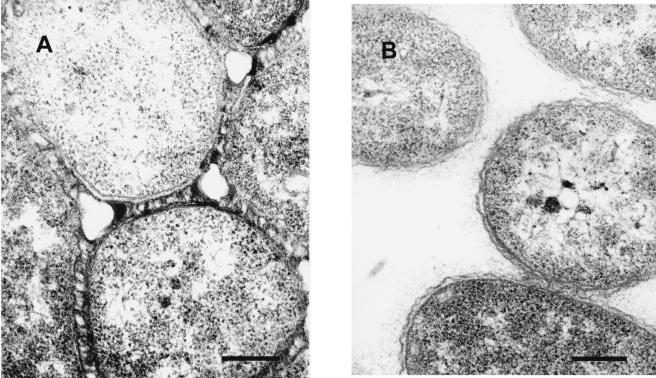
Five strains representative of the five major PFGE types were examined for capsule production. We did not examine all 19 isolates. Figure 2 shows the ultrastructural features of capsule expressed by an isolate stained by an Alcian Blue-lysine capsule-staining technique that was used in a previous study which investigated capsule expression by V. vulnificus (B. D. Tall, R. T. Gray, and D. B. Shah, Abstr. Annu. Meet. Micro. Soc. Amer., p. 378-379, 1993). Electron microscopy of individual V. fluvialis-like cells stained with this technique showed that the bacterial cell surface was covered with small, irregularly distributed, spikelike, Alcian Blue-lysine-laden, electron-dense deposits. These same cells were observed to form clusters of various sizes, held together by similar spikelike interdigitations between the cells. In contrast, unstained control cells either were barren of all surface structures or displayed a continuous, fine, lace-like coating (“fuzz”) of extracellular material.
FIG. 2.
Transmission electron photomicrograph of Vibrio fluvialis-like strain 1AMA stained with (A) and without (B) Alcian Blue and lysine. Note the thickened but extensive fibrous, electron-dense matrix formed by the condensation of the capsular glycocalyx when stained with Alcian Blue and lysine. Compare panel A with panel B to visualize the effect of Alcian Blue and lysine staining. Bars, 0.25 μm.
By culture, all of the isolates grew on MA; the colonies were 3 to 4 mm in size and were translucent to opaque in appearance. One difference noted was that isolate 6AlMA on MA grew as small (diameter, 1 to 2 mm) translucent colonies in contrast to 3- to 4-mm translucent to opaque colonies found for the other isolates, including isolate 1AMA (considered in this study to be the type strain due to the fact that it was included as a strain in all of the analyses undertaken). Only 18 of 19 isolates could grow on TCBS agar, of which only 7 were found to be sucrose positive (Table 1).
Biochemically, all of the V. fluvialis-like lobster isolates were positive for o-nitrophenyl-β-d-galactopyranoside, arginine dihydrolase activity, and indole production, and all produced acid from glucose and mannose. All of the isolates were sensitive to the vibriostatic compound 2,4-diamino-6,7-isopropyl pteridine phosphate (compound 0/129; 150 μg), and all possessed oxidase activity. All of the strains were negative for lysine decarboxylase activity, ornithine decarboxylase activity, H2S production, urea hydrolysis, Voges-Proskauer reaction, acid production from d-sorbitol, l-rhamnose, and production of poly-β-hyrdoxybutyrate storage granules. All of the isolates required supplementation with at least 1% NaCl in the growth medium.
All of the isolates were highly susceptible to apramycin, chloramphenicol, ciprofloxacin, cephalothin, gentamicin, imipenin, kanamycin, nalidixic acid, rifampin, streptomycin, sulfisoxazole, trimethoprim-sulfamethoxazole, tetracycline, trimethoprim, and norfloxacin. Six of the isolates were found to be resistant to erythromycin. Two other isolates were found to be resistant to ampicillin and carbenicillin and thus might possess a common β-lactamase resistance mechanism (29).
One interesting characteristic of these organisms was that all of the isolates grew better at 20°C than at higher temperatures. A thermal reduction time (D value) at 37°C was found to be 5.77 min for lobster V. fluvialis-like type strain 1AMA.
PFGE analysis.
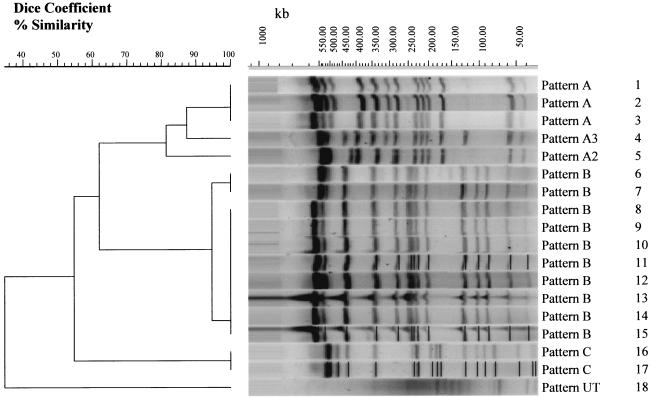
Of the three restriction enzymes used for PFGE analysis, i.e., ApaI, NotI, and SmaI, NotI restriction enzyme digestion produced DNA fragments suitable for analysis of the 19 lobster V. fluvialis-like isolates. As shown in Fig. 3, all but one strain produced patterns suitable for comparison with the method described above. The lone strain that produced a smear will require further evaluation. DNA fingerprinting patterns generated with endonuclease NotI and analyzed by PFGE revealed that 18 of the 19 isolates either shared a common DNA fingerprint pattern or possessed minor variants thereof. These results suggest that the isolates represent five closely related subgroups. PFGE fingerprint pattern subgroup A was found in three isolates from a single lobster, and PFGE fingerprint pattern subgroups A2 and A3 were each isolated from different animals. PFGE fingerprint pattern subgroup B was found in 11 isolates obtained from four different animals, and PFGE fingerprint pattern subgroup C was found in 2 isolates from a single animal. The single nontypeable isolate also came from the same animal from which the PFGE fingerprint pattern subgroup C isolates were obtained. There was one animal that was infected with both PFGE fingerprint pattern subgroups A and B. Table 1 lists the isolate names and PFGE group assignments. Approximately 12 to 17 NotI-digested DNA fragments were generated for each pattern, which ranged in size from less than 48.5 to approximately 500 kb. Isolates 1AMA (type strain), 6AlMA, and 15A4TSA presented identical banding patterns and represent banding pattern subgroup A. Cluster analysis showed that these isolates possessed a Dice index of 100%, signifying that they are essentially identical. Cluster analysis showed that PFGE subgroups A2 (strain DB6) and A3 (strain DB8) are tightly grouped together and are slightly more related to each other than to strains making up subgroup A. In comparison to subgroup A, there were three-band differences between subgroup fingerprint patterns A and A2 and two-band differences between subgroup fingerprint patterns A and A3. Dice coefficients for strains DB8 and DB6 were 89.7 and 85.7%, respectively. The PFGE subgroup B banding pattern was seen in 11 of the 19 isolates. Among these strains, Dice coefficients were 100%, suggesting shared uniqueness within subgroup B, even though these isolates were obtained from four different lobsters. The subgroup B banding pattern differed from that of subgroup A by the addition of six bands and the loss of three bands, giving a Dice coefficient of 62.1%. Finally, the PFGE subgroup C banding pattern was the most distinct pattern seen among all of the isolates. The Dice coefficient for these isolates was approximately 52%.
