Abstract
The costs and efforts for sample preparation of hundreds of individuals, their genomic enrichment for regions of interest, and sufficient deep sequencing bring a significant burden to next-generation sequencing-based experiments. We investigated whether pooling of samples at the level of genomic DNA would be a more versatile strategy for lowering the costs and efforts for common disease-associated rare variant detection in candidate genes or associated loci in a substantial patient cohort. We performed a pilot experiment using five pools of 20 abdominal aortic aneurysm (AAA) patients that were enriched on separate microarrays for the reported 9p21.3 associated locus and 42 additional AAA candidate genes, and sequenced on the SOLiD platform. Here, we discuss challenges and limitations connected to this approach and show that the high number of novel variants detected per pool and allele frequency deviations to the usually highly false positive cut-off region for variant detection in non-pooled samples can be limiting factors for successful variant prioritization and confirmation. We conclude that barcode indexing of individual samples before pooling followed by a multiplexed enrichment strategy should be preferred for detection of rare genetic variants in larger sample sets rather than a genomic DNA pooling strategy.
Keywords: Abdominal aortic aneurysm, Common disease, Rare variants, Targeted genomic enrichment, Genomic DNA pooling, SOLiD next-generation sequencing
Introduction
Abdominal aortic aneurysm (AAA) is classified as an increase in the aortic diameter of ≥50% or an infrarenal diameter of ≥30 mm and can be asymptomatic until a rupture event with a high chance of mortality occurs [1]. The main risk factors include advanced age, male gender, smoking, other cardiovascular disease, and positive family history [2–4]. AAA is therefore considered a multi-factorial disorder in which both environmental and genetic factors are believed to contribute to its pathogenesis [5]. In spite of a number of familial [6–9] and population studies [10–15], the major part of the heritability of AAA formation in general is not explained and a better understanding of genetic variants contributing to this phenotype is required.
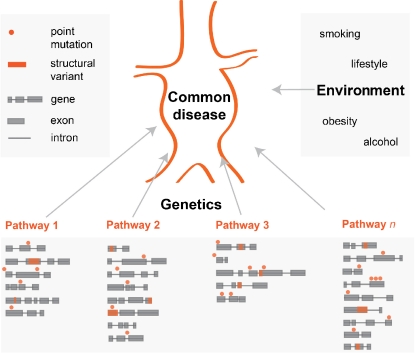
Although the impact of common–higher allele frequency variants was shown to be significant [16], genetic models of common diseases changed with recent discoveries and possibilities of next-generation sequencing (NGS). The field has widely accepted the common disease/common variant hypothesis but now tends more to common disease/rare variant hypothesis [17, 18]. Common diseases can be explained by a so-called “pathway model” (Fig. 1). This model assumes that disease of a specific organ or tissue can be caused by impairment of any pathway influencing the physiological function of the organ or tissue. Each pathway contains several genes and rare variants in relevant genes can contribute to the onset of the disease.
Fig. 1.
Schematic overview of the pathway model for common diseases. The pathway model of common diseases assumes that disease of a specific organ or tissue can be caused by impairment of any pathway influencing physiological function of the organ or tissue. Each pathway contains several genes and any point mutation or structural variation in relevant genes can contribute to the onset of the disease. Additionally, not only genetic but also intergenic mutations having a trans-effect on protein function should be considered. In addition to genetic factors, also environmental factors are expected to have a significant effect
Targeted genomic enrichment approaches with NGS enable deep sequencing of any complex genomic region of interest and may be a straightforward approach for detecting causal variants in common diseases and contribute to the understanding of their pathogenesis. However, the costs and efforts for library preparation and enrichment of larger sample sets necessary to discover enough rare variants to build pathway models for common disease are significant. Therefore, we explored a cost-reducing strategy by pooling of samples at the level of genomic DNA with subsequent library preparation, genomic enrichment, and sequencing of patient pools rather than individual samples. The identified candidate variants are subsequently confirmed by capillary sequencing of individual samples from a pool.
The strategy for the pooled DNA sequencing and its analysis differs from standard non-pooled NGS strategies. Accurate equimolar pooling of each genomic DNA is important for equal distribution of reads and the number of pooled samples should be balanced for successful variant detection. In this study, we focused on the discovery of rare variants in the replicable AAA genome-wide association study (GWAS) locus on 9p21 and 42 AAA candidate genes. We used a group of 100 well-characterized Dutch AAA patients that were divided into five pools of 20 individual genomic DNA samples. To detect a single heterozygote variant in the presence of 40 chromosomes, the variant calling thresholds for the non-reference allele percentage (NRA%) needed to be lowered to ~2.5%. We found that at this cut-off level, the majority of detected rare variants were false positives, indicating that lower pooling depths or individual indexing of samples is the preferred strategy.
Material and Methods
Patient Selection
The AAA samples in this study (n = 100) were collected during a recruitment effort in the years 2007–2009 by the Department of Medical Genetics, University Medical Center Utrecht, The Netherlands from eight large AAA-treating centers in the Netherlands. This sample collection was previously used for the discovery of common variants associated with AAA [10, 11, 13, 15]. Blood for genomic DNA isolation was obtained mainly when individuals visited their vascular surgeon in the polyclinic or, in rare cases, during hospital admission for elective or emergency AAA surgery. An AAA was diagnosed if the diameter of the infrarenal aorta was ≥30 mm. The sample set comprised 84% males, with a mean AAA diameter of 58.4 mm, 70% had received surgery, of which 2% was emergency. Of the patients, 59% reported a history of smoking, 29% had a familial history of AAA, 65% were diagnosed with hypertension, and 58% with other cardiovascular disease. The average age of patients was 76 years. Patients were divided per five pools of 20 according to sharing or absence of at least one major risk factor for AAA formation as follows: (1) never smoked, (2) family history of AAA, (3) other reported cardiovascular disease, (4) hypertension, and (5) mixed group (Table 1).
