Abstract
We performed an epidemiological study on Salmonella isolated from raw plant-based feed in Spanish mills. Overall, 32 different Salmonella serovars were detected. Despite its rare occurrence in humans and animals, Salmonella enterica serovar California was found to be the predominant serovar in Spanish feed mills. Different typing techniques showed that isolates of this serovar were genetically closely related, and comparative genomic hybridization using microarray technology revealed 23 S. enterica serovar Typhimurium LT2 gene clusters that are absent from serovar California.
Salmonella is one of the major bacterial agents that cause foodborne infections in humans worldwide (9). The principal source of human Salmonella infection is contaminated food of animal origin, and animal feed is one source of Salmonella for food-producing animals (19). The most prevalent serovars detected in feed products usually are not the same as those that cause disease in humans or animals, possibly because different strains survive in different environments (12). Nevertheless, feeds have been responsible for the infection of poultry with multidrug-resistant nontyphoid Salmonella in several industrialized countries (10, 19). Due to the possible importance of contamination in feed and the fact that information about feed as a gate for microbial entrance to the food chain is still lacking, we have analyzed the Salmonella isolated from feed mills in different regions of Spain.
For this study we used 231 isolates of Salmonella enterica obtained from raw feed of plant origin between May 1999 and May 2001 and 5 Salmonella strains supplied by the National Veterinary Laboratory of Spain as controls (7). The isolates were identified by conventional biochemical methods and serotyped at the National Microbiology Laboratory for Salmonella of Spain. We distinguished 32 different Salmonella enterica serovars (Table 1). We detected some feed-adapted S. enterica serovars (such as Senftenberg and Ohio) and S. enterica serovars implicated in infections (such as Enteritidis and Typhimurium), but the most prevalent S. enterica serovar was California (45% of all isolates), a serovar infrequently detected in animal and human infections.
TABLE 1.
Salmonella serovars isolated in feed from mills in different regions of Spain between May 1999 and May 2001
| Salmonella serovar | No. of strains isolated in different regions of Spain
|
Total no. of strains | ||
|---|---|---|---|---|
| Northwest | Northeast | Southeast | ||
| California | 10 | 94 | 104 | |
| Lexington | 3 | 9 | 6 | 18 |
| Enteritidis | 1 | 13 | 14 | |
| Poona | 1 | 11 | 12 | |
| Llandoff | 6 | 4 | 10 | |
| Anatum | 4 | 2 | 3 | 9 |
| Mbandaka | 1 | 8 | 9 | |
| Derby | 2 | 4 | 6 | |
| Tennessee | 3 | 1 | 2 | 6 |
| Kentucky | 4 | 4 | ||
| Rissen | 4 | 4 | ||
| Schwarzengrund | 4 | 4 | ||
| Senftenberg | 4 | 4 | ||
| Serotype I 3,10:−:1,6 | 1 | 1 | 1 | 3 |
| Tilburg | 3 | 3 | ||
| Typhimurium | 3 | 3 | ||
| Serotype I 4,12:−:− | 2 | 2 | ||
| Montevideo | 2 | 2 | ||
| Other serovarsa | 1 | 10 | 3 | 14 |
| Total | 34 | 170 | 27 | 231 |
Agona, Cubana, Essen, Hadar, Majdoiro, Mikawasima, Muenchen, Ndolo, Ohio, Oranienburg, subsp. I 4,12:g,m,t:z39, subsp. I 3,10:−:−, subsp. I 4,12:−:1,2, and subsp. I 4,5,12:i:−.
The antimicrobial susceptibilities of representative strains were determined by the disk diffusion technique according to National Committee for Clinical Laboratory Standards (NCCLS) standards, using ampicillin, amoxicillin-clavulanic acid, nalidixic acid, ciprofloxacin, chloramphenicol, co-trimoxazole, gentamicin, and tetracycline antimicrobial disks (Sanofi Diagnostics Pasteur, Marnes la Coquette, France). The strains were highly susceptible to all tested antimicrobial agents, even those serovars that were occasionally associated with multidrug resistance, such as S. enterica serovar Typhimurium or subsp. I 4,5,12:i:− strain (4). This lack of resistance could be related to an absence of antimicrobial pressure in a natural environment.