FIG. 3.
Electrophoretic migration patterns and similarity dendrogram of NotI-digested Vibrio fluvialis-like genomic DNA obtained by PFGE. Lanes 1 to 18 represent strains 1AMA, 6A1MA, 15A4TSA, DB6, DB8, 2E1MA, 3DMA, 4D119MA, 5E2MA, 8D122MA, 9DMA, 10C119MA, 11EMA, 18C110MA, DB7, 27F3MA, 28F4MA, and 31F7G, respectively. Each strain's PFGE pattern type is listed as well. DNA molecular weight scale was derived from XbaI-digested Salmonella newport AM01144 genomic DNA. V. fluvialis-like strain 7E1AMA is not shown in the picture but was shown to be a member of the PFGE subtype type B group by PFGE analysis.
Plasmid analysis.
Plasmid carriage results for the V. fluvialis-like isolates are shown in Table 1. Seventeen of 19 isolates possessed plasmids that were similar in size (a common band of approximately 12 kb was observed, based on linear DNA molecular mass standards), and 14 of the 19 isolates had more than one plasmid, suggesting that plasmid carriage is common among these strains. Attempts to cure the plasmid from type strain 1AMA were unsuccessful. It is noteworthy that the two isolates which lacked plasmids, 28F4MA and 31F7G, came from the same animal.
Infection studies.
Preliminary experimental infections of H. americanus lobsters were performed using V. fluvialis-like type stain 1AMA (plasmid positive) and V. fluvialis-like strain 31F7G (plasmid negative), both isolated from animals displaying limp lobster disease. The cumulative mortalities resulting from dorsal sinus injection of bacteria in lobsters demonstrated high virulence for the plasmid-bearing strain and suggested that V. fluvialis could be the etiological agent of limp lobster disease. Table 2 shows the results of the animal challenge studies. The LD50 for plasmid-bearing 1AMA was approximately 1 × 106 CFU per ml, with death occurring by 120 h after challenge. Higher doses (above 1 × 107 CFU per ml) showed mortality, with death occurring earlier, within 96 h and as early as 7 h postchallenge (108 CFU per ml). In comparison, the LD50 for plasmid-negative 31F7G was approximately 1.25 × 108 CFU per ml, with death occurring within 18 h after challenge. All animals challenged with 2.5 × 107 CFU per ml of the plasmid-negative strain survived, as did all control animals challenged with 2% NaCl. These results suggest that the presence of a plasmid may enhance the disease process. However, because these studies were not done with isogenic plasmid-bearing and -nonbearing strains, these results may also reflect interstrain variation in virulence. Tissue affinity studies demonstrated that the challenge microorganisms accumulated in heart and midgut tissues as well as in the hemolymph (Table 3). Microorganisms recovered from experimentally infected lobsters displaying limp lobster disease exhibited biochemical and PFGE profiles that were indistinguishable from those of the challenge strain (data not shown).
TABLE 2.
Infection studies with V. fluvialis-like organisms in lobsters: dose response
| Organism | Dose (CFU/ml) | No. of animals challenged | Mortality (%) | Time to death (h) |
|---|---|---|---|---|
| 1AMA (plasmid positive) | 109 | 6 | 100 | <14 |
| 4.5 × 108 | 12 | 100 | 7-18 | |
| 107 | 6 | 100 | 18-96 | |
| 4.5 × 106 | 18 | 77.7 | 96-120 | |
| 8 × 105 | 6 | 33 | 120 | |
| 31F7G (plasmid negative) | 2.5 × 108 | 6 | 100 | <18 |
| 2.5 × 107 | 6 | 0 | NDa |
No deaths after 120 h.
TABLE 3.
Infection studies with V. fluvialis-like organisms in lobsters: tissue affinity
| Infected or control | Tissue type and no. of V. fluvialis-like organisms recovered (CFU/ml)a
|
||
|---|---|---|---|
| Heart | Midgut | Hemolymph | |
| Infected | 8.4 × 104 | 3.8 × 105 | 2.9 × 104 |
| Saline control | 0b | 0 | 0 |
Lobsters were infected via the intradorsal sinus route. Dose was 108 CFU/ml of V. fluvialis-like organism strain 1AMA. Samples were taken 5 to 6 h postinfection.
Total plate count of 2.8×104 CFU/ml was recovered from midgut samples obtained from saline control animals; no V. fluvialis-like isolates were identified.
Hemolysins and enterotoxins.
Some of the V. fluvialis-like isolates grown on TSA-S with 5% sheep blood were found to be β-hemolytic (see Table 1). In addition, cell-free culture supernatants and polymyxin B lysates from three isolates, 1AMA, 9DMA, and 11EMA, caused the elongation of CHO cells, suggesting the presence of a hitherto unknown enterotoxin (data not shown). Furthermore, CHO cell activity of these samples was not abolished after the filtrates and lysates were heated to 56°C for 30 min (data not shown). These encouraging results led us to test three of the strains, 1AMA, 2E1MA, and 31F7G, in the 6-h suckling mouse assay. An inoculum of 108 CFU per ml caused a significant fluid accumulation response comparable to that produced by V. cholerae and V. parahaemolyticus (Table 4). The fluid accumulation responses seen in suckling mice infected with V. fluvialis-like strains were dependent on both dose and strain. As shown in Table 4, 50 and 10 optical density at 650 nm U (approximately 107, 108, and 109 CFU per ml, respectively) of strain 1AMA were sufficient to cause a significant fluid response, while doses of 50 optical density at 650 nm U (approximately 109 CFU per ml) of isolate 31F7G were required to produce an equivalent response. In all cases, no microorganisms were recovered from the intestinal contents.
TABLE 4.