Table 1.
Clinical information about the AAA patient set
| Patient pool | Diameter of aorta [mm]a | Gender f/m | Operated | Rupture | Ever smoked | Hypertension | Cardiovascular disease | Familial AAA |
|---|---|---|---|---|---|---|---|---|
| 1 | 59.9 | 5/15 | 16 | 0 | 0 | 13 | 10 | 8 |
| 2 | 61.2 | 4/16 | 14 | 0 | 14 | 14 | 8 | 20 |
| 3 | 60.8 | 0/20 | 15 | 0 | 20 | 8 | 20 | 0 |
| 4 | 51.4 | 2/18 | 11 | 0 | 12 | 20 | 11 | 1 |
| 5 | 60.6 | 2/18 | 14 | 2 | 13 | 10 | 9 | 0 |
aDiameter of infrarenal aorta. The cut-off for diagnosis of AAA was set at 30 mm.
Array Design
The list of genomic sequences for array design was divided into four groups: (1) the whole 9p21.3 locus detected by GWAS (the only replicable locus at the time of our array design), including full genes and intergenic regions, (2) coding sequences of pathway candidates from genes in the 9p21.3 locus, (3) coding sequences of genes from the TGF-beta pathway, and (4) coding sequences of additional candidate genes based on current literature (Table 2). The probe selection strategy was set using our previously described criteria [19]. The small target size allowed for a dense design where a capture probe starts on average every other base. For every 2-bp window, a single 60 bp long probe with the lowest penalty score was selected, resulting in average 30× probe coverage for both the negative and positive strand. To exclude potentially repetitive elements from the design, all probes were compared to the reference genome GRCh37/hg19 using BLAST and those returning more than one additional hit (as defined by >60% matching region) were discarded from the design. The final array design consisted of 425,892 uniquely mapping probes resulting into a design footprint of 498,658 bp. We define as “Request” the set of genomic positions of interest and “Design” as the genomic positions, which were covered by enrichment probes. Due to repetitive elements and low complexity in the intergenic and intronic regions of the 9p21.3 locus, 57.4% of all requested genomic positions and 99.6% of the requested protein-coding regions were covered by capture probes. Probes were synthesized on custom 1 Mb array (Agilent Technologies, CA, USA, five copies) with randomized positioning.
Table 2.
Overview of the locus and genes included into array design
| Gene name | Gene description | Ensemble gene name | GRCh37 location |
|---|---|---|---|
| 1. 9p21.3 locus | Chromosome 9: 21,750,000–22,400,000 | ||
| Containing genes | |||
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | ENSG00000147889 | Chromosome 9: 21,967,752–21,995,300 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | ENSG00000147883 | Chromosome 9: 22,002,902–22,009,280 |
| CDKN2BAS | CDKN2B antisense RNA (non-protein coding) | ENSG00000240498 | Chromosome 9: 21,994,790–22,121,096 |
| 2. CDK pathway | |||
| CDK1 | Cyclin-dependent kinase 1 | ENSG00000170312 | Chromosome 10: 62,538,101–62,554,610 |
| CDK2 | Cyclin-dependent kinase 2 | ENSG00000123374 | Chromosome 12: 56,360,556–56,366,565 |
| CDK3 | Cyclin-dependent kinase 3 | ENSG00000108504 | Chromosome 17: 73,975,312–74,002,080 |
| CDK4 | Cyclin-dependent kinase 4 | ENSG00000135446 | Chromosome 12: 58,142,005–58,146,164 |
| CDK5 | Cyclin-dependent kinase 5 | ENSG00000164885 | Chromosome 7: 150,750,899–150,754,996 |
| CDK6 | Cyclin-dependent kinase 6 | ENSG00000105810 | Chromosome 7: 92,234,239–92,465,941 |
| CDK7 | Cyclin-dependent kinase 7 | ENSG00000134058 | Chromosome 5: 68,530,622–68,573,256 |
| CDKN2C | Cyclin-dependent kinase inhibitor 2C | ENSG00000123080 | Chromosome 1: 51,426,417–51,440,309 |
| CDKN2D | Cyclin-dependent kinase inhibitor 2D | ENSG00000129355 | Chromosome 19: 10,677,139–10,679,655 |
| TP53 | Tumor protein p53 | ENSG00000141510 | Chromosome 17: 7,565,257–7,590,863 |
| MDM1 | Mdm1 nuclear protein homolog (mouse) | ENSG00000111554 | Chromosome 12: 68,688,346–68,726,161 |
| MDM2 | Mdm2 p53 binding protein homolog (mouse) | ENSG00000135679 | Chromosome 12: 69,201,980–69,234,214 |
| 3. TGFbeta pathway | |||
| TGFB1 | Transforming growth factor, beta 1 | ENSG00000105329 | Chromosome 19: 41,836,651–41,859,816 |
| TGFB2 | Transforming growth factor, beta 2 | ENSG00000092969 | Chromosome 1: 218,519,391–218,617,959 |
| TGFB3 | Transforming growth factor, beta 3 | ENSG00000119699 | Chromosome 14: 76,424,442–76,448,092 |
| FBN1 | Fibrillin 1 | ENSG00000166147 | Chromosome 15: 48,700,505–48,937,918 |
| LTBP1 | Latent transforming growth factor beta binding protein 1 | ENSG00000049323 | Chromosome 2: 33,359,706–33,624,576 |
| THBS1 | Thrombospondin 1 | ENSG00000137801 | Chromosome 15: 39,873,280–39,889,665 |