Due to the importance of the presence of S. enterica serovar California in Spanish feed mills, we have analyzed this serovar by epidemiological genotyping methods, including plasmid and pulsed-field gel electrophoresis (PFGE) profiling and comparative genomic hybridization using a Salmonella-specific microarray. Plasmid DNA was isolated with the Qiagen plasmid purification kit (Qiagen, Hilden, Germany) and subsequently digested with two different restriction enzymes (TaqI and HindIII). Most (91%) of the Salmonella serovar California strains showed the same plasmid profile characterized by a plasmid of approximately 3.5 kb in size. Two strains had two additional genetic elements of 10 and 20 kb, and two strains contained no plasmids. Macrorestriction by PFGE was performed as previously described (5) using restriction endonucleases SpeI and XbaI (Amersham Pharmacia Biotech, Buckinghamshire, England). Thiourea was added to the electrophoresis buffer to minimize degradation (11, 18). PFGE results were interpreted visually according to published guidelines (20, 21).
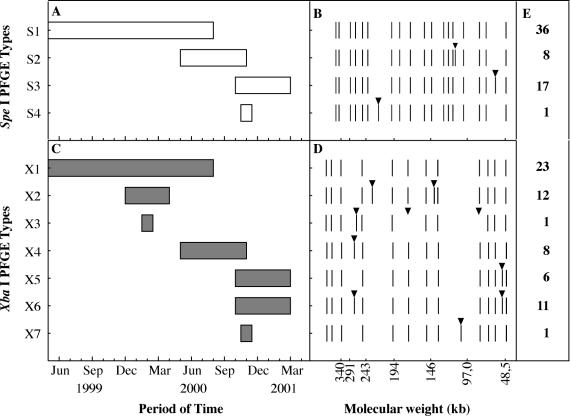
The PFGE typing data with SpeI and XbaI restriction enzymes showed that the Salmonella serovar California strains detected were closely related. The SpeI PFGE patterns of all the Salmonella serovar California strains were grouped into one type called S, which was subdivided into four subtypes (S1 to S4) with only one restriction fragment difference. The obtained XbaI patterns were also categorized in one type, called X, and seven subtypes (X1 to X7), with three or less different fragments (Fig. 1). The combination of the data from both enzymes did not improve the discrimination obtained with XbaI. Plasmid content and PFGE data suggested that a single strain was spreading in mills located in several regions of the country. As previously suggested (10), we hypothesized that this spread could be related to the import of Salmonella-contaminated plants, as there was no evidence for raw feed exchange between mills.
FIG. 1.
PFGE analysis of S. enterica serovar California strains identified in Spanish feed mills. Distribution of SpeI (A) and XbaI (C) PFGE profiles over time and the SpeI (B) and XbaI (D) PFGE profiles obtained (black arrowheads point to differences from profile S1 or X1). (E) Number of isolates for each PFGE pattern.
We detected a subtle evolution of the Salmonella serovar California genotypes over time. Some PFGE patterns were prevalent at the beginning of the study and were replaced by others during the study period (Fig. 1). At the beginning of our study (from May 1999 to August 2000), the prevalent PFGE profiles were S1 and X1, which were later replaced by S2 and S3 (for the SpeI enzyme) and X4, X5, and X6 (for the XbaI enzyme). Heir et al. showed that a change in the season resulted in replacement of a prevalent Salmonella macrorestriction profile by a very different one in Norwegian patients (8). However, in this study, patterns S1 and X1 were very similar to their derivative subtypes, supporting a genetic evolution hypothesis and suggesting a selection of the best-adapted strains to the ecological niche, as described for Pseudomonas fluorescens by Rainey and Travisano (17).