Fluid accumulation responses induced by different doses of V. cholerae O1 strain N16961, V. parahaemolyticus strain TX2103, and V. fluvialis-like strains 1AMA, 2E1MA, and 31F7G in suckling mice
| Sample and dosage (CFU/ml) | FA ratioa |
|---|---|
| Casamino acid-yeast extract broth (negative control) | 60.5 |
| V. parahaemolyticus TX2103, 108 | 71.8b |
| V. cholerae O1 N16961, 108 | 86.35b |
| V. fluvialis 1AMA, 109 | 76.0b |
| 108 | 76.0b |
| 107 | 71.0c |
| V. fluvialis 2E1MA, 108 | 77.0b |
| V. fluvialis 31F7G, 109 | 79.0b |
| 108 | 68.0c |
| 107 | 63.0c |
The FA ratios represent the mean ratios for six animals sacrificed 6 h after challenge. FA ratios induced by all the organism concentrations were significantly higher than those induced by buffer. FA, fluid accumulation.
Statistically significant by analysis of variance (P < 0.05).
Not a significant value.
Characterization of hemagglutinin and outer membrane complexes.
A nonproteolytic sheep hemagglutinin was found to be expressed by 94% of the isolates. The crude preparation, isolated from strain 1AMA, also possessed hemagglutination activity. This hemagglutinin was also capable of agglutinating chicken, eel, human O, and human O (tanned) erythrocytes, suggesting that a cell-associated adherence factor(s) might be present (Table 1). The crude preparation was found to be free of proteolytic activity by using a caseinolytic assay described by Kothary and Kreger (34), which suggests that the hemagglutinin is an adherence factor rather than a protease associated with the cell surface. SDS-PAGE analysis of the crude hemagglutinin revealed that it included over 30 protein bands ranging from 100 to less than 10 kDa. Three major protein bands with molecular masses of 15 to 20, 30, and 50 kDa were noted (data not shown).
Because hemagglutinins expressed by marine vibrios could be outer membrane proteins (OMPs), we decided to isolate OMP complexes from V. fluvialis 1AMA. For comparison, we also isolated OMP complexes from V. vulnificus and V. tubiashii. No hemagglutination activity or proteolytic activity could be found associated with these cellular components (data not shown). However, SDS-PAGE analysis (data not shown) revealed that the V. fluvialis OMP complexes are composed of more than eight proteins with four major protein bands (molecular masses, 43, 30, 20, and 14 kDa). In comparison, the SDS-PAGE-separated OMP complexes isolated from V. tubiashii possessed a single major protein band of 40 kDa, and the SDS-PAGE-separated OMP complexes isolated from V. vulnificus possessed two major protein bands of 35 and 40 kDa, respectively; both of these profiles differed from that of V. fluvialis, although a common protein band with similar molecular mass was noted (data not shown).
DISCUSSION
Characteristics of V. fluvialis-like bacteria isolated from lobsters with limp lobster disease.
Marine vibrios are ubiquitous in the marine environment; therefore, it is not surprising that many are pathogenic for various seafood hosts which are harvested for human consumption (14, 18, 50). Though V. fluvialis is capable of causing infrequent cases or outbreaks of human diarrhea worldwide (13, 32, 33), little is known about its ability to cause disease in other hosts. To date, only two reports have implicated V. fluvialis or V. fluvialis-like microorganisms with the causation of disease in a nonhuman host. Aguirre et al. (4) found V. fluvialis in Hawaiian green turtles (Chelonia mydas) afflicted with green turtle fibropapillomatosis. Though V. fluvialis was isolated from 47% of the turtles, its etiological relationship to green turtle fibropapillomatosis is not clear at this time. The isolation of V. fluvialis from diseased aquacultured European flounders (Pleuronectes platessa) by Pederson et al. (50) represents the first isolation of this species in Denmark and the first description of V. fluvialis linked to diseases of fish. The results described herein provide the first description of the isolation and characterization of a V. fluvialis-like organism from lobsters manifesting a newly described disease, called limp lobster disease.
Lobsters with this syndrome display weakness, lethargy, and slow or ineffectual responses to sensory stimuli. Vibrio fluvialis-like organisms were the predominant microorganisms isolated from the ill lobsters. Altogether, 19 strains were obtained. Other microorganisms isolated secondarily included Aeromonas species from two of the five culture-positive samples. However, these organisms were isolated only after MB enrichment followed by growth on TSA containing 10 μg of ampicillin per ml. This is not surprising since mixed Vibrio infections have been reported (29, 64). Of interest was the observation that only 18 of 19 isolates could grow on TCBS agar. The fact that some of the isolates could not grow on TCBS is also not surprising since other Vibrio species, such Vibrio hollisae, Vibrio damsela (now classified as Photobacterium damselae), and Vibrio metschnikovii, have also been reported not to grow or not to grow well on this medium (23, 24). Of further interest was the finding that 7 of the 18 isolates that could grow on TCBS were sucrose positive. Typically, human-derived V. fluvialis isolates are sucrose positive (23). The observation that less than half of the strains fermented sucrose is not unexpected among members of the genus Vibrio. For example, Farmer et al. (23) reported that as many as 99% of V. cholerae, but only 10 to 15% of V. vulnificus, 5% of V. damsela, and 1% of V. parahaemolyticus isolates from clinical specimens, are sucrose positive. Thus, sucrose fermentation is now being considered as a questionable taxonomic trait for the identification and isolation of V. vulnificus from TCBS (23).
Collectively, the results from the biochemical analyses suggest that the lobster V. fluvialis-like isolates taxonomically fall under Vibrio group 5 (arginine dihydrolase positive) as described by Farmer et al. (23). The presence of arginine dihydrolase activity is diagnostic for a number of species of Vibrio (12). Two other taxonomically important characteristics, production of poly-β-hyrdoxybutyrate storage granules and expression of an ensheathed flagellum, help to classify them in the genus Vibrio and exclude them from placement in the genera Photobacterium, Plesiomonas, and Aeromonas (12). The need for NaCl in the growth medium is another important trait found for a considerable number of marine vibrios and is one that defines a strict need for NaCl by these V. fluvialis-like isolates. However, the Na+ ion requirement for these organisms is currently unknown. All were able to grow in TSB supplemented with 1 to 2% NaCl, and all but one of the isolates could grow in the presence of 4% NaCl. Conversely, only 2 of the 19 isolates could grow in the presence of 8% NaCl. Salt tolerance historically has been used as a taxonomic trait to differentiate members of the genera Photobacterium and Vibrio (salt tolerant) from members of the related genera, Aeromonas and Plesiomonas (salt intolerant). The observation that all of the isolates were also sensitive to the vibriostatic compound 0/129 further separates them from members of the genus Aeromonas and from Vibrio furnissii (12).