| DCN | Decorin | ENSG00000011465 | Chromosome 12: 91,539,036–91,576,806 |
| ACVRL1 | Activin A receptor type II-like 1 | ENSG00000139567 | Chromosome 12: 52,301,202–52,317,145 |
| ACVR1B | Activin A receptor, type IB | ENSG00000135503 | Chromosome 12: 52,345,486–52,390,857 |
| TGFBR1 | Transforming growth factor, beta receptor I | ENSG00000106799 | Chromosome 9: 101,867,412–101,916,474 |
| TGFBR2 | Transforming growth factor, beta receptor II | ENSG00000163513 | Chromosome 3: 30,647,994–30,735,634 |
| TGFBR3 | Transforming growth factor, beta receptor III | ENSG00000069702 | Chromosome 1: 92,145,900–92,371,559 |
| ENG | Endoglin | ENSG00000106991 | Chromosome 9: 130,577,291–130,617,047 |
| SMAD2 | SMAD family member 2 | ENSG00000175387 | Chromosome 18: 45,359,466–45,456,926 |
| SMAD3 | SMAD family member 3 | ENSG00000166949 | Chromosome 15: 67,358,195–67,487,532 |
| SMAD4 | SMAD family member 4 | ENSG00000141646 | Chromosome 18: 48,556,583–48,611,415 |
| SMAD6 | SMAD family member 6 | ENSG00000137834 | Chromosome 15: 66,994,634–67,074,323 |
| SMAD7 | SMAD family member 7 | ENSG00000101665 | Chromosome 18: 46,446,224–46,477,081 |
| 4. Other candidate genes | |||
| ELN | Elastin | ENSG00000049540 | Chromosome 7: 73,442,119–73,484,237 |
| ACTA2 | Actin, alpha 2, smooth muscle, aorta | ENSG00000107796 | Chromosome 10: 90,694,831–90,751,147 |
| MYH11 | Myosin, heavy chain 11, smooth muscle | ENSG00000133392 | Chromosome 16: 15,796,992–15,950,890 |
| ACE | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | ENSG00000159640 | Chromosome 17: 61,554,432–61,599,209 |
| NOS | Nitric oxide synthase 3 (endothelial cell) | ENSG00000164867 | Chromosome 7: 150,688,083–150,711,676 |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) | ENSG00000087245 | Chromosome 16: 55,512,883–55,540,603 |
| MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | ENSG00000100985 | Chromosome 20: 44,637,547–44,645,200 |
| TIMP1 | TIMPmetallopeptidase inhibitor 1 | ENSG00000102265 | Chromosome X: 47,441,712–47,446,188 |
| MTHFR | Methylenetetrahydrofolate reductase (NAD(P)H) | ENSG00000177000 | Chromosome 1: 11,845,780–11,866,977 |
| ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | ENSG00000165029 | Chromosome 9: 107,543,283–107,690,518 |
| ABCB5 | ATP-binding cassette, sub-family B (MDR/TAP), member 5 | ENSG00000004846 | Chromosome 7: 20,654,830–20,816,658 |
| ABCC6 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 | ENSG00000091262 | Chromosome 16: 16,243,422–16,317,328 |
Library Preparation
Genomic DNA concentration of each sample was determined using the Quant-iT™ PicoGreen ® dsDNA method (Invitrogen). After measurements, 200 ng of each sample was used for subsequent equimolar pooling (five pools of 20 patients) at the level of genomic DNA and 4 μg of each pool was used as input material for preparation of five fragment libraries and targeted genomic enrichment on array for the SOLiD NGS platform as described previously [19]. In brief, each genomic DNA pool was fragmented using Covaris™ S2 System (Applied Biosystems) to 100–150 bp short fragments. After fragmentation, fragments were blunt-ended and phosphorylated at the 5′ end using End-It™ DNA End-Repair Kit (EpiCenter) for 30 min. at RT. Samples were then purified with the Agencourt AMPure XP system (Beckman Coulter Genomics) followed by ligation of double-stranded truncated versions of adaptors complementary to the SOLID next generation sequencing platform. Adaptors were prepared by hybridization of complementary HPLC-purified oligonucleotides. Ligation was performed using Quick Ligation Kit (New England BioLabs) for 10 min. at RT. After purification with the AMPure system, nick translation on non-phosphorylated and non-ligated 3′-ends and amplification in single polymerase chain reaction (PCR) reaction was done for each library separately using primer 1 complementary to adaptor 1 (5′-CTA TGG GCA GTC GGT GAT-3′) and primer 2 complementary to adaptor 2 (5′-GCT GTA CGG CCA AGG CG-3) at 72°C for 5 min for nick translation and followed by 95°C for 5 min, five cycles: 95°C for 15 s, 54°C for 15 s, 70°C for 1 min, 70°C for 4 min and 4°C hold. Intensity of library bands was checked on 2% agarose gel (Lonza FlashGel System). PCR products were purified with AMPure system to remove adaptor dimmers and heterodimmers. Amplified library fragments ranging 120–200 bp were size selected on 4% agarose gel and gel slices were purified using QIAquick Gel Extraction Kit (Qiagen). After size selection, additional 12 PCR cycles using aforementioned PCR program were performed to obtain enough material for array enrichment.