One representative strain of the spreading S. enterica serovar California was hybridized to a previously described Salmonella DNA microarray (6, 13, 15, 16), which contains almost all the coding DNA sequences (CDSs) from S. enterica serovar Typhimurium LT2 and the pSLT virulence plasmid. Overall, 311 CDSs of the 4,338 chromosomal open reading frames represented on the microarray were predicted to be absent or diverged and 280 CDSs were classified as uncertain. Table 2 summarizes regions of two or more genes of the serovar Typhimurium LT2 genome that are apparently absent from the serovar California strain investigated. Of these 23 gene clusters, 12 contained genes with no assigned function. In addition, the dgo operon involved in d-galacturonic acid metabolism, two phosphotransferase systems (PTS), some phoPQ-activated genes, a cluster containing phage remnants, one of the fimbrial operons (saf), and all four active prophages from the serovar Typhimurium LT2 genome were absent from serovar California. The serovar California isolate also contained a different hsdSMR restriction and modification system and lacked all pSLT genes, including the spv gene cluster, frequently associated with virulence (2). The major Salmonella pathogenicity islands and most of the flagellar genes appeared to be present in Salmonella serovar California, but the status of the gene encoding the major flagellin, fliC, was uncertain. Reliable hybridization of DNA to the target gene on the array occurs only if sequence identities are at least 97% over one 100-bp window within a gene (14) or if there is >90% DNA sequence identity over the entire gene. A previously sequenced Salmonella serovar California fliC gene (GenBank accession no. U05296) and the Salmonella serovar Typhimurium LT2 fliC gene share only a 64.5% similarity using the Matcher tool available at the Institute Pasteur website (http://bioweb.pasteur.fr). Therefore, the microarray correctly reported the gene to be divergent. The apparent absence of the highly conserved regulatory hin gene involved in flagellar phase switching explained the monophasic state of the California strain.
TABLE 2.
Gene clusters in the Salmonella serovar Typhimurium LT2 strain absent from the Salmonella serovar California strain
| STM numbera | Geneb | Function(s) |
|---|---|---|
| STM0030—0037 | Putative genes | |
| STM0290—0292 | Putative genes | |
| STM0299—0304 | safABCD-ybeJ-sinR | Fimbrial operon, transcriptional regulator |
| STM0325—0334 | Putative genes | |
| STM0571—0576 | Putative genes | |
| STM0716—0727 | Putative genes | |
| STM0854—0860 | Putative genes | |
| STM0893—0929 | Fels-1 prophage | |
| STM1005—1056 | Gifsy-2 prophage | |
| STM1860—1871 | pagOK | PhoPQ-activated, phage-related, and putative genes |
| STM2230—2244 | oafA sspH2 | Phage-related genes |
| STM2584—2636 | Gifsy-1 prophage | |
| STM2694—2772 | fljAB-hin | Fels-2 prophage, phase switching operon |
| STM3117—3123 | Putative genes | |
| STM3251—3256 | agaR | PTS |
| STM3277—3278 | Putative genes | |
| STM3752—3755 | sugR-rhuM | ATP binding protein |
| STM3780—3784 | Putative genes | |
| STM3827—3830 | dgoTAKR | d-Galacturonic acid metabolism |
| STM3844—3846 | Putative genes | |
| STM4109—4116 | talC-ptsA-frwBCD-pflCD | PTS |
| STM4488—4498 | Putative genes | |
| STM4523—4528 | yjiW-hsdSMR-mrr | Restriction and modification system |
STM, Salmonella serovar Typhimurium complete genome sequences.
If known.
S. enterica serovar California has been isolated from cases of human gastroenteritis (1, 3). However, the frequency of its involvement in human disease is insignificant compared to its apparent prevalence in feed. Therefore, it seems likely that neither humans nor the farm animals are the natural host for serovar California and that its natural host is yet to be elucidated. Alternatively, this strain may normally remain completely asymptomatic in a human or farm animal host.
In summary, we have detected the spread of a single Salmonella serovar California strain in raw plant-based feed samples from mills located in different regions of Spain. PFGE studies suggested genetic adaptation of the strain over time, while microarray analysis was used to further characterize the genetic repertoire present in this serovar. Epidemiological surveillance for Salmonella in local or imported supplies that will become part of animal feeds should be encouraged in order to prevent the introduction of this microorganism to animals and humans through this route.
Acknowledgments
This work was supported in part by Basque Government grant PI 1998/52,“Subvención general a Grupos de Investigación” UPV/EHU (2002-2005), and by NIH grant AI34829 (M.M.). Juan Alvarez and Ana Belén Vivanco were supported by a “Beca de Formación de Personal Investigador” from the Basque Government of Spain and a “Beca de Investigación Predoctoral” from the University of the Basque Country of Spain.
