Abstract
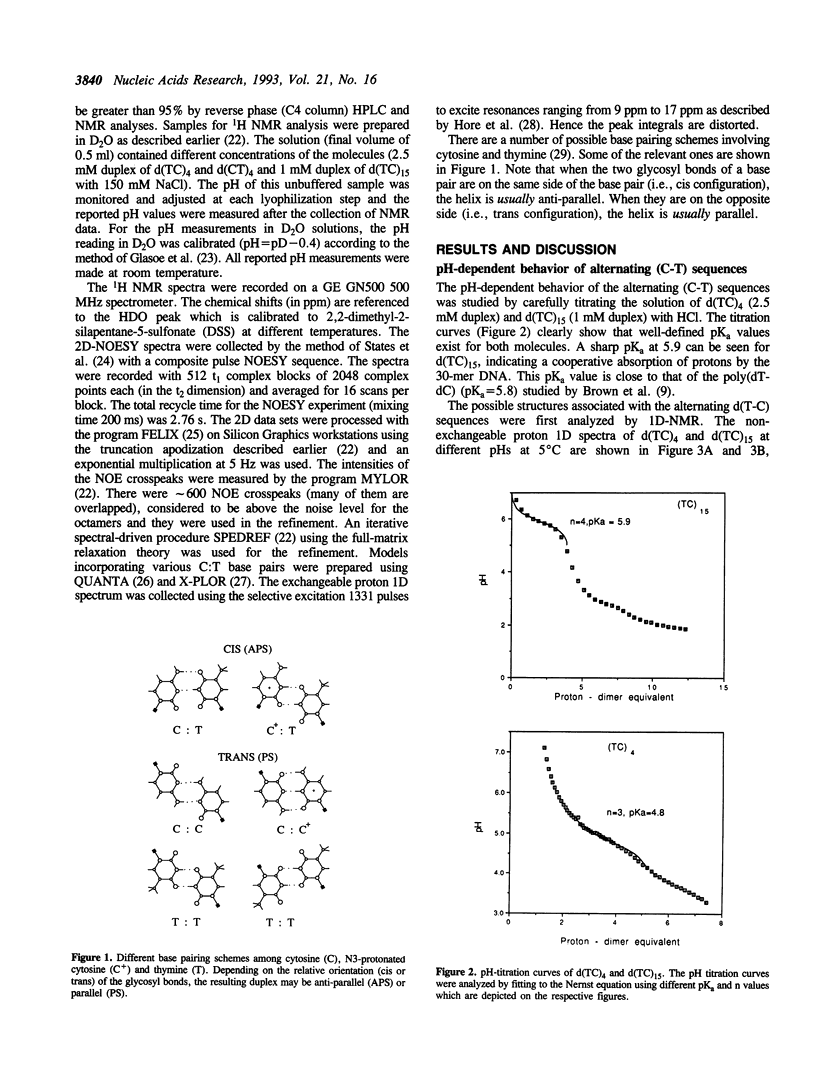
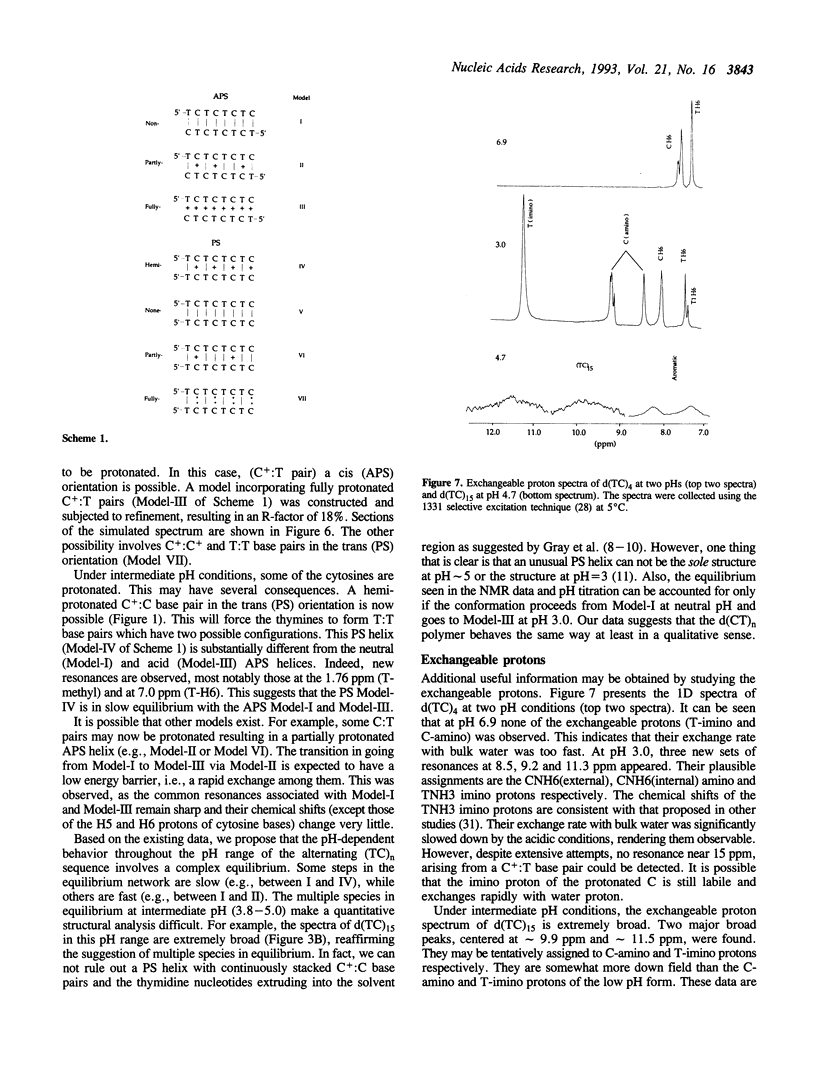
Alternating (C-T)n sequences are involved in the H-DNA structure associated with (GA)n.(CT)n sequences. Low pH values facilitate H-DNA formation. We have undertaken a detailed analysis of the structural consequences of the (C-T)n sequence as a function of pH. The structures of three DNA oligonucleotides, d(CT)4, d(TC)4 and d(TC)15, have been studied by NMR. We found that their conformations are polymorphic and pH dependent. There are at least three major conformational species: an antiparallel-stranded (APS) duplex with entirely C:T base pairs at pH 7, an antiparallel-stranded (APS) duplex with entirely C+:T base pairs at pH 3, and a possible parallel-stranded (PS) duplex with C+:C and T:T base pairs near pH 5. In the intermediate pH range, the APS duplex may have varying numbers of C+:T and C:T base pairs, and there may be a fast exchange going on between APS duplex species involving these two kinds of base pairs. However, the transition between the APS and PS duplexes is slow. Structural refinement of the two octamers, d(TC)4 and d(CT)4, at pH = 6.9 and pH = 3 using 2D-NOE data suggests that the molecules are likely in the duplex form at 5 degrees C. We lack evidence that the structure at pH 3 is a PS structure with T nucleotides residing in the exterior of the helix. Titration of the longer oligonucleotide, d(TC)15, showed a prominent pKa of approximately 6, approaching the value of 7.0 obtained from the titration of poly-(dC).
