Abstract
Computational chemists and structural biologists are often interested in characterizing ligand-receptor complexes for hydrogen-bond, hydrophobic, salt-bridge, van der Waals, and other interactions in order to assess ligand binding. When done by hand, this characterization can become tedious, especially when many complexes need be analyzed.
In order to facilitate the characterization of ligand binding, we here present a novel Python-implemented computer algorithm called BINANA (BINding ANAlyzer), which is freely available for download at http://www.nbcr.net/binana/. To demonstrate the utility of the new algorithm, we use BINANA to confirm that the number of hydrophobic contacts between a ligand and its protein receptor is positively correlated with ligand potency. Additionally, we show how BINANA can be used to search through a large ligand-receptor database to identify those complexes that are remarkable for selected binding features, and to identify lead candidates from a virtual screen with specific, desirable binding characteristics.
We are hopeful that BINANA will be useful to computational chemists and structural biologists who wish to automatically characterize many ligand-receptor complexes for key binding characteristics.
Keywords: BINANA, computer algorithm, ligand-binding analysis, computer-aided drug design, structural biology, virtual screening
1. Introduction
Great progress has been made in the fields of rational and computer-aided drug design in recent years. Three-dimensional models of ligand-receptor complexes, derived from X-ray crystallography, NMR, and homology modeling, are routinely used to help inform ligand optimization. A common task is to inspect a ligand-receptor complex using molecular visualization software in order to characterize ligand binding according to hydrogen-bond, hydrophobic, salt-bridge, van der Waals, and other interactions. While visual inspection of a few poses is feasible, characterizing the binding poses of many ligands can be tedious.
To automate the characterization of ligand-receptor binding, we have developed an algorithm called BINANA (BINding ANAlyzer). BINANA is implemented in Python and so is easily editable, customizable, and platform independent. Additionally, unlike many other similar software packages, BINANA is open source and freely available for download at http://www.nbcr.net/binana/.
2. Materials and Methods
2.1. The BINANA Algorithm
A novel algorithm called BINANA (BINding ANAlyzer) has been implemented in Python in order to characterize the binding of ligand-receptor complexes. The python implementation, which accepts as input a protein and ligand file in the PDBQT format [1], is freely available at http://www.nbcr.net/binana/.
2.1.1. BINANA: Close Contacts
BINANA begins by identifying all ligand and protein atoms that come within 2.5 Å of each other. These close-contact atoms are then characterized according to their respective AutoDock atom types, without regard for the receptor or ligand. The number of each pair of close-contact atoms of given AutoDock atom types is then tallied. For example, the program counts the number of times a hydrogen-bond accepting oxygen atom (atom type OA), either on the ligand or the receptor, comes within 2.5 Å of a polar hydrogen atom (atom type HD) on the corresponding binding partner, be it the receptor or the ligand. A similar list of atom-type pairs is tallied for all ligand and receptor atoms that come within 4.0 Å of each other.
2.1.2. BINANA: Electrostatic Interactions
For each atom-type pair of atoms that come within 4.0 Å of each other, as described above, a summed electrostatic energy is calculated using the Gasteiger partial charges assigned by AutoDockTools [1]:
where V(a,b) is the summed electrostatic interaction energy of all the atoms of types a and b, respectively, qai is the partial atomic charge of an atom of type a, is qbi the partial atomic charge of an atom of type b, and raibi is the distance separating these two atoms.
2.1.3. BINANA: Binding-Pocket Flexibility
BINANA also provides useful information about the flexibility of a binding pocket. Each receptor atom that comes within 4.0 Å of any ligand atom is characterized according to whether or not it belongs to a protein side chain or backbone. Additionally, the secondary structure of the corresponding protein residue of each atom, be it α helix, β sheet, or other, is also determined. Thus, there are six possible characterizations for each atom: α-sidechain, α-backbone, β-sidechain, β-backbone, other-sidechain, other-backbone. The number of close-contact receptor atoms falling into each of these six categories is tallied as a metric of binding-site flexibility.