Array-based Targeted Genomic Enrichment
For array-based targeted genomic enrichment of DNA libraries, we used an optimized protocol using enrichment arrays and elution settings from Agilent Technologies and NimbleGen hybridization and washing buffers and conditions [19, 20]. Of each library, 2,500 ng was mixed with 5× weight excess of human Cot-1 DNA (Invitrogen) for nonspecific hybridization, and concentrated using a speedvac to a final volume of 12.3 μl. DNA was mixed with NimbleGen Hybridization Kit and denatured at 95°C for 5 min. After denaturing, each pool library was hybridized on a separate enrichment array for 65 h at 42°C on a four-bay MAUI hybridization station using an active mixing MAUI AO chamber (BioMicro Systems). After hybridization, arrays were washed using the Nimblege Wash Buffer Kit (Roche) according to the user’s guide for aCGH hybridization. The temperature of Wash buffer I for library 2 was 42°C instead of room temperature. Elution was performed using 800 ml of elution buffer (10 mM Tris pH 8.0) in a DNA Microarray Hybridization Chamber–SureHyb (Agilent) at 95°C for 30 min. After 30 min, the chamber was quickly disassembled and elution buffer was collected into a separate 1.5 ml tube. Eluted ssDNA libraries were concentrated in a speedvac to a final volume of 30 μl. Each eluate has been used for post-hybridization PCR for introducing the full length SOLiD adaptor P1 and multiplex adaptor P2 with a barcode sequence (BC7, 9, 10, 11, 12). Primer 3 (5′-CCA CTA CGC CTC CGC TTT CCT CTC-3′) and Primer 4 (5′-CTG CCC CGG GTT CCT CAT TCT CTN NNN NNN NNN CTG CTG TAC GGC CAA GGC G-3′, where N represents unique sequence for each barcode) were used for 12 cycles in the above-mentioned PCR program. PCR products were purified with the MinElute PCR Purification Kit (Qiagen).
Sequencing, Variant Detection and Analysis
SOLiD sequencing was performed according to the SOLiD v3+ manual. Before emulsion PCR, all five pool libraries were quantified with the Quant-iT™ dsDNA HS Assay Kit (Invitrogen) to obtain equal amount of reads per pool. All five enriched pools were deposited on one sequencing slide and sequenced using SOLiD v3+ system to produce enough 50 bp reads to obtain sufficient coverage for a single allele. Color space reads were mapped against the GRCh37/hg19 reference genome for Human with BWA software [21]. Single nucleotide variants (SNVs) and small indels (≤7nt) were called by our custom analysis pipeline as described before [20] (all scripts available upon request). All common and rare polymorphisms present in Ensembl59 were tagged as known; other variants were considered to be novel. The criteria for variant detection were set as follows: minimally three independent reads on the positive and three on the negative strand, cut-off for coverage was set to 500 reads and cut-off for NRA% was set to 3%. To allow detection of one variant allele in a pool of 40 alleles the cut-off for NRA% should be lowered to theoretical 2.5%. However, this increased the number of detected variants exponentially and we empirically determined that 3% NRA gives a reasonable number of novel candidate variants. We included also a clonality filter that keeps maximally five clonal reads with the same start site and removes reads above this level. This clonality filter was already used in our previous reports [19, 20]. For each variant, location in genomic sequence, amino acid change, effect on the protein function, and conservation score were collected and subsequently used for prioritization for candidate variants. Confirmation of selected candidate mutations was performed by capillary sequencing and primer information is available under request.
Results
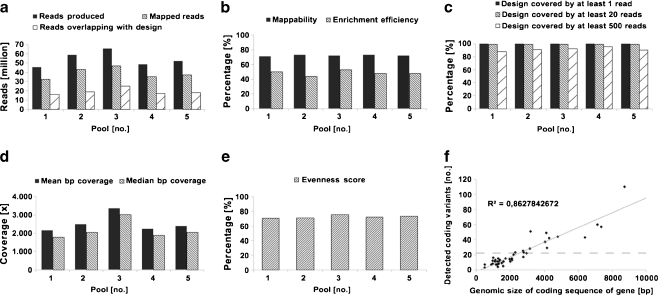
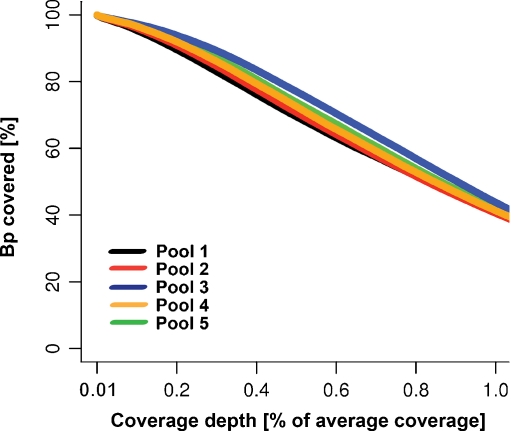
Sequencing of the five pools in a single slide SOLiD run resulted in 270,024,207 raw 50-mer reads (Fig. 2a) with even coverage distribution across the designed region: 99.9% of the target design was covered by at least one read, 99.3 ± 0.3% covered by at least 20 reads and 91.3 ± 3% covered by at least 500 reads (Fig. 2c). From all produced raw sequencing reads (270,024,207), 72.2 ± 0.8% mapped to the GRCh37/hg19 genome build for human and 48.6 ± 3.3% of all mapped reads overlapped with the enrichment design (Fig. 2b). The second percentage is often referred to as enrichment efficiency and correlates with our previous observations that design sizes smaller than 1 Mb (in our case ~0.5 Mb) have lower enrichment efficiency in comparison with designs larger than 1 Mb (up to 60–70%). For each patient pool, we obtained a mean bp coverage of 2,514 ± 481 reads and median bp coverage of 2,158 ± 497 reads and the small difference between these two values (14.6 ± 3.2%) can be used as indirect measure of even distribution (Fig. 2d). Evenness of coverage defined by an evenness score is similar in all five pools (72.4 ± 0.2) and falls into the normal range of 70–75% as described before [19] (Fig. 2e). More than 99% of all bp position from the designed regions was covered by at least 10% of mean coverage (Fig. 3).