We thank Cristina de Frutos (National Veterinary Laboratory of Spain) for providing some Salmonella serovar California isolates and Miguel Angel Usera (National Laboratory for Salmonella of Spain) for performing serotyping.
REFERENCES
- 1.Bellver, P., and M. García. 2000. Epidemiología de la salmonelosis no tifoidea en un hospital de Pontevedra (1994-1997). Enferm. Infecc. Microbiol. Clin. 18:125-132. [PubMed] [Google Scholar]
- 2.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, C., J. Cunningham, R. Ahmed, D. Woodward, K. Fonseca, S. Isaacs, A. Ellis, C. Anand, K. Ziebell, A. Muckle, P. Sockett, and F. Rodgers. 2001. Characterization of Salmonella associated with pig ear dog treats in Canada. J. Clin. Microbiol. 39:3962-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruchaga, S., A. Echeita, A. Aladueña, J. García-Peña, N. Frias, and M. A. Usera. 2001. Antimicrobial resistance in salmonellae from humans, food and animals in Spain in 1998. J. Antimicrob. Chemother. 47:315-321. [DOI] [PubMed] [Google Scholar]
- 5.Garaizar, J., N. López-Molina, I. Laconcha, D. L. Baggesen, A. Rementeria, A. Vivanco, A. Audicana, and I. Perales. 2000. Suitability of PCR fingerprinting, infrequent-restriction-site PCR, and pulsed-field gel electrophoresis, combined with computerized gel analysis, in library typing of Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 66:5273-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garaizar, J., S. Porwollik, A. Echeita, A. Rementeria, S. Herrera, R. M.-Y. Wong, J. Frye, M. A. Usera, and M. McClelland. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García, F. J., N. Frías, C. de Frutos, B. Martín, and C. López. 2001. Análisis de los serotipos de Salmonella spp. aislados en el año 2000 por los laboratorios de Sanidad Animal en España. Bol. Epidemiol. Sem. 9:287-288. [Google Scholar]
- 8.Heir, E., B.-A. Lindstedt, I. Nygård, T. Vardund, V. Hasseltvedt, and G. Kapperud. 2002. Molecular epidemiology of Salmonella Typhimurium isolates from human sporadic and outbreak cases. Epidemiol. Infect. 128:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herikstad, H., Y. Motarjemi, and R. V. Tauxe. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kariuki, S., G. Revathi, F. Gakuya, V. Yamo, J. Muyodi, and C. A. Hart. 2002. Lack of clonal relationship between non-typhi Salmonella strain types from humans and those isolated from animals living in close contact. FEMS Immunol. Med. Microbiol. 33:165-171. [DOI] [PubMed] [Google Scholar]
- 11.Liesegang, A., and H. Tschäpe. 2002. Modified pulsed-field gel electrophoresis method for DNA degradation-sensitive Salmonella enterica and Escherichia coli strains. Int. J. Med. Microbiol. 291:645-648. [DOI] [PubMed] [Google Scholar]
- 12.Lindqvist, N., S. Heinikainen, A. M. Toivonen, and S. Pelkonen. 1999. Discrimination between endemic and feedborne Salmonella Infantis infection in cattle by molecular typing. Epidemiol. Infect. 122:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 14.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 31:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porwollik, S., R. M.-Y. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porwollik, S., R. M.-Y. Wong, S. H. Sims, R. M. Schaaper, D. M. DeMarini, and M. McClelland. 2001. The ΔuvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat. Res. 483:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 18.Ray, T., A. Mills, and P. Dyson. 1995. Tris-dependent oxidative DNA strand scission during electrophoresis. Electrophoresis 16:888-894. [DOI] [PubMed] [Google Scholar]
- 19.Shirota, K., H. Katoh, T. Murase, T. Ito, and K. Otsuki. 2001. Monitoring of layer feed and eggs for Salmonella in Eastern Japan between 1993 and 1998. J. Food Prot. 64:734-737. [DOI] [PubMed] [Google Scholar]
- 20.Struelens, M. J., and the members of ESGEM. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 21.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]