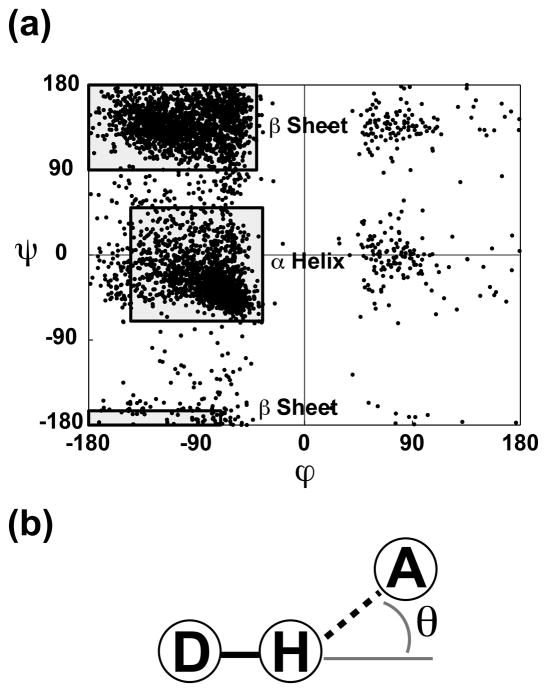
All protein atoms with the atom names “CA,” “C,” “O,” or “N” are assumed to belong to the backbone. All other receptor atoms are assigned side-chain status. Determining the secondary structure of the corresponding residue of each close-contact receptor atom is more difficult. First, preliminary secondary-structure assignments are made based on the ϕ and ψ angles of each residue. If ϕ ⊂ (−145, −35) and ψ ⊂ (−70, 50), the residue is assumed to be in the α-helix conformation. If ϕ ⊂ [−180, −40) and ψ ⊂ (90, 180], or ϕ ⊂ [−180, −70) and ψ ⊂ [−180, −165], the residue is assumed to be in the β-sheet conformation. Otherwise, the secondary structure of the residue is labeled “other” (Fig. 1a).
Fig. 1.
(a) A Ramachandran plot showing the ϕ and ψ angles of the protein residues of ten representative structures. The regions used to initially assign tertiary structure are shown as shaded boxes. (b) An illustration of the hydrogen-bond cutoff angle, θ. The circled D represents a hydrogen-bond donor, the circled H represents a hydrogen atom attached to that donor, and the circled A represents a hydrogen-bond acceptor.
Inspection of actual α-helix structures revealed that the Cα of an α-helix residue i is generally within 6.0 Å of the Cα of an α-helix residue three residues away (i + 3 or i − 3). Any residue that has been preliminarily labeled “α helix” that fails to meet this criterion is instead labeled “other.” Additionally, the residues of any α helix comprised of fewer than four consecutive residues are also labeled “other,” as these tend to belong to small loops rather than genuine helices.
True β strands hydrogen bond with neighboring β strands to form β sheets. Inspection of actual β strands revealed that the Cα of a β-sheet residue, i, is typically within 6.0 Å of the Cα of another β-sheet residue, usually on a different strand, when the residues [i − 2, i + 2] are excluded. Any residue labeled “β sheet” that does not meet this criterion is labeled “other” instead. Additionally, the residues of β strands that are less than three residues long are likewise labeled “other,” as these residues typically belong to loops rather than legitimate strands.
2.1.4. BINANA: Hydrophobic Contacts
To identify hydrophobic contacts, BINANA simply tallies the number of times a ligand carbon atom comes within 4.0 Å of a receptor carbon atom. These hydrophobic contacts are categorized according to the flexibility of the receptor carbon atom. There are six possible classifications: α-sidechain, α-backbone, β-sidechain, β-backbone, other-sidechain, other-backbone. The total number of hydrophobic contacts is simply the sum of these six counts.