V. fluvialis isolates were once grouped phenotypically into two biogroups (biotypes) based on the criteria put forth by Lee et al. (39). These researchers initially observed that organisms previously known as group F and group EF6 vibrios could be differentiated from each other based on two characteristic traits: gas production from the fermentation of glucose and the source or host from which they were isolated. Those organisms that were isolated from patients with diarrhea as well as from the environment and that did not produce gas during the fermentation of glucose were assigned to biogroup 1. Those organisms that were isolated only from environmental sources and also produced gas from the fermentation of glucose were placed in the biogroup 2 taxon. Brenner et al. (15) later used DNA-DNA hybridization analysis to demonstrate that strains from both biogroups were related but that strains assigned to biogroup 2 were sufficiently different from those in biogroup 1 to warrant status as a new species; they proposed that the biogroup 2 strains be named V. furnissii (15). Table 1 shows the results of the biochemical characterization of the lobster V. fluvialis-like isolates. We found that 15 of 19 lobster V. fluvialis-like isolates could produce gas from the fermentation of glucose, a trait which is exclusively positive for V. furnissii (originally biogroup 2), and that 18 of the 19 isolates could utilize salicin, a trait exclusively positive for V. fluvialis strains (originally biogroup 1). However, all of the lobster isolates could utilize neither d-glucuronate nor glutarate, which are positive traits for V. fluvialis biogroup 1 strains. Thus, on the basis of all of the foregoing biochemical test results, we propose that the lobster V. fluvialis-like isolates obtained in this study possibly embody a new biogroup for V. fluvialis. None of the isolates displayed bioluminescence as was previously described by Baumann and Schubert (12). Additionally, the organisms isolated in this study did not resemble those reported by Diggles et al. (22), which were isolated from phyllosoma larvae of the packhorse rock lobster, Jasus verreauxi, raised in an experimental culture facility. Finally, DNA-DNA hybridization experiments would help to taxonomically classify these organisms within the genus (15).
In general, antibiotic resistance is infrequently observed in Vibrio species compared with species of the family Enterobacteriaceae. Only the antibiotic susceptibilities of the clinically significant Vibrio species have been studied in detail (12, 23). Few of the V. fluvialis-like strains possessed antibiotic resistance. Similar findings were reported previously by Farmer et al. (23) and Farmer (24) for 25 strains of V. fluvialis obtained from human infections. Most (>84%) were susceptible to tetracycline, chloramphenicol, streptomycin, and kanamycin, while some were resistant to ampicillin and carbenicillin. The results reported herein are consistent with this study's findings. In comparison, Chowdhury et al. (19) tested 44 environmental and clinical strains of Vibrio mimicus and found that the environmental strains were resistant to streptomycin, kanamycin, and trimethoprim-sulfamethoxazole, while clinical strains were susceptible. They also found that environmental strains showed variable resistance to ampicillin (44%) but that clinical strains were susceptible; all strains tested were susceptible to chloramphenicol and gentamicin.
Morphologically, the V. fluvialis-like isolates possessed the typical ultrastructure of members of the family Vibrionaceae, including the expression of ensheathed polar flagella. An Alcian Blue-lysine staining technique was useful in determining the ultrastructural nature of an acidic mucopolysaccharide expressed by these organisms. These results also suggest that more than one type of capsule may be expressed as well. Many of the marine vibrios have been shown to express several different capsular types (5, 28). The chemical composition of the capsule expressed by the lobster V. fluvialis-like isolates is currently unknown. However, capsule expression by human-derived V. fluvialis strains has been known since 1983, when Shimada and Sakazaki showed the presence of a mucoid antigen which inhibited O agglutination in some strains isolated from humans (56). Though other marine vibrios, such as V. cholerae, can shift to a rugose colonial phenotype, which has been associated with expression of an amorphous exopolysaccharide promoting cell aggregation and greater resistance to antimicrobial agents (45, 46, 67). There was no evidence of the rugose colony phenotype expressed by the lobster isolates. However, strict nutrient response studies as described by Mizunoe et al. (45) and Wai et al. (67), which have been shown to induce the rugose colonial morphology, have not been performed.
Electron microscopy also showed that each cell expresses a polar ensheathed flagellum typical of that displayed by members of the genus Vibrio. Lateral flagella were not observed for these strains, although V. fluvialis isolates obtained from human clinical cases are known to possess such flagella (57). Cross-absorption analysis (58), which may reveal the extent to which the H antigens of the lobster V. fluvialis-like strains are similar to the H antigens of human V. fluvialis strains, is currently in progress.
None of the V. fluvialis-like strains grew at temperatures above 23°C, although rigid growth temperature studies were not performed. To our knowledge, this is the first report of a V. fluvialis-like bacterium whose growth resembles that of two recently described psychrophilic pathogenic marine Vibrio species, Vibrio viscosus and Vibrio wodanis (42). The latter are known to cause winter ulcers affecting salmonid fish raised in the cold coastal waters of Norway, Iceland, and Scotland (42). To date, all of the vibrios analyzed have been shown to be sensitive to heat, although wide ranges of thermal inactivation rates and conditions have been reported (49). However, with the exception of V. cholerae, V. vulnificus, and V. parahaemolyticus, relatively little is known about the susceptibility of any of the lesser-known vibrios to various food preservation methods. Cook and Ruple (20) reported that decimal reduction time of 78 s at 47°C was useful in decreasing the number of V. vulnificus in raw oysters and that heating oysters for 10 min in water at 50°C was sufficient to reduce V. vulnificus populations to undetectable levels. Thermal D values (times required to reduce the viable population of a given strain by 90%) and Z values (absolute values of the temperature required to reduce 1 log unit scale of D values) for encapsulated V. vulnificus cells were reported by Kim et al. (31) to be higher than those for unencapsulated cells. The results of both the heat-killed studies and the suckling mice challenge studies with the V. fluvialis-like strains support the strict growth temperature restriction data described above. They also suggest that these organisms probably would not survive at human body temperatures long enough to establish infection in humans.
In addition, heated and unheated cell-free culture supernatants and polymyxin B lysates from three isolates caused the elongation of CHO cells, suggesting the presence of a hitherto unknown enterotoxin (26, 37).
A crude preparation possessing hemagglutination activity was found to be free of proteolytic activity. Furthermore, heating the crude preparation at 100°C for 5 min abolished all hemagglutination activity, while exposing the hemagglutinin to trypsin did not. These results indicate that the hemagglutinin is a heat-sensitive, trypsin-resistant surface protein. Additionally, outer membrane complexes were shown to be free of proteolytic and hemagglutination activity. Together, these results suggest that the hemagglutinin is an adherence factor rather than a protease or an OMP.
Molecular characteristics of V. fluvialis-like bacteria.