Fig. 2.
Sequencing, mapping and enrichment statistics. The total number of reads produced, number of reads mapping to the reference genome and number of reads overlapping with the enrichment design (a), the percentage of mappability and enrichment efficiency (b), design footprint covered by ≥1 read, by ≥20 reads, and by ≥500 reads (c), mean and median bp coverage (d), and evenness score (e) show similar pattern for all five patient pools indicating the robustness of the method. The number of novel variants detected in all five patient pools correlates with the size of the coding sequence of genes (f) indicating a random distribution of detected variants. It is not possible to statistically test for enrichment of variants in genes since we did not include control genes or control samples (full line trend line, dashed line mean value line, R 2 trend line equation)
Fig. 3.
Distribution of sequencing coverage. The plot indicates the percentage of bases in the design that is covered by the depth of coverage normalized for the average coverage. Example: on average 99% of all bp is covered by at least 10-percentile of mean coverage (251.4 ± 48.1 per pool) and approximately 40% of all bp reached the mean coverage (2,514 ± 481 per pool)
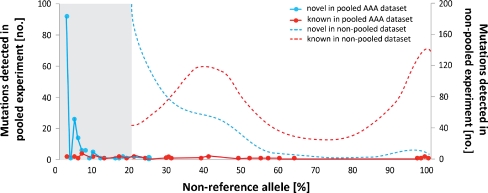
Using pre-defined settings with an allele cut-off >3%, we detected 2,236.8 ± 166.1 variants per pool. A detailed overview of the number of variants for each patient pool can be found in Table 3. The number of coding variants found per gene correlates with the size of its coding sequence as shown in Fig. 2f and suggests a random distribution of detected variants. Altogether, we detected 681 coding variants of which 608 were novel and 73 were present in dbSNP. This is deviating from previous observations by others, and us, as novel variants in non-pooled experiments usually represent 10–15% of all detected variants. The high percentage of potential novel variants detected in our study (89%) could be due to the very low threshold that had to be set for detection of single variant alleles in a pool of 40, complicating the distinction of true variants from noise. As expected, this led to a skewed NRA distribution in AAA pools, which is significantly deviating from the typical pattern observed in non-pooled experiments where a peak for heterozygous mutations at 30–50% NRA and homozygous mutations close to 100% NRA can be detected (Fig. 4). Usually, the region below 20% NRA contains the majority of false positive variants [19, 20], although this depends strongly on the depth of coverage. Within the range of 3–100%NRA, 77% of all detected coding variants had an allele frequency in one of the pools of ≤10% while the allele frequency for novel variants was ≤25%.
Table 3.
List of mutations detected in five pools
| Type of mutation | Patient pool | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Non-synonymous codinga | 229 | 193 | 265 | 239 | 236 |
| Stop-gained | 5 | 9 | 7 | 7 | 5 |
| Stop-lost | 1 | 1 | 0 | 0 | 0 |
| Essential splice site | 2 | 3 | 2 | 4 | 5 |
| Splice site | 9 | 11 | 7 | 11 | 14 |
| Synonymous coding | 61 | 56 | 61 | 57 | 64 |
| 5'UTR | 48 | 31 | 47 | 60 | 54 |
| 3'UTR | 191 | 185 | 201 | 201 | 222 |
| Intronic | 79 | 80 | 91 | 87 | 89 |
| Downstream | 222 | 228 | 251 | 276 | 255 |
| Upstream | 285 | 273 | 329 | 319 | 299 |
| Intergenic | 578 | 582 | 649 | 739 | 626 |
| Within non-coding gene | 357 | 352 | 380 | 403 | 406 |
| Small InDels | 28 | 31 | 29 | 30 | 27 |
| All mutations | 2,095 | 2,035 | 2,319 | 2,433 | 2,302 |
aEnsemble predicted variation consequences
Fig. 4.
Non-reference allele percentage distribution of detected variants in AAA patient pools. Non-reference allele (NRA) percentage distribution of detected variants in AAA pools is significantly deviating from the commonly observed pattern in non-pooled experiments with a clear peak for heterozygous mutations at 30–50% NRA and homozygous mutations close to 100% NRA. Dashed lines are indicative of a distribution of NRA% of novel and known variants from an exome sequencing of a healthy individual
Prioritization of candidate variants requires a substantial effort due to the high number of mutations discovered per pool and the fact that majority of them was novel and below 20% NRA. We decided to select 20 candidate novel coding variants with significant effect on protein and conservation score with NRA% ranging from 3% to 25% (Table 4). We performed capillary sequencing on each patient from a pool separately but failed to confirm any of the predicted variants, suggesting that the majority of candidate novel detected variants are false positives. This is indicative of a relatively high error rate in the sequencing process. Due to the high number of variants detected and the fact that the carriers of those variants in a pool are unknown until capillary sequencing confirmation, we did not perform a classical systematic verification of the false negative discovery rate. On the other hand, we were able to spot common variants previously detected in GWAS studies and candidate genes association studies using the same sample collection [10, 11, 13, 15] (data not shown).
Table 4.