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Leslie A. G. Structure of the single-stranded polyribonucleotide polycytidylic acid. J Mol Biol. 1976 Sep 25;106(3):735–748. doi: 10.1016/0022-2836(76)90262-x. [DOI] [PubMed] [Google Scholar]
- Brown D. M., Gray D. M., Patrick M. H., Ratliff R. L. Photochemical demonstration of stacked C.C+ base pairs in a novel DNA secondary structure. Biochemistry. 1985 Mar 26;24(7):1676–1683. doi: 10.1021/bi00328a016. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Coll M., Solans X., Font-Altaba M., Subirana J. A. Crystal and molecular structure of the sodium salt of the dinucleotide duplex d(CpG). J Biomol Struct Dyn. 1987 Apr;4(5):797–811. doi: 10.1080/07391102.1987.10507679. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Egert E., Kennard O., Sala G. B., Salisbury S. A., Viswamitra M. A. Self base pairing in a complementary deoxydinucleoside monophosphate duplex: crystal and molecular structure of deoxycytidylyl-(3'-5')-deoxyguanosine. Biochemistry. 1983 Apr 12;22(8):1833–1839. doi: 10.1021/bi00277a014. [DOI] [PubMed] [Google Scholar]
- Gehring K., Leroy J. L., Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993 Jun 10;363(6429):561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Thomas G. H., Elgin S. C. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989 Sep 29;245(4925):1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Vaughan M. Circular dichroism spectra show that repeating dinucleotide DNAs may form helices in which every other base is looped out. Nucleic Acids Res. 1980 Aug 25;8(16):3695–3707. doi: 10.1093/nar/8.16.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- He L., Kierzek R., SantaLucia J., Jr, Walter A. E., Turner D. H. Nearest-neighbor parameters for G.U mismatches: [formula; see text] is destabilizing in the contexts [formula; see text] and [formula; see text] but stabilizing in [formula; see text]. Biochemistry. 1991 Nov 19;30(46):11124–11132. doi: 10.1021/bi00110a015. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Il'ychova I. A., Lysov YuP, Chernyi A. A., Shchyolkina A. K., Gottikh B. P., Florentiev V. A. Parallel double helices of DNA. Conformational analysis of regular helices with the second order symmetry axis. J Biomol Struct Dyn. 1990 Feb;7(4):879–897. doi: 10.1080/07391102.1990.10508530. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- LANGRIDGE R., RICH A. Molecular structure of helical polycytidylic acid. Nature. 1963 May 25;198:725–728. doi: 10.1038/198725a0. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Gehring K., Kettani A., Guéron M. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry. 1993 Jun 15;32(23):6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- Luo J., Sarma M. H., Yuan R. D., Sarma R. H. NMR study of self-paired parallel duplex of d(AAAAACCCCC) in solution. FEBS Lett. 1992 Jul 20;306(2-3):223–228. doi: 10.1016/0014-5793(92)81005-7. [DOI] [PubMed] [Google Scholar]
- Manor H., Rao B. S., Martin R. G. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J Mol Evol. 1988;27(2):96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Opstelten R. J., Clement J. M., Wanka F. Direct repeats at nuclear matrix-associated DNA regions and their putative control function in the replicating eukaryotic genome. Chromosoma. 1989 Dec;98(6):422–427. doi: 10.1007/BF00292787. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Gao X., de los Santos C., Live D., Patel D. J. NMR structural studies of intramolecular (Y+)n.(R+)n(Y-)nDNA triplexes in solution: imino and amino proton and nitrogen markers of G.TA base triple formation. Biochemistry. 1991 Sep 17;30(37):9022–9030. doi: 10.1021/bi00101a016. [DOI] [PubMed] [Google Scholar]
- Robinson H., Wang A. H. 5'-CGA sequence is a strong motif for homo base-paired parallel-stranded DNA duplex as revealed by NMR analysis. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5224–5228. doi: 10.1073/pnas.90.11.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H., Wang A. H. A simple spectral-driven procedure for the refinement of DNA structures by NMR spectroscopy. Biochemistry. 1992 Apr 7;31(13):3524–3533. doi: 10.1021/bi00128a029. [DOI] [PubMed] [Google Scholar]
- Robinson H., van der Marel G. A., van Boom J. H., Wang A. H. Unusual DNA conformation at low pH revealed by NMR: parallel-stranded DNA duplex with homo base pairs. Biochemistry. 1992 Nov 3;31(43):10510–10517. doi: 10.1021/bi00158a014. [DOI] [PubMed] [Google Scholar]
- Sarma M. H., Gupta G., Sarma R. H. A cytosine . cytosine base paired parallel DNA double helix with thymine . thymine bulges. FEBS Lett. 1986 Sep 15;205(2):223–229. doi: 10.1016/0014-5793(86)80902-4. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Feigon J. Formation of a stable triplex from a single DNA strand. Nature. 1990 Jun 28;345(6278):836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]
- Weinreb A., Collier D. A., Birshtein B. K., Wells R. D. Left-handed Z-DNA and intramolecular triplex formation at the site of an unequal sister chromatid exchange. J Biol Chem. 1990 Jan 25;265(3):1352–1359. [PubMed] [Google Scholar]
- Yee H. A., Wong A. K., van de Sande J. H., Rattner J. B. Identification of novel single-stranded d(TC)n binding proteins in several mammalian species. Nucleic Acids Res. 1991 Feb 25;19(4):949–953. doi: 10.1093/nar/19.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos C., Rosen M., Patel D. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989 Sep 5;28(18):7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]