2.1.5. BINANA: Hydrogen Bonds
BINANA allows hydroxyl and amine groups to act as hydrogen-bond donors. Oxygen, nitrogen, and fluorine atoms can act as hydrogen-bond acceptors. Fairly liberal hydrogen-bond cutoffs are implemented in order to accommodate low-resolution crystal structures. A hydrogen bond is identified if the hydrogen-bond donor comes within 4.0 Å of the hydrogen-bond acceptor, and the angle formed between the donor, the hydrogen atom, and the acceptor, θ, is no greater than 40° (Fig. 1b). BINANA tallies the number of hydrogen bonds according to the secondary structure of the receptor atom, the side-chain/backbone status of the receptor atom, and the location (ligand or receptor) of the hydrogen-bond donor. Thus there are twelve possible categorizations: α-sidechain-ligand, α-backbone-ligand, β-sidechain-ligand, β-backbone-ligand, other-sidechain-ligand, other-backbone-ligand, α-sidechain-receptor, α-backbone-receptor, β-sidechain-receptor, β-backbone-receptor, other-sidechain-receptor, other-backbone-receptor.
2.1.6. BINANA: Salt Bridges
BINANA also identifies possible salt bridges binding the ligand to the receptor. First, charged functional groups are identified and labeled with a representative point to denote their location. For non-protein residues, BINANA searches for common functional groups or atoms that are known to be charged. Atoms containing the following names are assumed to be metal cations: MG, MN, RH, ZN, FE, BI, AS, AG. The identifying coordinate is centered on the metal cation itself. Sp3-hybridized amines (which could pick up a hydrogen atom) and quaternary ammonium cations are also assumed to be charged; the representative coordinate is centered on the nitrogen atom. Imidamides where both of the constituent amines are primary, as in the guanidino group, are also fairly common charged groups. The representative coordinate is placed between the two constituent nitrogen atoms.
Carboxylate groups are likewise charged; the identifying coordinate is placed between the two oxygen atoms. Any group containing a phosphorus atom bound to two oxygen atoms that are themselves bound to no other heavy atoms (e.g., a phosphate group) is also likely charged; the representative coordinate is centered on the phosphorus atom. Similarly, any group containing a sulfur atom bound to three oxygen atoms that are themselves bound to no other heavy atoms (i.e., a sulfonate group) is also likely charged; the representative coordinate is centered on the sulfur atom. Note that while BINANA is thorough in its attempt to identify charged functional groups on non-protein residues, it is not exhaustive. For example, one could imagine a protonated amine in an aromatic ring that, though charged, would not be identified as a charged group.
Identifying the charged functional groups of protein residues is much simpler. Functional groups are identified based on standardized protein atom names. Lysine residues have an amine; the representative coordinate is centered on the nitrogen atom. Arginine has a guanidino group; the coordinate is centered between the two terminal nitrogen atoms. Histidine is always considered charged, as it could pick up a hydrogen atom. The representative charge is placed between the two ring nitrogen atoms. Finally, glutamate and aspartate contain charged carboxylate groups; the representative coordinate is placed between the two oxygen atoms.
Having identified the location of all charged groups, BINANA is ready to predict potential salt bridges. First, the algorithm identifies all representative charge coordinates within 5.5 Å of each other. Next, it verifies that the two identified coordinates correspond to charges that are opposite. If so, a salt bridge is detected. These salt bridges are characterized and tallied by the secondary structure of the associated protein residue: α helix, β sheet, or other.
2.1.7. BINANA: π Interactions
A number of interactions are known to involve π systems. In order to detect the aromatic rings of non-protein residues, a recursive subroutine identifies all five or six member rings, aromatic or not. The dihedral angles between adjacent ring atoms, and between adjacent ring atoms and the first atom of ring substituents, are checked to ensure that none deviate from planarity by more than 15°. Planarity establishes aromaticity. For protein residues, aromatic rings are identified using standardized protein-atom names. Phenylalanine, tyrosine, and histidine all have aromatic rings. Tryptophan is assigned two aromatic rings.