List of novel variants selected for capillary sequencing confirmation
| Chr. | Positiona | Allele change | Amino acid change | Amino acid positionb | Gene name | Gerp1c | Gerp2d | Mutation effect | Patient pool NRA% | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||||||||
| 1 | 218520046 | G/A | MET/ILE | 1/443 | TGFB2 | 2.82 | 1.165 | Non-synonymous coding | 12 | 13 | 11 | 11e | 8 |
| 2 | 33477855 | G/C | CYS/SER | 378/1397 | LTBP1 | 2.82 | 2.171 | Non-synonymous coding | 8 | 16 | 13 | 15e | - |
| 3 | 30732960 | C/A | PRO/THR | 550/593 | TGFBR2 | 2.9 | 2.094 | Non-synonymous coding | 7e | – | – | – | – |
| 3 | 148459750 | A/T | LYS/stop | 310/360 | AGTR1 | No score | No score | Stop-gained | – | – | – | 3e | – |
| 5 | 68555715 | A/C | LYS/THR | 160/347 | CDK7 | 2.82 | 1.788 | Non-synonymous coding | – | 14 | – | 15e | – |
| 7 | 150698940 | A/G | THR/ALA | 512/1204 | NOS3 | 2.32 | 1.41 | Non-synonymous coding | 10e | 5 | – | 7 | 5 |
| 9 | 101891203 | T/G | PHE/CYS | 55/427 | TGFBR1 | 2.9 | 1.796 | Non-synonymous coding | 5 | 5 | 8 | 6e | 5 |
| 9 | 101891219 | G/C | GLU/ASP | 60/427 | TGFBR1 | 1.96 | 1.601 | Non-synonymous coding | – | – | 6e | 4 | – |
| 9 | 101894941 | C/T | SER/LEU | 165/504 | TGFBR1 | 2.9 | 1.614 | Non-synonymous coding | 5 | – | 4e | – | 3 |
| 12 | 52309230 | A/T | LYS/stop | 332/504 | ACVRL1 | 1.95 | 1.444 | Stop-gained | – | 6 | 5e | – | 3 |
| 12 | 68707507 | T/C | LYS/ARG | 509/715 | MDM1 | −1.15 | 0.841 | Non-synonymous coding | 18e | 19 | – | 15 | – |
| 15 | 48787411 | A/T | CYS/stop | 862/2872 | FBN1 | 1.66 | 1.497 | Stop-gained | 3 | 3 | 4 | 3e | – |
| 15 | 48807601 | A/T | LEU/HIS | 484/2872 | FBN1 | 2.9 | 1.508 | Non-synonymous coding | – | 3 | 3e | – | 3 |
| 15 | 48888508 | G/C | TYR/stop | 170/2872 | FBN1 | 1.11 | 1.114 | Stop gained | – | 3 | 4e | – | – |
| 15 | 48888520 | A/T | CYS/stop | 166/2872 | FBN1 | −0.67 | 0.961 | Stop-gained | 4 | 4 | 7e | – | 8 |
| 15 | 67073635 | A/G | ASP/GLY | 157/236 | SMAD6 | 1.87 | 1.144 | Non-synonymous coding | 5e | – | 4 | 3 | 6 |
| 16 | 15841741 | C/G | CYS/SER | 754/1946 | MYH11 | 2.82 | 2.118 | Non-synonymous coding | 6 | 8 | 11 | 13e | 11 |
| 16 | 15932031 | G/A | GLN/stop | 27/1946 | MYH11 | 2.82 | 2.017 | Stop-gained | – | – | – | 3e | – |
| 16 | 16282720 | A/G | PHE/LEU | 583/1504 | ABCC6 | 2.82 | 1.713 | Non-synonymous coding | 5e | 6 | – | 7 | 6 |
| 16 | 55530885 | T/C | LEU/PRO | 457/611 | MMP2 | 2.9 | 1.361 | Non-synonymous coding | 4 | 4 | 5 | 5e | 6 |
aEnsembl59 coordinate
bFirst number indicates the position of mutation, second total number of amino acids in a transcript
cEnsembl Genomic Evolutionary Rate Profiling (GERP) score
dEnsembl predicted variation consequences
ePool used for cappilary sequencing confirmation
Discussion
Here, we present our results with a genomic DNA pooling strategy for rare variant discovery on an NGS platform. This pilot experiment was designed to find an efficient method for detection of rare genetic variants in genomic regions of interest in large sample groups. Our primary aim was to test the hypothesis of the “pathway model” in our AAA sample collection. We planned to confirm the principle on a group of 5 × 20 patients and test detected variants in an additional collection of 1,400 patient samples and suitable control group. However, though we succeeded in obtaining a high average tiling density of our probe design, even coverage distribution and high mean bp coverage, this approach failed because low frequency alleles in the pool (e.g., a heterozygous variant in one sample in a pool of 20 samples) could not be reliably distinguished from noise. While decreasing the cut-off for NRA% to 2.5% increased the number of detected variants exponentially, we decided to first verify candidates under more stringent conditions reflecting two or more alleles in the pool (3% NRA). However, even under stringent conditions, verification of our biological candidates was unsuccessful.
Genomic DNA pooling strategies have a clear potential for discovery of variants that are common, as reported in several GWAS studies [22–24]. Bansal et al. could indentify 80–85% of low frequency single-nucleotide polymorphisms when using pools of 25 samples compared to results obtained with a non-pooled approach [25]. Recent NGS studies have shown that pooling of samples at the level of genomic DNA up to 20-fold and subsequent amplification of coding exons of candidate genes by conventional PCR does allow for the detection of novel variants in rare diseases [26–28]. However, these experiments did not use targeted genomic NGS enrichment as described in our study. On the other hand, the advantage of NGS targeted genomic enrichment in comparison to the conventional PCR assays is the size of genomic regions that can be assessed in a single experiment (one enrichment allows for up to 50 Mb of genomic target sequence). While diagnostic labs have successfully developed a wide range of amplicon assays for rare diseases, it could be argued that for research purposes of common diseases, it would be relatively laborious to develop such assays as compared to a universal NGS enrichment-based approach. The aforementioned studies also included control samples for discarding variants; we planned to perform the biological filtering by capillary sequencing of detected variants afterwards.