PFGE analysis revealed that 18 of the 19 isolates either shared a common DNA fingerprint pattern or possessed minor variants thereof. These results suggest that the isolates represent five closely related subgroups. The fact that the epidemiological picture of the limp lobster disease outbreak was due to a group of nonclonal, but highly related, strains is not too surprising in that several diseases caused by various pathogenic marine vibrio species do not seem to be clonal. For example, Ryang et al. (55) looked at the diversity of isolates obtained from primary V. vulnificus septicemia patients in Korea. They found that, of 22 strains evaluated, 4 could not be typed by PFGE in repeated trials. The PFGE patterns of the remaining 18 strains showed a remarkable polymorphism consisting of 12 to 19 fragments (fragment size range, 20 to 870 kb). For the most part, these results showed that V. vulnificus strains isolated from Korea are genetically diversified. Because the epidemiology of V. vulnificus is so closely tied to its ecology, Singer et al. (P. D. Singer, B. D. Tall, F. M. Khambaty, and D. B. Shah, Abstr. 94th Gen. Meet. Am. Soc. Microbiol. 1994, abstr. C-417, p. 564, 1994) examined an assembly of V. vulnificus strains by PFGE analysis to determine if genomic differences existed among strains that were obtained from different environmental and clinical sources. Their results also showed extensive diversity among the V. vulnificus isolates analyzed and suggested that strains in the environment that infect humans are not dominated by derivatives from a single clone. Similar results were found by Buchrieser et al. (17) and by Tamplin et al. (62). However, an emerging debate regarding whether or not eel-pathogenic V. vulnificus isolates constitute a distinct biogroup (biogroup 2) has come to the forefront. Recent data reported by Gutacker et al. (27) suggest that only eel-pathogenic biogroup 2 strains are clonal, while eel-pathogenic V. vulnificus biogroup 1 strains are not. However, a definitive study in which PFGE analysis has been combined with other definitive population-based molecular tools, such as multilocus enzyme electrophoresis analysis, has not been done. Taken together, the results obtained in the present study indicate that limp lobster disease is probably caused by a cohort of highly related, strictly halophilic, psychrophilic V. fluvialis-like strains.
Antibiotic resistance in Vibrio species is presumably an innate rather than an acquired trait through plasmid transfer or antibiotic selection (23). Seventeen of the 19 V. fluvialis-like strains were found to possess plasmids (Table 1). Some possessed multiple plasmid bands. However, linkage of the presence of plasmids in these organisms to antibiotic resistance has not been determined. Those marine vibrios that are known to possess plasmids (drug resistance, P sex plasmids, and transferable R plasmids) include O1 and non-O1 V. cholerae, V. fluvialis, V. anguillarum, and V. parahaemolyticus strains (6-9, 11, 43). The fish pathogen V. anguillarum also harbors a virulence plasmid, pJM1, that encodes an 86-kDa OMP, OM2 (1-3). These studies showed that the OMP is necessary for iron transport and regulation and is inducible under conditions of iron limitation. Furthermore, Davidson et al. showed that plasmids were carried by 12% of 42 clinical and environmental V. vulnificus isolates (21). In contrast, these authors found that 20 of 32 (62.5%) unidentified lactose-fermenting Vibrio spp. possessed plasmids with molecular masses ranging from 2.1 to 150 MDa. There have also been cases of antibiotic resistance found in outbreaks of V. cholerae, which demonstrated a global resistance pattern to multiple antibiotics (25); Pedersen et al. (51) found that most V. anguillarum O1 strains were resistant to colistin and sensitive to ampicillin and cephalothin, while most V. anguillarum O2 strains were sensitive to colistin but resistant to ampicillin and cephalothin. There are recent reports indicating that withdrawal of antibiotics (or selective pressure) during a cholera epidemic can lead to loss of the antibiotic resistance phenotype (66). Though tetracycline or oxytetracycline is used to treat lobsters showing signs of gaffkemia, there was no evidence of any resistance to this antibiotic in the lobster V. fluvialis-like isolates. Even though initial anecdotal evidence (R. Bayer, unpublished data) that oxytetracycline could abate limp lobster disease in ill animals obtained from lobster pounds suggested early in the investigation of the outbreak that the causative agent was bacterial in character, it should be noted that the use of these drugs for mitigating this disease has not been approved. However, it was the effectiveness of these preliminary antibiotic susceptibility studies that spurred the sending of the hemolymph samples to the FDA for preliminary analyses.
Animal challenge studies.
To determine the pathogenic mechanism involved and to satisfy Koch's postulates, lobster challenge studies were performed. Results reported herein showed that V. fluvialis-like organisms could be recovered 6 h postinfection from lobsters given a dose of approximately 108 CFU/ml from both pooled heart and midgut samples at levels of 8.4 × 104 CFU/g and 3.8 × 105 CFU/g, respectively. The mean geometric peak number of organisms recovered from hemolymph was ∼3 × 104 CFU/ml. All animals presented clinical signs of limp lobster disease. Even though the reduced numbers of organisms recovered from these samples were unexpected given the relatively high inoculum size, the overall geometric mean number of organisms recovered from these samples is similar to what others have found with other marine vibrios. For example, after the oral administration of 107 CFU of V. cholerae O139 to 10 human volunteers, only 1.4 × 104 CFU/g of stool and 1.6 × 103 CFU/ml of duodenal fluid, respectively, were recovered (60). Additionally, the immune status of the lobsters used in these preliminary studies was not known. In hindsight, it would have been interesting to test whether other pathogenic marine vibrios, such as V. cholerae and V. parahaemolyticus, would have caused a similar disease. Future studies are warranted.
Production of hemolysins or cytotoxins by human-derived V. fluvialis was first reported by Lockwood et al. (41). Studies in our lab showed that a human-derived V. fluvialis strain can produce an enterotoxigenic El Tor-like cytotoxin (37). Purified preparations of such toxins have not been obtained for any of the lobster isolates. Farmer et al. (23) pointed out that only 20% of human V. fluvialis isolates yielded positive rabbit ileal loop test results for enterotoxin. However, Nishibuchi et al. (48) were able to discriminate pathogenic and toxigenic clinical strains from nonpathogenic, nontoxigenic environmental V. fluvialis strains by using the suckling mouse assay. Thus, the suckling mouse assay seems to be a sensitive assay for determining enterotoxigenicity and for discriminating between toxigenic and nontoxigenic vibrio strains. The fluid accumulation responses seen in suckling mice infected with V. fluvialis-like strains were dependent on both dose and strain. Additionally, V. fluvialis-like organisms were not recovered from intestinal contents of infected mice. These data support the 37°C thermal reduction data and suggest that the mechanism resulting in fluid accumulation in mice differs from the disease process observed in lobsters by requiring neither the persistence of viable microorganisms nor the presence a plasmid. Incidentally, there were no human cases of occupational or lobster-related illness reported during the time of the outbreak. Though the causation of human disease through intoxication is theoretically possible, the possibility is very remote and would probably be dose dependent and would most likely involve eating raw or severely undercooked lobster products.