Once an aromatic ring is identified, it must be fully characterized. First, a plane is defined that passes through three ring atoms, preferably the first, third, and fifth atoms. The center of the ring is calculated by averaging the coordinates of all ring atoms, and the radius is given to be the maximum distance between the center point and any of these atoms. From this information, a ring disk can be defined that is centered on the ring center point, oriented along the ring plane, and has a radius equal to that of the ring plus a small buffer (0.75 Å).
Having identified and characterized all aromatic rings, the algorithm next attempts to identify π-π stacking interactions. First, every aromatic ring of the ligand is compared to every aromatic ring of the receptor. If the centers of two rings are within 7.5 Å of each other, the angle between the two vectors normal to the planes of each ring is calculated. If this angle suggests that the two planes are within 30° of being parallel, then each of the ring atoms is projected onto the plane of the opposite ring by identifying the nearest point on that plane. If any of these projected points fall within the ring disk of the opposite ring, the two aromatic rings are said to participate in a π-π stacking interaction. We note that it is not sufficient to simply project the ring center point onto the plane of the opposite ring because π-π stacking interactions are often off center [2].
To detect T-stacking or edge-face interactions, every aromatic ring of the ligand is again compared to every aromatic ring of the receptor. If the centers of two rings are within 7.5 Å of each other, the angle between the two vectors normal to the planes of each ring is again calculated. If this angle shows that the two planes are within 30° of being perpendicular, a second distance check is performed to verify that the two rings come within 5.0 Å at their nearest point. If so, each of the coordinates of the ring center points is projected onto the plane of the opposite ring by identifying the nearest point on the respective plane. If either of the projected center points falls within the ring disk of the opposite ring, the two aromatic rings are said to participate in a T-stacking interaction.
Finally, BINANA also detects cation-π interactions between the ligand and the receptor. Each of the representative coordinates identifying positively charged functional groups is compared to each of the center points of the identified aromatic rings. If the distance between any of these pairs is less than 6.0 Å, the coordinate identifying the charged functional group is projected onto the plane of the aromatic ring by identifying the nearest point on that plane. If the projected coordinate falls within the ring disk of the aromatic ring, a cation-π interaction is identified.
π-π stacking, T-stacking, and cation-π interactions are tallied according to the secondary structure of the receptor residue containing the associated aromatic ring or charged functional group: α helix, β sheet, or other.
2.1.8. BINANA: Ligand Atom Types and Rotatable Bonds
BINANA also tallies the number of atoms of each AutoDock type present in the ligand as well as the number of ligand rotatable bonds identified by the AutoDockTools scripts [1] used to generate the input PDBQT files.
2.2. Compiling a Database of Experimentally Derived Ligand-Receptor Complexes
The database of ligand-receptor complexes used in the current study was derived from a database that has been described previously [3]. In brief, crystallographic and NMR structures that had Kd values listed in the PDBbind-CN [4, 5] and MOAD [6] databases were downloaded from the Protein Data Bank [7]. Hydrogen atoms were added to the ligands and receptors of the database using the Schrodinger Maestro (Schrodinger) and AutoDockTools 1.5.1 [1] computer programs, respectively. Both the ligand and receptor were further processed with AutoDockTools to merge nonpolar hydrogen atoms with parent atoms and to assign AutoDock atom types and Gasteiger charges. Ligand-receptor complexes containing atom types that were not compatible with the default AutoDock force field were discarded. Finally, an in-house script was used to optimize the geometry of the hydrogen bonds between ligand and receptor atoms.