The reason for failure of our approach is that the single heterozygote allele frequency is close to the sequencing noise level in a pool of 20 samples. This noise could arise during any step of library preparation, enrichment, sequencing, or mapping. We do not have data supporting or excluding the possibility that any other NGS platform would perform differently from the SOLiD platform. There are no indications that specific errors are introduced due to biases in SOLiD sequencing, making it impossible to systematically exclude specific errors from the lists of detected variants by biological filtering. Furthermore, the library preparation and enrichment method we used was developed as an improved method for array enrichment on the SOLiD sequencing platform, which performed better than the commercially available methods [19]. It could be possible that pre and post-enrichment PCR cycles may contribute to higher rates of noise as compared to for example whole genome sequencing. These levels are not problematic for non-pooled approaches [19, 20] but may become a problem for pooled approaches as described here. We minimized the number of PCR cycles during library preparation and enrichment to five cycles during library preparation, 12 cycles prior to enrichment, and 12 cycles after enrichment.
Possible solutions for the improvement of this pooling approach would be decreasing the number of pooled samples or decreasing the noise introduced by library preparation, enrichment, sequencing, and mapping. With a decreasing number of pooled samples, the cut-off of NRA% for the detection of a single allele can be increased and the difference between noise and a single heterozygote call in the pool would be much larger and the call more distinguishable. However, this would require the generation of more pools, which would not be attractive for the analysis of large cohorts. We performed the experiment described here before we introduced an alternative multiplexed enrichment approach with pre-barcoded samples [20]. Using multiplexed enrichment sequencing results can be split per sample, resulting in efficient variant detection with novel variant confirmation rates of 50–90% (in contrast to 0% described in the study described here) without biases for allele frequency and with no adverse effects on the NRA% distribution in comparison to the non-pooled experiments. Although multiplexed enrichment gives rise to an increased burden of library preparation, this step can easily be automated and therefore, we recommend using barcoded multiplexed enrichment rather that non-indexed genomic DNA pooling for rare variant detection in common diseases on current versions of next-generation sequencing platforms.
Acknowledgments
Collection of the AAA cohort was sponsored by a grant from the Novartis foundation for cardiovascular excellence. A.F. Baas was supported by a grant from the Dr. E. Dekker Program from the Netherlands Heart Foundation (2009 T001).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Weintraub NL. Understanding abdominal aortic aneurysm. The New England Journal of Medicine. 2009;361(11):1114–1116. doi: 10.1056/NEJMcibr0905244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johansen K, Koepsell T. Familial tendency for abdominal aortic aneurysms. Journal of the American Medical Association. 1986;256(14):1934–1936. doi: 10.1001/jama.256.14.1934. [DOI] [PubMed] [Google Scholar]
- 3.Crawford CM, Hurtgen-Grace K, Talarico E, Marley J. Abdominal aortic aneurysm: an illustrated narrative review. Journal of Manipulative and Physiological Therapeutics. 2003;26(3):184–195. doi: 10.1016/S0161-4754(02)54111-7. [DOI] [PubMed] [Google Scholar]
- 4.Cornuz J, Sidoti Pinto C, Tevaearai H, Egger M. Risk factors for asymptomatic abdominal aortic aneurysm: systematic review and meta-analysis of population-based screening studies. European Journal of Public Health. 2004;14(4):343–349. doi: 10.1093/eurpub/14.4.343. [DOI] [PubMed] [Google Scholar]
- 5.Lindsay J., Jr Diagnosis and treatment of diseases of the aorta. Current Problems in Cardiology. 1997;22(10):485–542. doi: 10.1016/S0146-2806(97)80004-7. [DOI] [PubMed] [Google Scholar]
- 6.Wahlgren CM, Larsson E, Magnusson PK, Hultgren R, Swedenborg J. Genetic and environmental contributions to abdominal aortic aneurysm development in a twin population. Journal of Vascular Surgery. 2010;51(1):3–7. doi: 10.1016/j.jvs.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 7.Ogata T, MacKean GL, Cole CW, Arthur C, Andreou P, Tromp G, Kuivaniemi H. The lifetime prevalence of abdominal aortic aneurysms among siblings of aneurysm patients is eightfold higher than among siblings of spouses: an analysis of 187 aneurysm families in Nova Scotia, Canada. Journal of Vascular Surgery. 2005;42(5):891–897. doi: 10.1016/j.jvs.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibamura H, Olson JM, van Vlijmen-Van KC, Buxbaum SG, Dudek DM, Tromp G, Ogata T, Skunca M, Sakalihasan N, Pals G, Limet R, MacKean GL, Defawe O, Verloes A, Arthur C, Lossing AG, Burnett M, Sueda T, Kuivaniemi H. Genome scan for familial abdominal aortic aneurysm using sex and family history as covariates suggests genetic heterogeneity and identifies linkage to chromosome 19q13. Circulation. 2004;109(17):2103–2108. doi: 10.1161/01.CIR.0000127857.77161.A1. [DOI] [PubMed] [Google Scholar]