Why did this particular disease outbreak appear? The answer to this question still remains a mystery. One possibility may be the effect of global climatic patterns on the cyclic nature of infectious diseases. Recently, a review by Lipp et al. (40) addressed many of these issues and concerns. Global climatic patterns, such as El Niño and the North Atlantic oscillation (NAO), have been shown to influence the abundance and ecology of pathogens naturally present in the environment (10, 63). Data from ice cores found in Greenland have demonstrated that the NAO has caused intermittent climatic undulations characterized by temporally active and passive phases which may affect the abundance of marine fish and zooplankton (10). It is unclear whether this effect ascends through trophic levels to top-dwelling, long-lived marine predators (63).
Other global weather patterns which have been shown to affect the marine environment include the carriage of iron-laden dust from one area of the globe to another. Data reported by Garrison et al. (25a) show that dust, heavily laden with iron and arising from the desert regions of Africa, can be transported by upper air currents created by the NAO. The dust, transported from Africa, is deposited in the Caribbean waters, ultimately stimulating planktonic blooms. Satellite data indicate that African dust plumes can reach the Eastern coast of North America and travel as far north as Greenland. A case is also made by Garrison et al. that these planktonic blooms influence the occurrence of coral bleaching disease. Furthermore, we know from the studies reported by Romalde et al. (54) that marine vibrios predominately associate with plankton species during such plankton blooms. Could a similar set of circumstances have occurred in the Gulf of Maine during the summers of 1997 and 1998? Such links between the occurrence of limp lobster disease and global weather patterns have not been established. However, to appreciate the complexity of the environmental parameters associated with the occurrence of limp lobster disease, one also needs to look at the traditional harvesting practices of Maine lobstermen. The lobster season in Maine occurs from late May through October and November. During August, the lobstermen begin to send their catch to lobster pounds. Lobsters housed in pounds (essentially a marine aquaculture technique) seem to be more susceptible to infections, possibly due to confinement and stress (52, 59). It is also typical for the lobstermen in Maine to set their traps according to the natural migration patterns of lobsters within the Gulf of Maine. By the end of July, the lobstermen have set the maximum number of traps that is feasibly manageable. Because of the large number of traps that are set (in some cases several hundred per lobsterman), the traps tend to remain in the water longer between harvest dates, essentially setting up a confined aquaculture situation similar to that found in the lobster pounds and thus creating the optimal conditions for the transmission and sustenance of disease. This scenario could explain why limp lobster disease was first observed in lobsters housed in pounds and later in lobsters harvested directly from traps. It is not clear at this time if conditions optimal for the V. fluvialis-like pathogen to establish the disease were first present in the pounds or could have existed in the wild.
Is there a correlation between the pathogenesis, emergence, and persistence of this strictly halophilic marine vibrio and its stenothermal growth temperature requirements? Were there other human activities associated with the occurrence of the epidemic? What is the natural life history of the disease? Are other hosts involved? All of these questions remain unresolved.
In summary, we have biochemically and molecularly characterized a group of V. fluvialis-like isolates obtained from ill lobsters afflicted with a newly described systemic disease. Analysis of the isolates by PFGE revealed five closely related subgroups. We have also provided evidence for the satisfaction of Koch's postulates at both the organismal and molecular levels, supporting the hypothesis that this microorganism is the etiological agent responsible for this disease, now known as limp lobster disease. Although V. fluvialis has emerged as a pathogen capable of infecting fish and humans, we now report for the first time its ability to cause disease in crustaceans, a finding that poses a significant threat to animal and economic health, thereby meriting additional studies. Understanding how this organism is able to overcome species barriers and adapt to new hosts is crucial in producing disease-free seafoods.
Acknowledgments
We thank Maya Crosby of the University of Maine at Orono for help in obtaining the initial hemolymph samples. We also thank Manashi Dey for help with the initial PFGE analysis.
REFERENCES
- 1.Actis, L. A., S. A. Potter, and J. H. Crosa. 1985. Iron-regulated outer membrane protein Om2 of Vibrio anguillarum is encoded by virulence plasmid Pjm1. J. Bacteriol. 161:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1995. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 17:197-204. [DOI] [PubMed] [Google Scholar]
- 3.Actis, L. A., M. E. Tolmasky, D. H. Farrell, and J. H. Crosa. 1988. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J. Biol. Chem. 263:2853-2860. [PubMed] [Google Scholar]
- 4.Aguirre, A. A., G. H. Balazs, B. Zimmerman, and T. R. Spraker. 1994. Evaluation of Hawaiian green turtles (Chelonia mydas) for potential pathogens associated with fibropapillomas. J. Wildl. Dis. 30:8-15. [DOI] [PubMed] [Google Scholar]
- 5.Amako, K., K. Okada, and S. Miake. 1984. Evidence for the presence of a capsule in Vibrio vulnificus. J. Gen. Microbiol. 130:2741-2743. [DOI] [PubMed] [Google Scholar]
- 6.Aoki, T. 1988. Drug-resistant plasmids from fish pathogens. Microbiol Sci. 5:219-223. [PubMed] [Google Scholar]
- 7.Aoki, T., T. Arai, and S. Egusa. 1976. R factors detected from Vibrio anguillarum and marine Vibrio spp., p. 223-228. In S. Mitsuhashi and J. Hashimoto (ed.), Microbial drug resistance. University Park Press, Baltimore, Md.
- 8.Aoki, T., T. Kitao, S. Watanabe, and S. Takeshita. 1984. Drug resistance and R plasmids in Vibrio anguillarum isolated in cultured ayu (Plecoglossus altivelis). Microbiol. Immunol. 28:1-9. [DOI] [PubMed] [Google Scholar]
- 9.Aoki, T., T. Satoh, and T. Kitao. 1987. New tetracycline resistance determinant on R plasmids from Vibrio anguillarum. Antimicrob. Agents Chemother. 31:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appenzeller, C., T. F. Stocker, and M. Anklin. 1998. North Atlantic oscillation dynamics recorded in Greenland ice cores. Science 282:446-449. [DOI] [PubMed] [Google Scholar]
- 11.Barja, J. L., Y. Santos, I. Huq, R. R. Colwell, and A. E. Toranzo. 1990. Plasmids and factors associated with virulence in environmental isolates of Vibrio cholerae nOn-O1 in Bangladesh. J. Med. Microbiol. 33:107-114. [DOI] [PubMed] [Google Scholar]
- 12.Baumann, P., and R. H. W. Schubert. 1984. Family II. Vibrionaceae, p. 516-544. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams and Wilkins, Baltimore, Md.