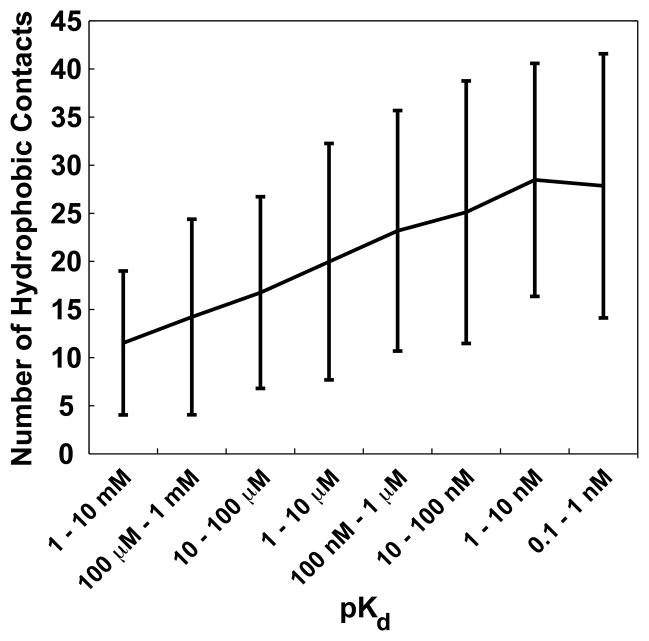
To generate Fig. 2, subsets of complexes were defined comprised of ligands with affinities [100 μM, 1 mM), [1 mM, 10 mM), [10 μM, 100 μM), [100 nM, 1 μM), [1 μM, 10 μM), [10 nM, 100 nM), [1 nM, 10 nM), and [0.1 nM, 1 nM), containing 274, 113, 389, 456, 407, 429, 288, and 149 complexes, respectively.
Fig. 2.
The average number of ligand-receptor hydrophobic contacts per complex for ligands of varying potency. Error bars range from mean − standard deviation to mean + standard deviation.
2.3. Virtual Screening
A model of Trypanosoma brucei RNA editing ligase 1 (TbREL1) was downloaded from the Protein Data Bank [7] (PDBID: 1XDN [8]). Selenomethionine residues were converted to methionine residues, and rotamers, ligand residues, and water residues were removed. Hydrogen atoms were added to the crystal structure using the PDB2PQR server [9, 10], with the pH set to 7.0. AutoDockTools [11] was used to merge non-polar hydrogen atoms with their parent atoms and to assign Gasteiger charges [12] and AutoDock atom types.
Four low-micromolar TbREL1 inhibitors were identified from the literature: the two most potent inhibitors found by Amaro et al. (S1 and S5) [13], and the two most potent inhibitors found by Durrant et al. (V2 and V4) [14]. The geometry of these ligands was optimized using Schrodinger maestro (Schrodinger). AutoDockTools [11] was used to merge non-polar hydrogen atoms with parent atoms, to assign Gasteiger charges [12] and AutoDock atom types, and to identify rotatable bonds. Ninety-six additional small molecules were selected from the NCI Diversity Set II at random to serve as decoys.
AutoDock Vina [15] was used to dock these 100 ligands into the TbREL1 active site. The ligands were ordered by the Vina score and assigned a corresponding rank (RankVina). A recently developed neural-network scoring function, NNScore, was then used to rescore the best-scoring Vina pose of each small-molecule model. The average output of the 24 default neural networks provided with the program was used. The ligands were reordered by this neural-network score and assigned a second rank, RankNNScore. A composite Vina-NNScore consensus score was calculated by averaging RankVina and RankNNScore.
2.4. Electrostatic Calculations
The electrostatic potential of the inositol-hexakisphosphate binding site of Vibrio cholerae RTX toxin RtxA (PDBID: 3EEB [16]) was calculated using the Adaptive Poisson-Boltzmann Solver (APBS) [17]. The potential was mapped onto a SURF molecular representation [18].
2.5. Molecular Visualization
Visual Molecular Dynamics (VMD) 1.8.7 was used for all molecular visualizations [19]. Final images were rendered using Tachyon Parallel/Multiprocessor Ray Tracer, version 0.98.7.
3. Results and Discussion
The purpose of the current work is to present a novel algorithm called BINANA (BINding ANAlyzer). Computational chemists and structural biologists are often faced with the task of using molecular visualization software to characterize ligand-receptor complexes according to hydrogen-bond, hydrophobic, salt-bridge, van der Waals, and other interactions. While visual inspection of a few poses is feasible, characterizing the binding poses of many ligands can be tedious.