- 9.Van Vlijmen-Van Keulen CJ, Rauwerda JA, Pals G. Genome-wide linkage in three Dutch families maps a locus for abdominal aortic aneurysms to chromosome 19q13.3. European Journal of Vascular and Endovascular Surgery. 2005;30(1):29–35. doi: 10.1016/j.ejvs.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 10.Baas AF, Medic J, van't Slot R, de Vries JP, van Sambeek MR, Geelkerken BH, Boll BP, Grobbee DE, Wijmenga C, Ruigrok YM, Blankensteijn JD. Association study of single nucleotide polymorphisms on chromosome 19q13 with abdominal aortic aneurysm. Angiology. 2010;61(3):243–247. doi: 10.1177/0003319709354752. [DOI] [PubMed] [Google Scholar]
- 11.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nature Genetics. 2008;40(2):217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 12.Elmore JR, Obmann MA, Kuivaniemi H, Tromp G, Gerhard GS, Franklin DP, Boddy AM, Carey DJ. Identification of a genetic variant associated with abdominal aortic aneurysms on chromosome 3p12.3 by genome wide association. Journal of Vascular Surgery. 2009;49(6):1525–1531. doi: 10.1016/j.jvs.2009.01.041. [DOI] [PubMed] [Google Scholar]
- 13.Gretarsdottir S, Baas AF, Thorleifsson G, Holm H, den Heijer M, de Vries JP, Kranendonk SE, Zeebregts CJ, van Sterkenburg SM, Geelkerken RH, van Rij AM, Williams MJ, Boll AP, Kostic JP, Jonasdottir A, Walters GB, Masson G, Sulem P, Saemundsdottir J, Mouy M, Magnusson KP, Tromp G, Elmore JR, Sakalihasan N, Limet R, Defraigne JO, Ferrell RE, Ronkainen A, Ruigrok YM, Wijmenga C, Grobbee DE, Shah SH, Granger CB, Quyyumi AA, Vaccarino V, Patel RS, Zafari AM, Levey AI, Austin H, Girelli D, Pignatti PF, Olivieri O, Martinelli N, Malerba G, Trabetti E, Becker LC, Becker DM, Reilly MP, Rader DJ, Mueller T, Dieplinger B, Haltmayer M, Urbonavicius S, Lindblad B, Gottsater A, Gaetani E, Pola R, Wells P, Rodger M, Forgie M, Langlois N, Corral J, Vicente V, Fontcuberta J, Espana F, Grarup N, Jorgensen T, Witte DR, Hansen T, Pedersen O, Aben KK, de Graaf J, Holewijn S, Folkersen L, Franco-Cereceda A, Eriksson P, Collier DA, Stefansson H, Steinthorsdottir V, Rafnar T, Valdimarsson EM, Magnadottir HB, Sveinbjornsdottir S, Olafsson I, Magnusson MK, Palmason R, Haraldsdottir V, Andersen K, Onundarson PT, Thorgeirsson G, Kiemeney LA, Powell JT, Carey DJ, Kuivaniemi H, Lindholt JS, Jones GT, Kong A, Blankensteijn JD, Matthiasson SE, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nature Genetics. 2010;42(8):692–697. doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saratzis A, Abbas A, Kiskinis D, Melas N, Saratzis N, Kitas GD. Abdominal aortic aneurysm: a review of the genetic basis. Angiology. 2010 doi: 10.1177/0003319710373092. [DOI] [PubMed] [Google Scholar]
- 15.Baas AF, Medic J, van't Slot R, de Kovel CG, Zhernakova A, Geelkerken RH, Kranendonk SE, van Sterkenburg SM, Grobbee DE, Boll AP, Wijmenga C, Blankensteijn JD, Ruigrok YM. Association of the TGF-beta receptor genes with abdominal aortic aneurysm. European Journal of Human Genetics. 2010;18(2):240–244. doi: 10.1038/ejhg.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International HIV Controllers Study. Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nature Genetics. 2008;40(6):695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, Del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nature Genetics. 2010;42(12):1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 19.Mokry M, Feitsma H, Nijman IJ, de Bruijn E, van der Zaag PJ, Guryev V, Cuppen E. Accurate SNP and mutation detection by targeted custom microarray-based genomic enrichment of short-fragment sequencing libraries. Nucleic Acids Research. 2010;38(10):e116. doi: 10.1093/nar/gkq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nijman IJ, Mokry M, van Boxtel R, Toonen P, de Bruijn E, Cuppen E. Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nature Methods. 2010;7(11):913–915. doi: 10.1038/nmeth.1516. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krumbiegel M, Pasutto F, Schlotzer-Schrehardt U, Uebe S, Zenkel M, Mardin CY, Weisschuh N, Paoli D, Gramer E, Becker C, Ekici AB, Weber BH, Nurnberg P, Kruse FE, Reis A. Genome-wide association study with DNA pooling identifies variants at CNTNAP2 associated with pseudoexfoliation syndrome. European Journal of Human Genetics. 2010 doi: 10.1038/ejhg.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirov G, Zaharieva I, Georgieva L, Moskvina V, Nikolov I, Cichon S, Hillmer A, Toncheva D, Owen MJ, O'Donovan MC. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Molecular Psychiatry. 2009;14(8):796–803. doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- 24.Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, Williams J, Owen MJ, Kirov G. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Medical Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bansal V. A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics. 2010;26(12):i318–324. doi: 10.1093/bioinformatics/btq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, Redman MC, Wiltshire E, Wilson CJ, Altshuler D, Gabriel SB, Daly MJ, Thorburn DR, Mootha VK. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nature Genetics. 2010;42(10):851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen S, Ramaswami G, Davis EE, Hurd T, Airik R, Kasanuki JM, Van Der Kraak L, Allen SJ, Beales PL, Katsanis N, Otto EA, Hildebrandt F. Mutation analysis in Bardet–Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Human Genetics. 2010 doi: 10.1007/s00439-010-0902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto EA, Ramaswami G, Janssen S, Chaki M, Allen SJ, Zhou W, Airik R, Hurd TW, Ghosh AK, Wolf MT, Hoppe B, Neuhaus TJ, Bockenhauer D, Milford DV, Soliman NA, Antignac C, Saunier S, Johnson CA, Hildebrandt F. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. Journal of Medical Genetics. 2010 doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]