- 13.Bellet, J., B. Klein, M. Altieri, and D. Ochsenschlager. 1989. Vibrio fluvialis, an unusual pediatric enteric pathogen. Pediatr. Emerg. Care. 5:27-28. [DOI] [PubMed] [Google Scholar]
- 14.Binta, M. G., and P. N. Nyaga. 1982. The distribution of Vibrio parahaemolyticus serotypes in Kenyan seafish, shellfish, marine water and sediment. Trans. R. Soc. Trop. Med. Hyg. 76:497-499. [DOI] [PubMed] [Google Scholar]
- 15.Brenner, D. J., F. W. Hickman-Brenner, J. V. Lee, A. G. Steigerwalt, G. R. Fanning, D. G. Hollis, J. J. Farmer, R. E. Weaver, S. W. Joseph, and R. J. Seidler. 1983. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J. Clin. Microbiol. 18:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brinkley, A. W., F. A. Rommel, and T. W. Huber. 1976. The isolation of Vibrio parahaemolyticus and related vibrios from moribund aquarium lobsters. Can. J. Microbiol. 22:315-317. [DOI] [PubMed] [Google Scholar]
- 17.Buchrieser, C., V. V. Gangar, R. L. Murphree, M. L. Tamplin, and C. W. Kaspar. 1995. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl. Environ. Microbiol. 61:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan, K. Y., M. L. Woo, L. Y. Lam, and G. L. French. 1989. Vibrio parahaemolyticus and other halophilic vibrios associated with seafood in Hong Kong. J. Appl. Bacteriol. 66:57-64. [DOI] [PubMed] [Google Scholar]
- 19.Chowdhury, M. A., K. M. Aziz, Z. Rahim, and B. A. Kay. 1986. Antibiotic resistance patterns of Vibrio mimicus isolated from human and environmental sources in Bangladesh. Antimicrob. Agents Chemother. 30:622-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook, D. W., and A. D. Ruple. 1992. Cold storage and mild heat treatment as processing aids to reduce the numbers of Vibrio vulnificus in raw oysters. J. Food Prot. 55:985-989. [DOI] [PubMed] [Google Scholar]
- 21.Davidson, L. S., and J. D. Oliver. 1986. Plasmid carriage in Vibrio vulnificus and other lactose-fermenting marine vibrios. Appl. Environ. Microbiol. 52:211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diggles, B. K., G. A. Moss, J. Carson, and C. D. Anderson. 2000. Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Dis. Aquat. Organ. 43:127-137. [DOI] [PubMed] [Google Scholar]
- 23.Farmer, J. J., F. W. Hickman-Brenner, and M. T. Kelly. 1985. Vibrio, p. 282-301. In E. H. Lennette, A. Ballows, W. J. Hausler, Jr., and H. J. Shadomy (ed.), Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, D.C.
- 24.Farmer, J. J., III. 1992. The family Vibrionaceae, p. 2938-2951. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 24a.Fassel, T. A., J. R. Sanger, and C. E. Edminston. 1993. Lysine effect on Ruthenium Red and Alcian Blue preservation of the staphylococci glycocalyx, p. 374-375. In G. W. Bailey, and C. L. Rieder (ed.), Proceedings of the 51st Annual Meeting of the Microscopy Society of America, San Francisco Press, San Francisco, Calif.
- 25.Finch, M. J., J. G. Morris, Jr., J. Kaviti, W. Kagwanja, and M. M. Levine. 1988. Epidemiology of antimicrobial-resistant cholera in Kenya and East Africa. Am. J. Trop. Med. Hyg. 39:484-490. [DOI] [PubMed] [Google Scholar]
- 25a.Garrison, V., W. Foreman, M. Majewski, C. Holmes, E. Shinn, R. Smith, and M. Ranneberger. 2002. Timbuktu meets coral reefs: chemical contaminants in African dust, p. 31. In R. C. Cipriano (ed.), Proceedings of the 27th Annual Eastern Fish Health Workshop. U.S. Geological Survey, Kearneysville, W.Va.
- 26.Guerrant, R. L., L. L. Brunton, T. C. Schnaitman, L. L. Rebhun, and A. G. Gilman. 1974. Cyclic adenosine monophosphate and alteration of Chinese hamster ovary cell morphology: a rapid, sensitive in vitro assay for the enterotoxins of Vibrio cholerae and Escherichia coli. Infect. Immun. 10:320-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutacker, M., N. Conza, C. Benagli, A. Pedroli, M. V. Bernasconi, L. Permin, R. Aznar, and J. C. Piffaretti. 2003. Population genetics of Vibrio vulnificus: identification of two divisions and a distinct eel-pathogenic clone. Appl. Environ. Microbiol. 69:3203-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayat, U., G. P. Reddy, C. A. Bush, J. A. Johnson, A. C. Wright, and J. G. Morris, Jr. 1993. Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J. Infect. Dis. 168:758-762. [DOI] [PubMed] [Google Scholar]
- 29.Hickman-Brenner, F. W., D. J. Brenner, A. G. Steigerwalt, M. Schreiber, S. D. Holmberg, L. M. Baldy, C. S. Lewis, N. M. Pickens, and J. J. Farmer. 1984. Vibrio fluvialis and Vibrio furnissii isolated from a stool sample of one patient. J. Clin. Microbiol. 20:125-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston, K. H., K. K. Holmes, and E. C. Gotschlich. 1976. The serological classification of Neisseria gonorrhoeae. I. Isolation of the outer membrane complex responsible for serotypic specificity. J. Exp. Med. 143:741-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, C. M., K. C. Jeong, J. H. Rhee, and S. H. Choi. 1997. Thermal-death times of opaque and translucent morphotypes of Vibrio vulnificus. Appl. Environ. Microbiol. 63:3308-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klontz, K. C., and J. C. Desenclos. 1990. Clinical and epidemiological features of sporadic infections with Vibrio fluvialis in Florida, USA. J. Diarrhoeal Dis. Res. 8:24-26. [PubMed] [Google Scholar]
- 33.Kolb, E. A., S. C. Eppes, and J. D. Klein. 1997. Vibrio fluvialis: an underrecognized enteric pathogen in infants? South. Med. J. 90:544-545. [DOI] [PubMed] [Google Scholar]
- 34.Kothary, M. H., and A. S. Kreger. 1987. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J. Gen. Microbiol. 133:1783-1791. [DOI] [PubMed] [Google Scholar]
- 35.Kothary, M. H., R. B. Delston, B. A. McCardell, S. K. Curtis, and B. D. Tall. 2001. Purification and characterization of a vulnificolysin-like cytolysin produced by Vibrio tubiashii. Appl. Environ. Microbiol. 67:3703-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kothary, M. H., and S. H. Richardson. 1987. Fluid accumulation in infant mice caused by Vibrio hollisae and its extracellular enterotoxin. Infect. Immun. 55:626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kothary, M. H., H. Lowman, B. A. McCardell, and B. D. Tall. 2003. Purification and characterization of enterotoxigenic El Tor-like hemolysin produced by Vibrio fluvialis. Infect. Immun 71:3213-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 39.Lee, J. V., P. Shread, A. L. Furniss, and T. N. Bryant. 1981. Taxonomy and description of Vibrio fluvialis sp. nov. (synonym group F vibrios, group EF6). J. Appl. Bacteriol. 50:73-94. [DOI] [PubMed] [Google Scholar]