BINANA automates the characterization of ligand-receptor binding. BINANA is implemented in Python and so is easily editable, customizable, and platform independent. Additionally, unlike many other similar software packages, BINANA is open source and freely available for download at http://www.nbcr.net/binana/.
3.1. Ligand-Receptor Hydrophobic Contacts and Ligand Potency
In order to demonstrate the utility of the BINANA algorithm, we characterized 2 505 ligand-receptor complexes with known Kd values according to the number of hydrophobic contacts per complex by using BINANA to count the number of ligand-receptor carbon-atom pairs separated by less than 4.0 Å. The average number of hydrophobic contacts per complex was calculated for ligands of varying Kd values, revealing that the number of hydrophobic contacts was positively correlated with potency (Fig. 2).
This result is not surprising, as others have noted the important role that hydrophobic contacts play in ligand binding [20, 21]. That BINANA was able to identify this general trend is evidence of its utility. Hydrophobic ligands are energetically unfavorable in solution, as they disrupt the water-to-water hydrogen-bond network and significantly stabilize the water molecules in ligand solvation shells, thereby decreasing the total entropy of the system. Upon binding, the hydrophobic portions of a well-designed ligand are placed in hydrophobic regions within the binding pocket. Thus, an energetically unfavorable state in solution is replaced by an energetically favorable state upon binding, resulting in a net gain of free energy.
3.2. Identifying Ligand-Receptor Complexes with Remarkable Binding Features
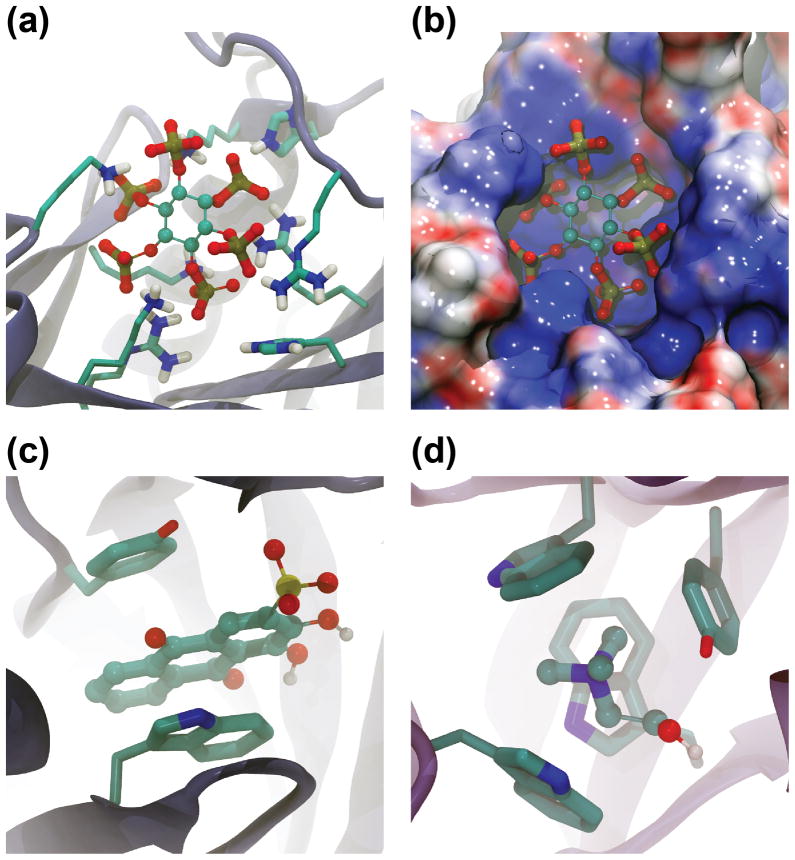
BINANA can also be used to identify ligand-receptor complexes with remarkable selected binding features from a large database of complexes. For example, of the 2 505 complexes in the database described above, BINANA identified the complex formed by inositol hexakisphosphate bound to the RTX toxin RtxA from Vibrio cholerae (PDBID: 3EEB [16], Fig. 3a and Fig. 3b) as the complex with the greatest number of salt bridges, i.e., simple electrostatic interactions between oppositely charged functional groups. The negatively charged inositol hexakisphosphate with its six phosphate groups sits in a positively charged pocket on the protein surface, where it interacts electrostatically with R29, K54, H55, R85, R171, K183, K195, K200, and possibly R182 (Fig. 3a and Fig. 3b). Given these extensive salt-bridge interactions, it is not surprising that the Kd of binding is 1.3 μM [16].