- 40.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lockwood, D. E., A. S. Kreger, and S. H. Richardson. 1982. Detection of toxins produced by Vibrio fluvialis. Infect. Immun. 35:702-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lunder, T., H. Sorum, G. Holstad, A. G. Steigerwalt, P. Mowinckel, and D. J. Brenner. 2000. Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with ‘winter ulcers.' Int. J. Syst. Evol. Microbiol. 50:427-450. [DOI] [PubMed] [Google Scholar]
- 43.Matsushita, S., S. Yamada, Y. Kudoh, and M. Ohashi. 1987. Drug-resistance and transferable R plasmids in Vibrio cholerae O-1 and non O-1, Vibrio fluvialis and Vibrio parahaemolyticus recently isolated from human sources. Kansenshogaku Zasshhi 61:109-117. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy, S. A., and F. M. Khambaty. 1994. International dissemination of epidemic Vibrio cholerae by cargo ship ballast and other nonpotable waters. Appl. Environ. Microbiol. 60:2597-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizunoe, Y., S. N. Wai, A. Takade, and S. I. Yoshida. 1999. Isolation and characterization of rugose form of Vibrio cholerae O139 strain MO10. Infect. Immun. 67:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174:1364-1368. [DOI] [PubMed] [Google Scholar]
- 47.National Committee for Clinical Laboratory Standards. 1999. NCCLS document M100-S9. Performance standards for antimicrobial susceptibilities testing—9th informational supplement. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 48.Nishibuchi, M., R. J. Seidler, D. M. Rollins, and S. W. Joseph. 1983. Vibrio factors cause rapid fluid accumulation in suckling mice. Infect. Immun. 40:1083-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliver, J. D., and J. B. Kaper. 1997. Vibrio species, p. 228-264. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology fundamentals and frontiers. ASM Press, Washington, D.C.
- 50.Pedersen, K., B. Austin, D. A. Austin, and J. L. Larsen. 1999. Vibrios associated with mortality in cultured plaice Pleuronectes platessa fry. Acta Vet. Scand. 40:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen, K., T. Tiainen, and J. L. Larsen. 1995. Antibiotic resistance of Vibrio anguillarum, in relation to serovar and plasmid contents. Acta Vet. Scand. 36:55-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabin, H. 1965. Studies on gaffkemia, a bacterial disease of the American lobster, Homarus americanus (Milne-Edwards). J. Invertebr. Pathol. 7:391-397. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds, E. S. 1963. The use of lead citrate at high Ph as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romalde, J. L., J. L. Barja, and A. E. Toranzo. 1990. Vibrios associated with red tides caused by Mesodinium rubrum. Appl. Environ. Microbiol. 56:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryang, D. W., S. B. Koo, M. G. Shin, J. H. Shin, and S. P. Suh. 1999. Molecular typing of Vibrio vulnificus isolated from clinical specimens by pulsed-field gel electrophoresis and random amplified polymorphic DNA analysis. Jpn. J. Infect. Dis. 52:38-41. [PubMed] [Google Scholar]
- 56.Shimada, T., and R. Sakazaki. 1983. Serological studies on Vibrio fluvialis. Jpn. J. Med. Sci. Biol. 36:315-323. [DOI] [PubMed] [Google Scholar]
- 57.Shinoda, S., N. Nakahara, and H. Kane. 1984. Lateral flagellum of Vibrio fluvialis: a species-specific antigen. Can. J. Microbiol. 30:1525-1529. [DOI] [PubMed] [Google Scholar]
- 58.Simonson, J., and R. J. Siebeling. 1986. Rapid serological identification of Vibrio vulnificus by anti-H coagglutination. Appl. Environ. Microbiol. 52:1299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart, J. E., B. Arie, B. M. Zwicker, and J. R. Dingle. 1969. Gaffkemia, a bacterial disease of the lobster, Homarus americanus: effects of the pathogen, Gaffkya homari, on the physiology of the host. Can. J. Microbiol. 15:925-932. [DOI] [PubMed] [Google Scholar]
- 60.Tacket, C. O., R. K. Taylor, G. Losonsky, Y. Lim, J. P. Nataro, J. B. Kaper, and M. M. Levine. 1998. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect. Immun. 66:692-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tall, B. D., M. H. Kothary, M. D. Miliotis, O. L. McGrane, and D. B. Shah. 1997. Expression of fibrillae by Vibrio vulnificus which resembles the pH 6-antigen of Yersinia pestis, p. 265-273. In G. T. Keusch and M. Kawakami (ed.), Cytokines, cholera and the gut. IOS Press/Ohmaha, Ltd., Burke, Va.
- 62.Tamplin, M. L., J. K. Jackson, C. Buchrieser, R. L. Murphree, K. M. Portier, V. Gangar, L. G. Miller, and C. W. Kaspar. 1996. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, P. M., and J. C. Ollason. 2001. Lagged effects of ocean climate change on fulmar population dynamics. Nature 413:417-420. [DOI] [PubMed] [Google Scholar]
- 64.Tokoro, M., M. Kato, K. Goto, M. Watanabe, F. Yamada, T. Sako, K. Otsuka, O. Sugiyama, M. Furukawa, and S. Niwa. 1984. Community outbreaks of mixed food-borne infection with Vibrio parahaemolyticus and Vibrio fluvialis. Kansenshogaku Zasshhi 58:1038-1045. [DOI] [PubMed] [Google Scholar]
- 65.Traub, W. H., B. Leonhard, and D. Bauer. 1998. Antibiotic susceptibility of Stenotrophomonas (Xanthomonas) maltophilia: comparative (NCCLS criteria) evaluation of antimicrobial drugs with the agar dilution and the agar disk diffusion (Bauer-Kirby) tests. Chemotherapy 44:164-173. [DOI] [PubMed] [Google Scholar]
- 66.Urassa, W., E. Lyamuya, and F. Mhalu. 1997. Recent trends on bacterial resistance to antibiotics. East Afr. Med. J. 74:129-133. [PubMed] [Google Scholar]
- 67.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber, K., J. R. Pringle, and M. Osborn. 1972. Measurement of molecular weights by electrophoresis on SDS-acrylamide gel. Methods Enzymol. 26:3-27. [DOI] [PubMed] [Google Scholar]