Fig. 3.
Examples of remarkable ligand-receptor complexes. (a) The complex formed by inositol hexakisphosphate bound to the RTX toxin RtxA from Vibrio cholerae is remarkable for the number of ligand-receptor salt bridges (PDBID: 3EEB). (b) An electrostatic calculation confirms that the inositol-hexakisphosphate binding site is remarkably positively charged. (c) The complex formed by an IgE antibody and the hapten alizarin red is remarkable for the number of ligand-receptor π-π stacking interactions (PDBID: 1OAR). (d) The complex formed by a putative ABC transporter from Rhizobium meliloti bound to choline is remarkable for the number ligand-receptor cation-π interactions (PDBID: 2REG).
A similar BINANA-based search revealed that the ligand-receptor complex formed by an IgE antibody and the hapten alizarin red forms more ligand-receptor π-π stacking interactions than any other complex in the database (PDBID: 1OAR [22], Fig. 3c). In these stacking interactions, a non-covalent bond is thought to arise when the orbitals of parallel “stacked” aromatic rings overlap [23]. Alizarin red is an anthracene compound comprised of three fused aromatic rings and so contains a large conjugated π system. The ligand is sandwiched between two aromatic receptor residues that likely contribute to the ligand’s overall measured potency of 40 nM [22].
As a final demonstration of BINANA-based searching, the database of 2 505 complexes was probed for complexes characterized by many cation-π interactions. In these interactions, a π-system quadrupole, and particularly the electron-rich π system of an aromatic-ring face, is thought to interact with a nearby cation primarily through electrostatic means. It is not surprising that the complex that had the most cation-π interactions was a choline-binding protein, a putative ABC transporter from Rhizobium meliloti bound to choline itself (PDBID: 2REG [24], Fig. 3d). Choline-binding proteins are often cited as classic examples of ligand binding mediated by cation-π interactions [25, 26]. A positively charged ligand moiety, often a quaternary amine, is positioned within an “aromatic nest,” wherein it is surrounded by multiple aromatic protein side chains. Given that some studies have suggested that the energy of a cation-π interaction can rival that of a hydrogen bond [27], it is not surprising that choline binds this ABC transporter from Rhizobium meliloti with a Kd value of 2.7 μM [24].
3.3. Using BINANA to Select Virtual-Screening Candidates to Test Experimentally
To demonstrate how BINANA can be used to improve virtual-screening hit rates, we performed a small, illustrative virtual screen against Trypanosoma brucei RNA editing ligase 1 (TbREL1). Four known low-micromolar inhibitors were selected: the two most potent inhibitors found by Amaro et al. (S1 and S5) [13], and the two most potent inhibitors found by Durrant et al. (V2 and V4) [14]. Ninety-six additional small molecules were selected from the NCI Diversity Set II at random to serve as decoys.
These 100 compounds were docked into a crystal structure of TbREL1 [8] using AutoDock Vina [15] and were subsequently rescored using a new neural-network scoring function called NNScore [3]. When the compounds were reranked by a Vina-NNScore consensus score, as described previously [3], the four known inhibitors ranked 1st, 2nd, 5th, and 9th.
The most potent TbREL1 inhibitors found to date contain naphthalene cores that are thought to interact via π-π stacking interactions with TbREL1 F209, just as the adenine ring of the native ligand, ATP, does. As naphthalene consists of two fused aromatic rings, we expect naphthalene-based inhibitors to participate in at least two π-π stacking interactions, one for each aromatic ring of the naphthalene moiety. Indeed, when BINANA was used to identify and eliminate all docked compounds that formed fewer than two π-π stacking interactions with the receptor, the ranking of the four known inhibitors improved to 1st, 2nd, 4th, and 6th out of 100. Interestingly, the decoy compound that ranked 3rd also contained a naphthalene core that formed two π-π stacking interactions with F209.
4.1. Conclusion
We here presented a novel Python-implemented computer algorithm called BINANA (BINding ANAlyzer), which is freely available for download at http://www.nbcr.net/binana/. BINANA automates the potentially tedious task of characterizing ligand-receptor complexes for binding characteristics like hydrogen-bond, hydrophobic, salt-bridge, van der Waals, and other interactions.
To demonstrate the utility of the algorithm, we used BINANA to confirm the known positive correlation between the number of ligand-receptor hydrophobic contacts and ligand potency. Additionally, BINANA was used to search through a large ligand-receptor database to identify complexes remarkable for selected binding features, and to identify lead candidates with specific, desirable binding characteristics from among the compounds that scored well in an illustrative virtual screen.
We are hopeful that BINANA will be useful to computational chemists and structural biologists who wish to automatically characterize many ligand-receptor complexes for key binding characteristics.
Acknowledgments
This work was carried out with funding from NIH GM31749, NSF MCB-0506593, and MCA93S013 to JAM. Additional support from the Howard Hughes Medical Institute, the National Center for Supercomputing Applications, the San Diego Supercomputer Center, the W.M. Keck Foundation, the National Biomedical Computational Resource, and the Center for Theoretical Biological Physics is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 2.McGaughey GB, Gagne M, Rappe AK. pi-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 3.Durrant JD, McCammon JA. NNScore: A Neural-Network-Based Scoring Function for the Characterization of Protein-Ligand Complexes. J Chem Inf Model. 2010;50:1865–1871. doi: 10.1021/ci100244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 5.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind database: methodologies and updates. J Med Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 6.Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Proteins. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 7.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J Mol Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 9.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanner MF. A component-based software environment for visualizing large macromolecular assemblies. Structure. 2005;13:447–462. doi: 10.1016/j.str.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity--a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 13.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proceedings of the National Academy of Sciences. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durrant JD, Hall L, Swift RV, Landon M, Schnaufer A, Amaro RE. Novel Naphthalene-Based Inhibitors of Trypanosoma brucei RNA Editing Ligase 1. PLoS Negl Trop Dis. 2010;4:e803. doi: 10.1371/journal.pntd.0000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322:265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varshney A, Brooks FP, Wright WV. Linearly Scalable Computation of Smooth Molecular Surfaces. IEEE Comp Graphics and Applications. 1994;14:19–25. [Google Scholar]
- 19.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 20.Davis AM, Teague SJ. Hydrogen Bonding, Hydrophobic Interactions, and Failure of the Rigid Receptor Hypothesis. Angewandte Chemie International Edition. 1999;38:736–749. doi: 10.1002/(SICI)1521-3773(19990315)38:6<736::AID-ANIE736>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 21.Young T, Abel R, Kim B, Berne BJ, Friesner RA. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc Natl Acad Sci U S A. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 23.Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 24.Oswald C, Smits SH, Hoing M, Sohn-Bosser L, Dupont L, Le Rudulier D, et al. Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J Biol Chem. 2008;283:32848–32859. doi: 10.1074/jbc.M806021200. [DOI] [PubMed] [Google Scholar]
- 25.Ma JC, Dougherty DA. The cation-pi interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 26.Hibbs RE, Sulzenbacher G, Shi J, Talley TT, Conrod S, Kem WR, et al. Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor. EMBO J. 2009;28:3040–3051. doi: 10.1038/emboj.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dougherty DA. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]